Abstract
Translation of m7G-capped cellular mRNAs is initiated by recruitment of ribosomes to the 5′ end of mRNAs via eukaryotic translation initiation factor 4F (eIF4F), a heterotrimeric complex comprised of a cap-binding subunit (eIF4E) and an RNA helicase (eIF4A) bridged by a scaffolding molecule (eIF4G). Internal translation initiation bypasses the requirement for the cap and eIF4E and occurs on viral and cellular mRNAs containing internal ribosomal entry sites (IRESs). Here we demonstrate that eIF4E availability plays a critical role in the switch from cap-dependent to IRES-mediated translation in picornavirus-infected cells. When both capped and IRES-containing mRNAs are present (as in intact cells or in vitro translation extracts), a decrease in the amount of eIF4E associated with the eIF4F complex elicits a striking increase in IRES-mediated viral mRNA translation. This effect is not observed in translation extracts depleted of capped mRNAs, indicating that capped mRNAs compete with IRES-containing mRNAs for translation. These data explain numerous reported observations where viral mRNAs are preferentially translated during infection.
Recruitment of ribosomes to mRNA is the rate-limiting step in translation initiation and a frequent target for translational control (22). Two different mechanisms of ribosome binding exist in mammalian cells. Cap-dependent translation is mediated by the mRNA 5′ cap structure (m7GpppN, where N is any nucleotide), and represents the standard mode of translation used by most cellular mRNAs. Cap-independent translation is utilized by some plus-stranded RNA viruses, including picornaviruses and hepatitis C virus, as well as by some cellular mRNAs, and involves the binding of ribosomes to an mRNA structural element termed an internal ribosomal entry site (IRES) (20, 41).
The eukaryotic translation initiation factor 4F (eIF4F) mediates 40S ribosomal subunit binding to the 5′end of capped mRNA. eIF4F is a complex containing three proteins: eIF4E, the cap-binding subunit; eIF4A, an RNA-dependent ATPase/ATP-dependent RNA helicase; and eIF4G, a high-molecular-weight protein that acts as a scaffold for binding eIF4E and eIF4A. In addition, eIF4G interacts with the 40S ribosome binding factor eIF3 and the poly(A)-binding protein, thereby establishing a critical link between mRNA and the ribosome (reviewed in references 14, 21, and 22). The various eIF4F subunits are expressed to remarkably different levels in most cell types, with the eIF4E subunit being the least abundant (22).
Importantly, formation of the eIF4F complex is dynamic and tightly regulated (44). In particular, eIF4E availability for participation in eIF4F formation is modulated by a family of small translation repressor molecules, the eIF4E-binding proteins (4E-BPs) (31, 38). While hypophosphorylated 4E-BPs interact strongly with eIF4E, hyperphosphorylated 4E-BPs do not (15). 4E-BP phosphorylation levels are modulated by many types of extracellular stimuli. In particular, hormonal or nutritional stimulation tends to increase 4E-BP1 phosphorylation levels, while environmental or nutritional stress elicits 4E-BP dephosphorylation (15, 43). Thus, a binary subcomplex consisting of eIF4G and eIF4A (eIF4G/4A) appears to exist in a dynamic equilibrium with eIF4F. This equilibrium may be shifted to increase or decrease eIF4F formation in response to nutrients, hormonal stimulation, or stress (43).
Internal translation initiation on most IRES-containing mRNAs, such as encephalomyocarditis virus (EMCV) mRNA, requires the same canonical eIFs that are required for translation of capped mRNAs, except for eIF4E (1, 39, 40). In contrast to typical eIF4E-mediated ribosomal recruitment, the initial step in recruitment of the ribosome to the EMCV IRES is the eIF4A-dependent high-affinity binding of the central domain of eIF4G to the J-K stem-loop of the IRES (27, 32). Subsequent addition of the 40S ribosomal subunit, presumably via the eIF4G-eIF3-40S interaction, and the 60S subunit completes the assembly of the initiation complex.
EMCV and other picornavirus infections are accompanied by a shutoff of host cell protein synthesis (10). In cells infected with poliovirus (PV), human rhinovirus, and foot-and-mouth disease virus, the primary event responsible for this shutoff is the cleavage of the eIF4G isoforms by virus-specific proteases (11, 47). The C-terminal cleavage fragment of eIF4G can efficiently support IRES-dependent, but not cap-dependent translation, as it retains the binding sites for IRES, eIF4A and eIF3, but cannot bind eIF4E. This situation is akin to a net increase of eIF4G/4A at the expense of the eIF4F complex. While infection of cells with EMCV also inhibits host cell protein synthesis, this inhibition develops more slowly than that caused by PV and is not mediated by cleavage of eIF4G (25, 35).
Although eIF4G is not cleaved in EMCV-infected cells, it is highly likely that the ratio of eIF4G/4A to eIF4F is also increased during EMCV infection. We previously described the dephosphorylation and activation of 4E-BP1 following EMCV infection (16). Inasmuch as 4E-BP1 dephosphorylation coincides with the shutoff of host mRNA translation in EMCV-infected cells, we hypothesized that these two events are causally related (16). Dissociating eIF4F may also favor viral protein synthesis, as suggested by experiments employing rapamycin and wortmannin, two inhibitors of 4E-BP1 phosphorylation (5, 51). Upon forced dephosphorylation of 4E-BPs by treating cells with rapamycin and wortmannin at the beginning of EMCV infection, viral protein synthesis and viral titers were higher than in untreated control cells (5, 51). However, because rapamycin and wortmannin also have other cellular targets (15) and because these in vivo studies were merely correlative, it was critical to directly assess the function of eIF4F in the translation of EMCV mRNA.
We were recently able to reconstitute EMCV translation and replication in Krebs-2 cell extract (55). This system enabled us to address the importance of eIF4F subunit composition in viral protein synthesis and replication. Here we report that when EMCV mRNA is translated in competition with cellular mRNAs (i.e., in extracts that are not treated with nuclease), addition of 4E-BPs significantly augments viral protein synthesis. In contrast, addition of eIF4E dramatically inhibits viral protein synthesis. Furthermore, when eIF4F is converted to the eIF4G/4A subcomplex by eIF4E knockdown, the onset of viral protein synthesis in EMCV- or PV-infected cells is markedly accelerated and the viral yield is higher. These findings demonstrate that active eIF4E functions as a negative modulator of IRES-mediated translation by increasing competition from capped mRNAs for the eIF4F complex.
MATERIALS AND METHODS
Materials.
mRNAs encoding luciferase were transcribed from pT3luc(A)+ and pT7EMCVluc(A)+ using T3 or T7 RNA polymerase, respectively (52). EMCV mRNA was isolated from purified virus by extraction with a mixture of phenol and chloroform (51). Total poly(A)-containing RNA [poly(A)+ mRNA] was isolated from the cytoplasm of Krebs-2 cells and purified by two cycles of chromatography on oligo(dT)-cellulose (3, 49). Globin mRNA was purchased commercially (Gibco Invitrogen Corporation [discontinued]). The components used to prepare Krebs-2 cell extracts and to supplement translation reactions were those specified previously (56). Recombinant glutathione S-transferase (GST)-4E-BP1, GST-4E-BP1Δ4E (4E-BP1Δ54-63), GST-4E-BP2, GST-eIF4E, GST-eIF4EW73A, eIF4E, eIF4A, and eIF4G-Ct were described previously (12, 13, 33, 52-54). Protein expression was performed in Escherichia coli BL21 (DE3) cells according to the manufacturer's instructions (GE Healthcare). To assess purity, proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. 2Apro was a kind gift of H.-D. Liebig (52). Mouse monoclonal antibody 8D10 against recombinant mengovirus protein 3Dpol (8) was kindly provided by Ann Palmenberg (University of Wisconsin, Madison, WI). Mouse monoclonal antibodies against eIF4E and β-actin were purchased from BD Biosciences and Sigma, respectively. Rabbit polyclonal antibodies against eIF4GI and eIF4AI were previously described (30, 34). Secondary horseradish peroxidase (HRP)-conjugated sheep anti-mouse and donkey anti-rabbit antibodies were obtained from GE Healthcare. HeLa S3 and BHK-21 cells were obtained from the American Type Culture Collection (ATCC numbers CCL-2.2 and CCL-10, respectively). Dulbecco's modified modified Eagle's medium (DMEM), Lipofectamine 2000, and OPTIMEM were from Invitrogen.
Krebs-2 cell extract preparation.
Krebs-2 ascites cell propagation in mice and the preparation of extracts were done as previously described (56). Before homogenization, cells were suspended in methionine-free DMEM and incubated at 37°C for 2 h with gentle agitation. The cells were broken with a Dounce homogenizer, and a postmitochondrial supernatant (S10) was obtained by a high-speed centrifugation (18,000 × g, 4°C, 20 min). Where indicated, the extracts were treated with micrococcal nuclease in the presence of CaCl2 (56).
In vitro assays for EMCV mRNA translation, RNA replication, and virion synthesis.
EMCV mRNA translation and replication reaction mixtures (30 μl) that contained either untreated or nuclease-treated Krebs-2 S10 extract were programmed with EMCV mRNA (4 μg/ml), as described previously (55). For protein labeling, reaction mixtures were supplemented with [35S]methionine. After incubation at 32°C for 1.5 to 3 h, reactions were stopped with Laemmli sample buffer. Protein products were resolved by SDS-PAGE (15% gels), electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane, and detected by autoradiography. Western blotting for 3Dpol was performed as described below. RNA replication and virion production were assayed in the reaction mixtures that contained unlabeled methionine as described previously (55). For RNA labeling, [α-32P]CTP was added to the reaction mixtures after 4 h of incubation. One hour later, RNA was extracted and RNA products were analyzed by native 1% agarose gel electrophoresis and autoradiography (55). To assay for EMCV synthesis, reaction mixtures were incubated at 32°C for 20 h and treated with a mixture of RNase A and T1 (55). Plaque assays were performed using serial dilutions of samples as described below.
siRNA transfection.
Target sequences for small interfering RNA (siRNA) were designed using the Dharmacon web-based criteria and were purchased from Dharmacon. The positions and sequences of the siRNAs used in this study are listed in Table 1. HeLa S3 cells were seeded in a 24-well culture dish at a density of 7 × 105 cells per well. siRNA transfection was performed using Lipofectamine 2000 as described previously (7).
TABLE 1.
Positions and sequences of the siRNAs used to knock down gene expression
| Synthetic siRNA | Positions in open reading frame | siRNA sequencea |
|---|---|---|
| eIF4E | 331-350 | 5′-GGACGAUGGCUAAUUACAUdTdT-3′ |
| 3′-dTdTCCUGCUACCGAUUAAUGUA-5′ | ||
| Controlb | 935-953 | 5′-CGUACCGUGGAAUAGUUCCdTdT-3′ |
| 3′-dTdTGCAUGGCACCUUAUCAAGG-5′ |
dT, deoxyribosylthymine.
siRNA against 4E-T (inverted).
Virus infections and metabolic radiolabeling.
Forty-eight hours after siRNA transfection, HeLa S3 cells were infected with EMCV or PV at a multiplicity of infection of 5 PFU per cell. Virus adsorption was at room temperature for 30 min. The medium was then replaced with methionine-free DMEM, and the incubation was continued at 37°C. At various times postinfection (see figure legends), the media were replaced with media containing 35S-protein labeling mix (10 μCi/ml). After 30 min of labeling, the cell monolayers were washed with phosphate-buffered saline and lysed with Laemmli sample buffer. Radiolabeled proteins were resolved by SDS-PAGE (15% gels), transferred to a PVDF membrane, and detected by autoradiography. The same membrane was used for Western blotting.
Western blotting.
PVDF membranes were blocked with Tris-buffered saline/0.1% (vol/vol) Tween 20 containing 5% nonfat dry milk and probed with the indicated antibodies. The antibodies against eIF4E, eIF4GI, eIF4AI, and β-actin were used diluted 1:500, 1:1,000, 1:1,000, and 1:5,000, respectively. The antibody against mengovirus protein 3Dpol was used at a dilution of 1:1,500. After washing, the membrane was incubated with HRP-conjugated anti-mouse or anti-rabbit antibody, as appropriate (diluted 1:5,000). HRP was detected using the Western Lightning chemiluminescence kit as recommended by the manufacturer (Perkin-Elmer Life Sciences, Inc.).
Plaque assays.
Plaque assays were performed as previously described using confluent monolayers of either BHK-21 cells (for EMCV) or HeLa R19 cells (for PV) in 60-mm-diameter plates (45). Virus-infected cells from 24-well dishes were lysed in 500 μl DMEM per well by three cycles of freezing and thawing. Cell debris was removed by centrifugation (10,000 × g, 4°C, 5 min), and the supernatants were diluted with DMEM containing 2% fetal bovine serum. Cells were infected with 250 μl of serially diluted lysates. Plaques were allowed to develop under semisolid agar for 26 h (EMCV) or 36 h (PV) at 37°C and were detected by staining with 1% crystal violet.
RESULTS
Stimulation of EMCV IRES-mediated translation by 4E-BPs in an untreated cell-free translation system.
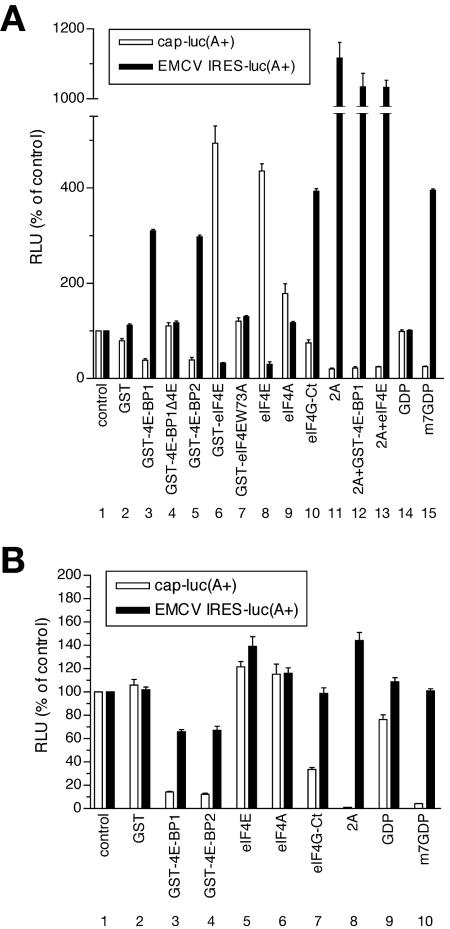
We sought direct evidence of a role of the eIF4G/4A subcomplex in the regulation of EMCV IRES-directed translation in vitro. Previously, we added recombinant 4E-BPs to a nuclease-treated rabbit reticulocyte lysate to sequester eIF4E and prevent its incorporation into eIF4F; these conditions did not result in stimulation of EMCV IRES-mediated translation (38, 53). These data are in contrast to observations from EMCV-infected cells in vivo, where the prevention of eIF4E incorporation into eIF4F by rapamycin and wortmannin stimulated viral protein synthesis (5, 51). We hypothesized that cellular mRNAs present in virus-infected cells sequester eIF4F, making this factor limiting for viral translation. Generation of eIF4G/4A, which does not bind capped mRNAs efficiently, would be expected to relieve this competition. We therefore mimicked in vivo conditions by performing assays under conditions of mRNA competition—that is, in extracts in which endogenous mRNAs were not destroyed by pretreatment with a nuclease. We utilized a translation system derived from Krebs-2 cells to study the effects of 4E-BPs on the translation of capped [cap-luc(A+)] or EMCV IRES-containing [EMCV IRES-luc(A+)] polyadenylated luciferase mRNAs. In this system, both 4E-BP1 and 4E-BP2 inhibited translation of cap-luc(A+) mRNA by three- to fourfold, similar to the cap analog, m7GDP, which was used as a positive control (Fig. 1A; compare open bars 3, 5, and 15 with bar 1). Importantly, and in contrast to observations in nuclease-treated RRL, 4E-BPs and m7GDP stimulated translation of EMCV IRES-luc(A+) mRNA (∼threefold; compare black bars 3, 5, and 15 with bar 1). Both cap-dependent translation and IRES-dependent translation were unaffected by 4E-BP1Δ4E, a 4E-BP1 mutant protein lacking the eIF4E binding site (bars 4) (12, 33). Exogenous recombinant eIF4E, either with or without a GST tag, stimulated cap-dependent translation (four- to fivefold) but inhibited EMCV IRES-directed translation (three- to fourfold; Fig. 1A; compare black bars 6 and 8 with bar 1 for IRES-driven translation). The effects of eIF4E on translation were exerted via the eIF4E/4G complex, as they were negated by the W73A mutation in eIF4E, which abolishes this complex formation (bars 7) (13).
FIG. 1.
Regulation of cap-dependent and cap-independent translation by effectors of eIF4F function in Krebs-2 cell extracts. (A) Translation in untreated extract. Cap-luc(A+) and EMCV IRES-luc(A+) mRNAs (5 μg/ml) were translated in 12.5-μl reaction mixtures at 32°C for 90 min in the presence of unlabeled methionine (52). Prior to the additions of mRNA, the extracts were preincubated at 32°C for 2 min with either control buffer (control) or the following components: GST (20 μg/ml), GST-4E-BP1, GST-4E-BP1Δ4E, GST-4E-BP2, GST-eIF4E, GST-eIF4EW73A (40 μg/ml each), eIF4E (16 μg/ml), eIF4A (80 μg/ml), eIF4G-Ct (40 μg/ml), GDP, or m7GDP (0.5 mM), as indicated in the figure. Where indicated (2A), the reaction mixtures contained extract treated with 2Apro (25 μg/ml, 32°C, 5 min) (52). (B) Translation of cap-luc(A+) and EMCV IRES-luc(A+) mRNAs in nuclease-treated extract. Protein additions and translation conditions were as described in panel A, except for S10, which was nuclease treated. Luciferase activity (relative light units [RLU]) was determined as previously described (56) and is shown as a percentage of that of the control sample. Data represent the average of three independent determinations. Error bars indicate the standard deviation from the mean.
Neither cap-dependent nor IRES-driven translation was significantly affected by eIF4A (Fig. 1A, bars 9). However, eIF4G-Ct, the C-terminal portion of eIF4G that does not contain the eIF4E-binding site and corresponds to the C-terminal picornavirus protease cleavage fragment (28), stimulated IRES-mediated translation approximately fourfold (Fig. 1A; compare black bar 10 with bar 1). An even more striking (>10-fold) enhancement of EMCV IRES-directed translation was observed upon cleavage of eIF4G by rhinovirus protease 2A (2Apro; Fig. 1A; compare black bar 11 with bar 1). Addition of 4E-BP1 did not further potentiate this effect. Also, eIF4E did not influence IRES-mediated translation when eIF4G was cleaved. Overall, there was an inverse correlation between the efficiencies of cap-dependent and IRES-dependent translation, suggesting that they are oppositely regulated by eIF4F. These results also demonstrate that stimulation of EMCV IRES-directed translation by 4E-BPs can be reproduced in vitro.
To prove that the relative excess of the eIF4G/4A subcomplex as compared with the intact eIF4F complex indirectly stimulates EMCV IRES-directed translation by decreasing competition from cellular mRNAs, we performed assays similar to those above using an extract in which endogenous cellular mRNAs were degraded by nuclease treatment (Fig. 1B). EMCV IRES-directed translation was enhanced (∼threefold) in the nuclease-treated extract (data not shown), demonstrating that competing cellular mRNAs in the untreated extract indeed had an inhibitory effect on EMCV IRES activity. Our results were consistent with those reported for nuclease-treated rabbit reticulocye lysate (38, 53). Although addition of 4E-BPs and m7GDP strongly inhibited cap-dependent translation (8- to 20-fold), these components did not stimulate EMCV IRES activity in the nuclease-treated extract; in fact, 4E-BPs had a slightly adverse effect on EMCV IRES activity (Fig. 1B; compare black bars 3 and 4 with bar 1). Nuclease treatment also abolished the ability of eIF4E to inhibit translation from the EMCV IRES (Fig. 1B; compare black bars 5 and 1). In contrast to observations in the untreated extract, 2Apro treatment or eIF4G-Ct addition did not substantially stimulate EMCV IRES activity in the nuclease-treated extract (Fig. 1B; compare black bars 8, 7, and 1). Taken together, these findings suggest that a relative excess of the free eIF4G/4A subcomplex, compared with eIF4F, upregulates EMCV IRES-driven translation only in the presence of competing cellular mRNAs.
The eIF4G/4A subcomplex is essential and limiting for EMCV replication in untreated extract.
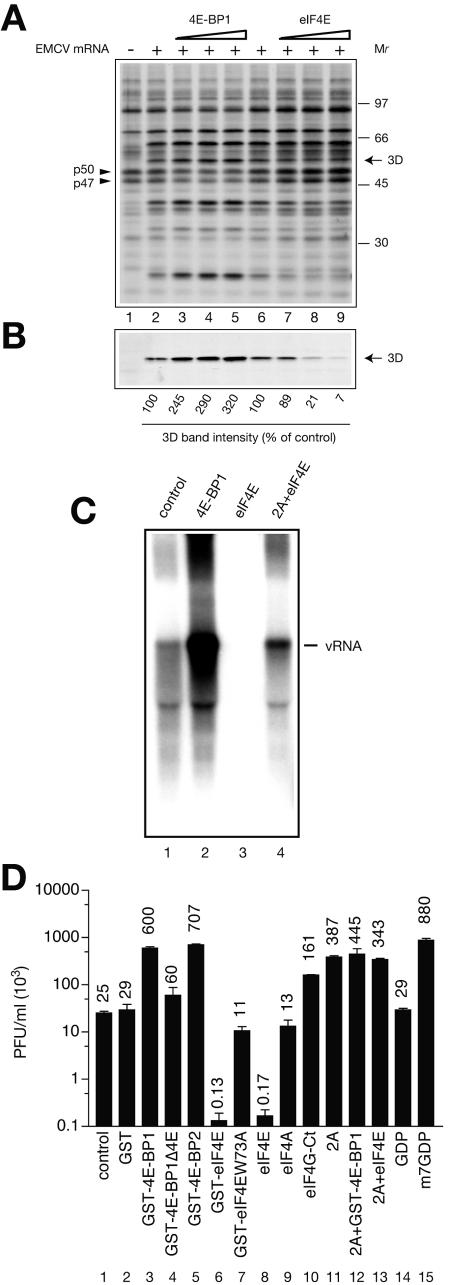
We next examined whether 4E-BP1 and eIF4E modulate translation from the EMCV IRES when the full-length EMCV mRNA is used in untreated extract. EMCV mRNA was translated in the presence of increasing concentrations of 4E-BP1 or eIF4E. Translation products were resolved by SDS-PAGE, transferred to a PVDF membrane, and detected by autoradiography. [35S]methionine incorporation into polypeptides in the untreated extract primarily reflected elongation of preexisting polypeptide chains. However, consistent with the contribution from de novo translation initiation, incorporation of [35S]methionine into cellular proteins was inhibited by 4E-BP1 and stimulated by eIF4E (Fig. 2A; note the corresponding changes in the intensities of two prominent cellular proteins, p47 and p50). The latter observation indicates that eIF4E is limiting for translation of endogenous cellular mRNAs. It was difficult to discern virus-specific polypeptides on this autoradiograph due to the high degree of labeling of endogenous cellular proteins. We therefore assessed the efficiency of EMCV mRNA translation by Western blotting using an antibody against the nonstructural protein 3Dpol (an RNA-dependent RNA polymerase). The addition of 4E-BP1 stimulated (up to 3.2-fold) 3Dpol synthesis (Fig. 2B; compare lanes 5 and 2). In contrast, eIF4E dramatically inhibited 3Dpol synthesis by up to 14-fold (Fig. 2B; compare lanes 9 and 6). Thus, in the untreated extract, translation of full-length EMCV mRNA was upregulated by 4E-BP1 and downregulated by eIF4E. Thus, the effects of 4E-BP1 and eIF4E on the translation of full-length EMCV mRNA were similar to those measured using the surrogate template, EMCV IRES-luc(A+) mRNA.
FIG. 2.
EMCV mRNA translation, RNA replication, and virus yield in the untreated EMCV mRNA-programmed S10 extracts. (A) Effects of 4E-BP1 and eIF4E concentration on protein synthesis in untreated EMCV mRNA-programmed Krebs-2 cell extract. [35S]methionine labeling of proteins was performed in a 20-μl total reaction volume in the absence (lane 1) or presence (lanes 2 to 9) of EMCV mRNA (4 μg/ml). Prior to the addition of mRNA, the extracts were preincubated with the indicated proteins, as described in the legend to Fig. 1. GST-4E-BP1 was used at 15, 30, and 60 μg/ml (lanes 3, 4, and 5, respectively). eIF4E was used at 3, 6, and 12 μg/ml (lanes 7, 8, and 9, respectively). Translation products were separated by SDS-PAGE and transferred to a PVDF membrane. The autoradiograph of the membrane is shown. The positions of two abundant cellular proteins (p47 and p50; arrowheads), the EMCV-specific protein 3Dpol (arrow), and the [14C]methylated protein molecular weight markers (GE Healthcare) are indicated. (B) Western blotting analysis of EMCV-specific protein 3Dpol synthesis. The middle portion of the membrane from panel A was probed with anti-3Dpol as described in Materials and Methods. 3Dpol band intensities in different lanes were compared using NIH Image version 1.63 software. The values obtained from reactions performed in the absence of added proteins (lanes 2 and 6) were defined as 100%. (C) EMCV RNA replication was assayed in 30-μl reaction mixtures containing untreated extract, EMCV mRNA (4 μg/ml), and other components as described in Materials and Methods. Before mRNA addition, reaction mixtures were preincubated with control buffer (lane 1), GST-4E-BP1 (lane 2), eIF4E (lane 3), or a combination of eIF4E and 2Apro (lane 4), as described for Fig. 1A. The RNA products were pulse-labeled with [α-32P]CTP after 4 to 5 h of incubation at 32°C and analyzed by agarose gel electrophoresis and autoradiography. The position of the intact EMCV mRNA is indicated (vRNA). (D) Reaction mixtures (30 μl) preincubated with either control buffer or the indicated components (as described for Fig. 1A) and programmed with EMCV mRNA (4 μg/ml) were incubated for 20 h at 32°C. The samples were then treated with a mixture of RNases A and T1 and assayed for infectivity after appropriate dilution, as described in Materials and Methods. Values represent the average of three independent titer determinations. Error bars indicate the standard deviation from the mean.
To determine whether the stimulation of viral protein synthesis by 4E-BP1 is sufficient to affect EMCV RNA replication, we pulse-labeled the reaction mixtures with [α-32P]CTP 4 h after the beginning of incubation with 4E-BP1. The newly synthesized RNA was extracted and analyzed by agarose gel electrophoresis and autoradiography. EMCV RNA synthesis was stimulated approximately 15-fold in the presence of 4E-BP1 (Fig. 2C; compare lanes 2 and 1). Conversely, addition of eIF4E reduced RNA synthesis to below detectable levels (Fig. 2C; compare lanes 3 and 1). Consistent with the importance of intact eIF4G for the eIF4E-mediated inhibition of translation, cleavage of eIF4G by 2Apro restored viral RNA synthesis (Fig. 2C; compare lane 4 with lanes 3 and 1).
We then examined the effects of 4E-BP1, 4E-BP2, eIF4E, eIF4A, and eIF4G-Ct on EMCV yield. EMCV titers in reaction mixtures supplemented with different factors were determined after a 20-h incubation (Fig. 2D). Strikingly, 4E-BP1 and 4E-BP2, as well as m7GDP, stimulated EMCV synthesis 24- to 35-fold (compare bars 3, 5, and 15 with 1), whereas 4E-BP1Δ4E, which cannot bind eIF4E, had only a marginal effect. Conversely, eIF4E, but not the eIF4E W73A mutant, dramatically decreased the viral titer by 150- to 200-fold (Fig. 2D; compare bars 6 and 8 with 1). Addition of the C-terminal portion of eIF4G or 2Apro potently stimulated infectivity (7- and 15-fold, respectively; compare bars 10 and 11 with 1), and this enhancement was not influenced by coaddition of eIF4E or 4E-BP1. Overall, EMCV titers under different conditions covaried with luciferase expression from EMCV IRES-luc(A+) mRNA (compare Fig. 2D with Fig. 1A). However, the magnitude of the changes in viral titer was greater than that measured for translation efficiency.
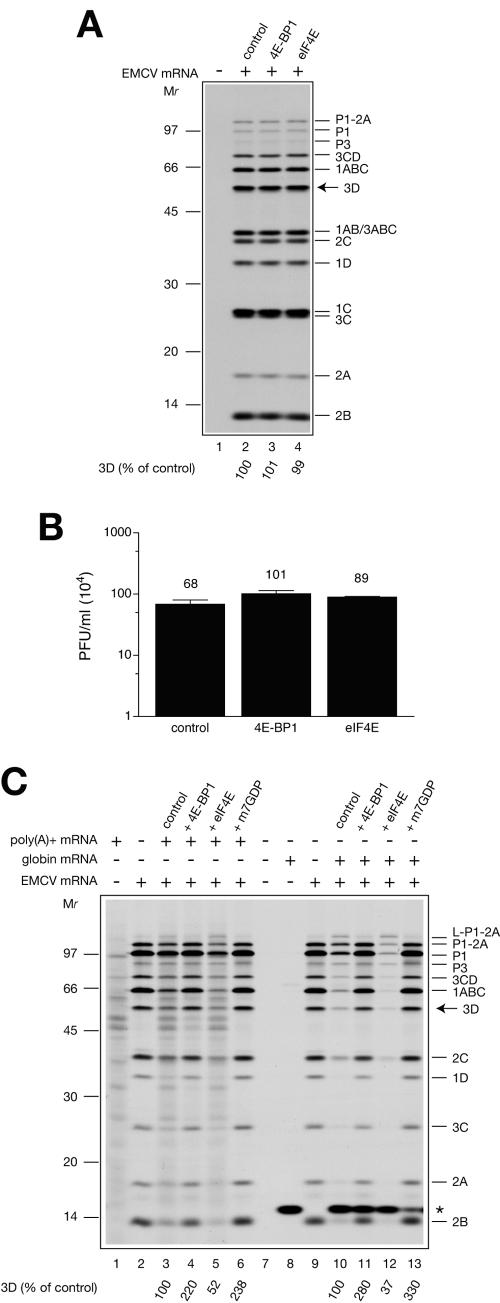
Importantly, and in agreement with a role for mRNA competition in the regulation of viral RNA translation, addition of eIF4E to the EMCV mRNA-programmed or nuclease-treated extract neither inhibited EMCV mRNA translation (as judged by the accumulation of the viral protein 3D) nor changed the expression pattern of virus-specific polypeptides (Fig. 3A; compare lane 4 with lane 2). Also, eIF4E had no effect on EMCV synthesis (Fig. 3B). These results rule out the possibility that contaminating bacterial proteins, which may be present in the eIF4E preparation, adversely affected EMCV replication. Nuclease treatment also abolished the stimulatory effect of 4E-BP1 on EMCV translation and replication (Fig. 3A and B). Although these negative controls argued for the importance of mRNA competition in the regulation of viral protein expression by eIF4F, they did not rule out an alternative possibility. Specifically, the detrimental effect of nuclease treatment might be a consequence of destruction or inactivation of some labile regulatory components of the extract. To address this possibility, we restored mRNA competition by adding saturating concentrations of capped mRNAs [either total poly(A)+ mRNA isolated from the cytoplasm of Krebs-2 cells or globin mRNA] along with EMCV mRNA to the nuclease-treated extract and examined the effects of 4E-BP1 and eIF4E on viral protein expression. Translation of poly(A)+ mRNA alone yielded heterogeneous polypeptides similar to the products of endogenous mRNA translation in the untreated extract (Fig. 3C, lane 1). Globin mRNA translation yielded a 15-kDa polypeptide as expected (lane 8). When EMCV mRNA was translated in the presence of poly(A)+ or globin mRNA, the expression of viral proteins was reduced two- to threefold (compare lane 3 with 2 and lane 10 with 9). In parallel, 18S rRNA was used as a negative control and found not to inhibit viral translation (data not shown). (It should be noted that a molar excess of capped mRNAs over EMCV mRNA was used in these experiments. This was to mimic the initial stage of infection when viral mRNA constitutes a minor fraction of total mRNA.) Addition of 4E-BP1 to the system programmed with a mixture of EMCV mRNA and capped mRNAs stimulated virus protein expression two- to threefold (compare lane 4 with 3 and lane 11 with 10), similar to the addition of m7GDP (compare lane 6 with 3 and lane 13 with 10). In contrast, eIF4E markedly inhibited viral protein synthesis (1.9- to 2.7-fold inhibition; compare lane 5 with 3 and lane 12 with 10). Thus, the addition of capped mRNAs to the EMCV mRNA-programmed nuclease-treated extract rescues the regulation of viral protein expression by eIF4F.
FIG. 3.
4E-BP1 and eIF4E have no effect on EMCV protein synthesis and replication in nuclease-treated Krebs-2 cell extract. (A) Products of EMCV mRNA translation. Reaction mixtures contained the nuclease treated Krebs-2 cell extract but otherwise were identical to that described in Fig. 2A. Reaction mixtures were preincubated with control buffer, GST-4E-BP1 (60 μg/ml), or eIF4E (12 μg/ml) where indicated. Translation was performed at 32°C for 3 h. Aliquots (5 μl) of the translation reaction mixtures were analyzed by SDS-PAGE. An autoradiogram of the dried gel is shown. [35S]methionine incorporation into EMCV-specific protein 3Dpol was quantified using a Fuji BAS2000 phosphorimager. The value obtained from the reaction performed in the absence of added protein (lane 2, control) was defined as 100%. (B) EMCV yields. Reaction mixtures (30 μl, unlabeled) were preincubated with control buffer, GST-4E-BP1, or eIF4E and programmed with EMCV mRNA, as specified above. Plaques were scored following incubation for 20 h at 32°C and RNase treatment. Data represent the average of three determinations. Error bars indicate the standard deviation from the mean. (C) Coaddition of capped mRNA competitors rescues the regulation of translation of EMCV mRNA by 4E-BP1 and eIF4E in nuclease-treated extract. EMCV mRNA (4 μg/ml) was translated at 32°C for 90 min in the absence (lanes 2 and 9) or presence of either total Krebs-2 cell poly(A)+ mRNA (40 μg/ml, lanes 3 to 6) or globin mRNA (10 μg/ml, lanes 10 to 13). Reaction mixtures were preincubated with control buffer, GST-4E-BP1 (60 μg/ml), eIF4E (12 μg/ml), or m7GDP (0.5 mM) where indicated. Products of translation of Krebs-2 cell poly(A)+ mRNA or globin mRNA alone are shown in lanes 1 and 8, respectively. No mRNA was added to the reaction mixture analyzed in lane 7. Relative values for [35S]methionine incorporation into EMCV-specific protein 3Dpol were determined as in panel A. On panels A and C, the assignment of EMCV polypeptides was as described previously (55). The positions of the 14C-methylated protein molecular weight markers (GE Healthcare) are also shown. An asterisk on panel C indicates the position of globin.
Specific stimulation of EMCV and PV translation in cells treated with siRNA against eIF4E.
To provide evidence that the eIF4G/4A subcomplex concentration determines the rate of EMCV protein synthesis in vivo, we used RNA interference to specifically deplete eIF4E (58). Such depletion would be expected to decrease competition from cellular mRNAs by increasing the amount of the eIF4G/4A complex available for viral mRNA translation. The selected siRNA elicited strong (∼85%) knockdown of eIF4E (Fig. 4B; also see Fig. S1A and S2B in the supplemental material). eIF4E knockdown did not lead to a decrease in the overall abundance of eIF4GI or eIF4AI, which accounts for the majority of total eIF4G and eIF4A (6, 50), but dramatically decreased the amount of eIF4F, as determined by cap-column pull-down assays (see Fig S1A in the supplemental material) (data not shown). To determine whether down-regulation of eIF4E decreases the rate of translation initiation, polyribosomes isolated from the control and eIF4E knockdown cells were fractionated by sucrose density gradient centrifugation. eIF4E knockdown cells displayed a higher 80S monosome/polyribosome ratio compared to the control. In addition, a small shift of the polyribosome distribution in favor of lighter polysomes was evident (see Fig. S1B in the supplemental material). The reduction in polyribosome loading after eIF4E depletion is consistent with the inhibition of cellular mRNA translation initiation under these conditions.
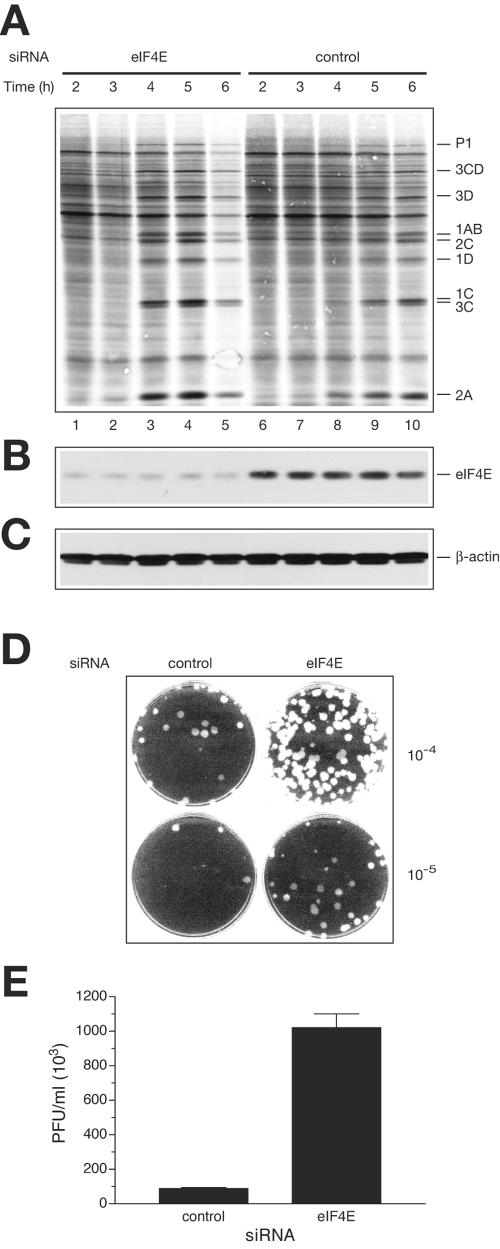
FIG. 4.
eIF4E knockdown stimulates translation and replication of EMCV in vivo. (A) Time course of protein synthesis in EMCV-infected cells. siRNA against eIF4E or a nonspecific siRNA (con- trol) was transfected into HeLa S3 cells. eIF4E knockdown or control cells were infected with EMCV, and protein synthesis was examined by pulse-labeling with [35S]methionine at the indicated time points. After labeling, polypeptides were resolved by SDS-PAGE and transferred to a PVDF membrane. The autoradiograph of the membrane is shown. The positions of the major EMCV-specific proteins are indicated on the right. (B) eIF4E levels in cells, as analyzed by Western blotting. The membrane from panel A was probed with anti-eIF4E, and signals were quantified as described in Materials and Methods. The average level of eIF4E depletion for lanes 1 to 5 (versus lanes 6 to 10) was 86%. (C) β-Actin detection by Western blotting (a loading control). (D) Plaque assays of the indicated dilutions of the lysates from control and eIF4E knockdown cells 4 h after infection. (E) EMCV yield, as affected by eIF4E siRNA treatment. EMCV-infected cells (eIF4E knockdown or control, unlabeled) were lysed at 4 h postinfection. Viral titer was measured as described in the legend to Fig. 2D.
To examine the effect of eIF4E knockdown on viral protein synthesis, cells were pulse-labeled with [35S]methionine at various times after EMCV infection. At 4 h postinfection, only trace amounts of viral proteins could be detected in cells pretreated with the control siRNA (Fig. 4A, lane 8). In contrast, in cells treated with siRNA against eIF4E, viral protein synthesis was robust by 4 h postinfection (lane 3). The stimulatory effect of eIF4E knockdown on viral protein synthesis was also observed after 5 h of infection (compare lane 4 to lane 9). After 6 h of infection, the eIF4E knockdown cells, but not control cells, exhibited morphological changes, indicative of virus-induced cytopathic effect, accompanied by a general decline in protein-synthesizing capacity (Fig. 4A, lane 5) (data not shown). These data suggest that knocking down eIF4E expression accelerates EMCV protein synthesis. We also analyzed viral titers recovered from control and eIF4E knockdown cells at 4 h postinfection. An approximately 10-fold increase in infectious virus production was associated with eIF4E knockdown (Fig. 4D and E). It is worth mentioning that this elevation in the virus titer, although significant, is less than that exerted by the sequestration of eIF4E by 4E-BPs in vitro (24- to 28-fold stimulation [Fig. 2D]). We attribute this to the fact that some residual eIF4E (10 to 20%), and by inference some eIF4F, is present in eIF4E knockdown cells. On the contrary, sequestering of eIF4E by 4E-BP excess in vitro would completely disrupt eIF4F.
The abundance of the eIF4G/4A subcomplex could also play a role at an early stage of enterovirus infection when cleavage of eIF4G is not yet accomplished. To test this hypothesis, we examined the effect of eIF4E knockdown on PV infection. HeLa cells transfected with either control siRNA or siRNA directed against eIF4E were infected with PV, and the kinetics of viral protein synthesis were analyzed by [35S]methionine pulse-labeling. As with EMCV, eIF4E knockdown significantly shortened the eclipse phase of infection, during which no virus proteins can be detected (see Fig. S2A in the supplemental material). Higher rates of PV protein synthesis were evident at 3, 4, and 5 h postinfection in eIF4E knockdown cells as compared to control cells (see Fig. S2A in the supplemental material). Consistent with these results, cells depleted of eIF4E exhibited a PV-induced cytopathic effect earlier than control cells and produced more PV (see Fig. S2D in the supplemental material) (data not shown). Thus, eIF4E appears to be a general, rather than EMCV mRNA-specific, inhibitor of IRES-mediated translation. An unlikely possibility that cannot be rigorously excluded is that eIF4E depletion primarily stimulates viral RNA replication and that the enhancement of viral protein accumulation is a secondary effect.
DISCUSSION
Because eIF4E is not required for translation by internal ribosome entry, it has been generally assumed that this factor does not play a role in the regulation of IRES activity. Here we demonstrate that eIF4E is, in fact, a negative regulator of EMCV mRNA translation under conditions of competition with cellular mRNAs. Saturation of the eIF4G/4A subcomplex with eIF4E to generate eIF4F dramatically decreases EMCV mRNA translation and virus yield in untreated extracts. In contrast, sequestration of eIF4E by 4E-BPs in vitro or depletion of eIF4E in vivo stimulates EMCV mRNA translation and increases viral titer. These results imply that the intact eIF4F complex is unable to support efficient IRES function because it is sequestered by capped mRNA. Freeing the eIF4G/4A subcomplex relieves competition from cellular mRNAs, thereby favoring viral mRNA translation. Our findings support the idea that the cytoplasmic concentration of active eIF4F is less than the concentration of total cellular mRNAs (1, 57). The idea that a discriminatory initiation factor regulates competition between cellular and viral mRNAs in EMCV infection was first proposed three decades ago (17, 29, 49). However, the limiting step and limiting components in translation were not defined. Our results clearly demonstrate that eIF4F plays this discriminatory role through the availability of its cap-binding subunit, eIF4E. The abundance of functional eIF4E, which is regulated by 4E-BPs, thus acts as a switch between cap-dependent and IRES-mediated translation.
How does eIF4E dissociation from eIF4F enhance virus-specific translation? eIF4E dissociation is believed to cause a conformational change in eIF4GI that can be detected by its slower rate of cleavage by picornavirus proteases (19, 37). However, several lines of evidence suggest that this conformational change cannot account for the stimulatory effect of the eIF4G/4A subcomplex on virus-specific translation. First, UV cross-linking experiments suggest that eIF4G binds efficiently to the EMCV IRES as a component of the eIF4F complex (42). Second, the cap analog m7GDP, which inhibits the cap-binding activity of eIF4F but does not alter eIF4F assembly, stimulates EMCV IRES activity and viral production in a manner similar to 4E-BPs (Fig. 1A and 2D). Finally, and most importantly, 4E-BP1 and eIF4E have no effect on viral RNA translation in a nuclease-treated extract (Fig. 3A). Thus, in the reconstituted system or in nuclease-treated extract, EMCV IRES appears to interact with the eIF4F or eIF4G/4A complexes with comparable efficiency. We therefore conclude that competition from cellular mRNAs for eIF4F is required for the regulation of EMCV synthesis by eIF4E and 4E-BPs.
Luciferase translation from the EMCV IRES is enhanced in response to eIF4G cleavage or upon addition of the eIF4G C-terminal protein fragment to the extract. Similar results have been reported for PV IRES-mediated translation (4, 52). If this effect were to be influenced by mRNA competition, it should be more pronounced in the presence of competing cellular mRNAs. Consistent with this prediction, we found that 2Apro treatment stimulated EMCV IRES activity much more potently in untreated than in nuclease-treated extracts (11-fold versus 1.4-fold; compare Fig. 1A, black bar 11, and B, black bar 8). Likewise, cellular mRNA competition was required for eIF4G-Ct- or m7GDP-mediated stimulation of IRES activity, as this stimulation occurred exclusively in untreated extracts.
RNA replication and virus yield correlated with EMCV mRNA translation efficiency, indicating that translation is the limiting step in virus replication. Strikingly, the magnitude of modulation of RNA replication and virion formation by 4E-BP1 and eIF4E was substantially higher than the magnitude of their effect on EMCV mRNA translation. Thus, effects associated with competition for translation factors are amplified at subsequent steps of the infectious cycle. Interestingly, eIF4F complex dissociation is also beneficial for PV gene expression, inasmuch as eIF4E-depleted or rapamycin-treated cells supported viral protein synthesis to a higher level than the respective control cells (see Fig. S2A in the supplemental material) (5). Presumably, this stimulation occurs early in infection—prior to eIF4G cleavage, when the viral RNA must compete with cellular mRNAs for the limiting pool of intact eIF4F. It remains to be determined whether eIF4F dissociation stimulates infectious processes induced by other picornaviruses.
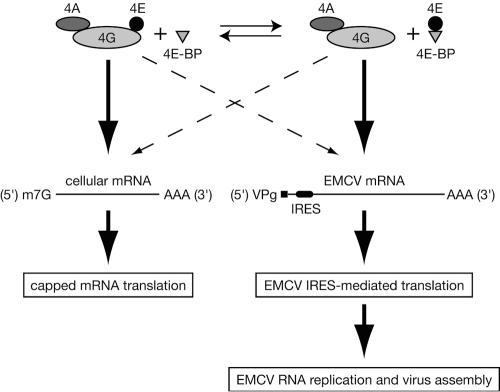
A model illustrating the regulation of EMCV replication by eIF4F is shown in Fig. 5. Central to this model is the fact that EMCV IRES does not compete efficiently with capped cellular mRNAs for eIF4F unless the cap-binding subunit eIF4E is sequestered in a complex with the 4E-BPs, and the relative abundance of the eIF4G/4A subcomplex is increased. As there is no cap-binding subunit within the eIF4G/4A subcomplex, one can assume that it is not recruited efficiently by cellular mRNAs. Indeed, in the presence of eIF4A, the binding affinity of eIF4G for β-globin mRNA is lower than that for EMCV IRES by up to 100-fold (32). However, the concentration of the eIF4G/4A subcomplex in HeLa cells is limiting for translation of picornavirus RNAs, since eIF4E knockdown significantly augments the expression of viral proteins in EMCV- and PV-infected cells. Because eIF4A is not tightly associated with eIF4G and recycles during translation (39), some eIF4G may also exist outside the eIF4F complex or the binary eIF4G/4A subcomplex. However, this “free” eIF4G is not expected to bind EMCV IRES with high affinity (32) and is therefore not shown in the model.
FIG. 5.
Model explaining eIF4F regulation of mRNA competition in EMCV-infected cells. It is presumed that there is equilibrium between eIF4F (eIF4E/4G/4A) and a binary subcomplex comprised of eIF4G and eIF4A (eIF4G/4A) and that EMCV mRNA competes with capped cellular mRNA for the recruitment of eIF4G shared by these complexes. Saturation of the eIF4G/4A subcomplex with eIF4E to generate eIF4F increases its recruitment by capped cellular mRNAs and dramatically inhibits EMCV translation and replication (Fig. 1 and 2). Hence, EMCV mRNA encounters strong competition from cellular mRNA when it binds to eIF4G within the ternary eIF4F complex. By default, EMCV mRNA uses eIF4G within the binary eIF4G/4A subcomplex, which is recruited inefficiently by capped mRNA (32, 36). Dephosphorylated (active) 4E-BP1 and 4E-BP2 (designated as 4E-BP) trigger the expulsion of eIF4E from the eIF4F ternary complex. Elevation of the concentration of the eIF4G/4A subcomplex, resulting from either 4E-BP activation (5, 51) or eIF4E knockdown (Fig. 4), stimulates EMCV IRES-directed translation and downstream virus-specific processes. Thick solid and thin dashed arrows designate efficient and inefficient pathways, respectively. m7G and AAA denote the cap structure and the poly(A) tail of the mRNA, respectively. VPg denotes the genome-linked protein of EMCV.
The cellular tropism and pathogenesis of picornaviruses may be determined in part by the availability of IRES trans-acting factors, which bind specifically to picornavirus IRESs and regulate their function (4). An intriguing idea is that tropism and pathogenesis are also influenced by the concentration of free eIF4G/4A, which is dependent on the expression of the eIF4F components and regulation of the abundance and phosphorylation state of 4E-BPs. Host permissiveness for virus translation could be limited by exposure of cells to extracellular stimuli that activate mTOR through phosphatidylinositol 3-kinase signaling and increase the phosphorylation of 4E-BPs (43). Hence, it is likely that both cell-type-specific and environmental factors affect the ability of the virus to establish an efficient infection. It is also possible that pathological conditions that affect the concentration of eIF4F components will affect the outcome of a viral infection. For example, several cancers are associated with increased levels of eIF4F subunits, in particular eIF4E and eIF4G (2, 23). Under conditions of eIF4G excess, cellular mRNAs may not compete with picornavirus mRNAs for translation (or compete in another fashion for another limiting component), and these cells would be expected to be highly susceptible to infection. In this regard, it is noteworthy that picornaviruses preferentially kill malignant cells over normal cells (18, 46). However, it is not known whether the tumor cells used in these studies contained higher levels of eIF4G than normal cells.
In addition to its role in the expression of virus genomes, the ratio between the different eIF4G complexes may regulate cellular proliferation, survival, and death, as IRES elements are often found in the mRNAs of genes controlling these processes (20, 24, 26, 48). Our results suggest that IRES-mediated translation of cellular mRNAs should not only be resistant to eIF4F dissociation but stimulated by it. The following examples of selective translation conform to this notion. Despite a reduction in overall protein synthesis, the X-linked inhibitor of apoptosis mRNA, which possesses an IRES, is translated more efficiently under serum starvation, which decreases 4E-BP phosphorylation (24). Elevated levels of X-linked inhibitor of apoptosis are thought to delay the onset of apoptosis and allow the cell to survive under stress conditions. A rapid inhibition of translation—as a consequence of 4E-BP1 dephosphorylation and eIF2α phosphorylation by PERK—develops during hypoxia, which is common in many human diseases such as stroke, heart disease, and cancer (24). However, at least two proteins (HIF1α and vascular endothelial growth factor) involved in cell survival are upregulated during hypoxia, presumably via their synthesis by IRES-dependent translation. It is also noteworthy that in neurons from the mollusk Aplysia californica the switch from cap-dependent to IRES-dependent translation is believed to be triggered by dephosphorylation of eIF4E (9). Knowing how selective translation allows cells to adapt to environmental and physiological stresses, such as hypoxia, heat shock, toxins, and drug exposure, is important for understanding many human disorders and may lead to the development of new therapeutic approaches for such conditions.
Supplementary Material
Acknowledgments
We thank Ann Palmenberg for the antibody against mengovirus protein 3Dpol, Hiroaki Imataka for pT7EMCVluc(A)+, Sandra Perreault and Colin Lister for excellent technical assistance, and Wayne Sossin for critical reading of the manuscript.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) to N.S., who is a CIHR Distinguished Scientist and a Howard Hughes Medical Institute International Scholar.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anthony, D. D., and W. C. Merrick. 1991. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J. Biol. Chem. 266:10218-10226. [PubMed] [Google Scholar]
- 2.Avdulov, S., S. Li, V. Michalek, D. Burrichter, M. Peterson, D. M. Perlman, J. C. Manivel, N. Sonenberg, D. Yee, P. B. Bitterman, and V. A. Polunovsky. 2004. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5:553-563. [DOI] [PubMed] [Google Scholar]
- 3.Aviv, H., and P. Leder. 1972. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. USA 69:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Beretta, L., Y. V. Svitkin, and N. Sonenberg. 1996. Rapamycin stimulates viral protein synthesis and augments the shutoff of host protein synthesis upon picornavirus infection. J. Virol. 70:8993-8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conroy, S. C., T. E. Dever, C. L. Owens, and W. C. Merrick. 1990. Characterization of the 46,000-dalton subunit of eIF-4F. Arch. Biochem. Biophys. 282:363-371. [DOI] [PubMed] [Google Scholar]
- 7.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duque, H., and A. C. Palmenberg. 1996. Epitope mapping of monoclonal antibodies raised to recombinant Mengo 3D polymerase. Virus Genes 13:159-168. [DOI] [PubMed] [Google Scholar]
- 9.Dyer, J. R., S. Michel, W. Lee, V. F. Castellucci, N. L. Wayne, and W. S. Sossin. 2003. An activity-dependent switch to cap-independent translation triggered by eIF4E dephosphorylation. Nat. Neurosci. 6:219-220. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenfeld, E. 1996. Initiation of translation by picornavirus RNAs, p. 549-573. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 11.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 12.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 15.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 16.Gingras, A. C., Y. Svitkin, G. J. Belsham, A. Pause, and N. Sonenberg. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA 93:5578-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golini, F., S. S. Thach, C. H. Birge, B. Safer, W. C. Merrick, and R. E. Thach. 1976. Competition between cellular and viral mRNAs in vitro is regulated by a messenger discriminatory initiation factor. Proc. Natl. Acad. Sci. USA 73:3040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromeier, M., S. Lachmann, M. R. Rosenfeld, P. H. Gutin, and E. Wimmer. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA 97:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 21.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 22.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Hershey, J. W. B., and S. Miyamoto. 2000. Translational control and cancer, p. 637-654. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Jen, G., B. M. Detjen, and R. E. Thach. 1980. Shutoff of HeLa cell protein synthesis by encephalomyocarditis virus and poliovirus: a comparative study. J. Virol. 35:150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolupaeva, V. G., I. B. Lomakin, T. V. Pestova, and C. U. T. Hellen. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 23:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamphear, B. J., R. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200-19203. [PubMed] [Google Scholar]
- 29.Lawrence, C., and R. E. Thach. 1974. Encephalomyocarditis virus infection of mouse plasmacytoma cells. I. Inhibition of cellular protein synthesis. J. Virol. 14:598-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Q., H. Imataka, S. Morino, G. W. Rogers, Jr., N. J. Richter-Cook, W. C. Merrick, and N. Sonenberg. 1999. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19:7336-7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, T. A., X. Kong, T. A. Haystead, A. Pause, G. Belsham, N. Sonenberg, and J. C. J. Lawrence. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653-656. [DOI] [PubMed] [Google Scholar]
- 32.Lomakin, I. B., C. U. T. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707-716. [DOI] [PubMed] [Google Scholar]
- 34.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosenkis, J., S. Daniels-McQueen, S. Janovec, R. Duncan, J. W. B. Hershey, J. A. Grifo, W. C. Merrick, and R. E. Thach. 1985. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eucaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J. Virol. 54:643-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novoa, I., and L. Carrasco. 1999. Cleavage of eukaryotic translation initiation factor 4G by exogenously added hybrid proteins containing poliovirus 2Apro in HeLa cells: effects on gene expression. Mol. Cell. Biol. 19:2445-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohlmann, T., V. M. Pain, W. Wood, M. Rau, and S. J. Morley. 1997. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E-binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 16:844-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pause, A., G. J. Belsham, A. C. Gingras, O. Donzé, T. A. Lin, J. C. J. Lawrence, and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 39.Pause, A., N. Méthot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova, T. V., C. U. T. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova, T. V., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raught, B., A.-C. Gingras, and N. Sonenberg. 2000. Regulation of ribosomal recruitment in eukaryotes, p. 245-293. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Richter, J. D., and N. Sonenberg. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477-480. [DOI] [PubMed] [Google Scholar]
- 45.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 46.Shafren, D. R., G. G. Au, T. Nguyen, N. G. Newcombe, E. S. Haley, L. Beagley, E. S. Johansson, P. Hersey, and R. D. Barry. 2004. Systemic therapy of malignant human melanoma tumors by a common cold-producing enterovirus, coxsackievirus A21. Clin. Cancer Res. 10:53-60. [DOI] [PubMed] [Google Scholar]
- 47.Skern, T., B. Hampölz, A. Guarné, I. Fita, E. Bergmann, J. Petersen, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 48.Stoneley, M., and A. E. Willis. 2004. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23:3200-3207. [DOI] [PubMed] [Google Scholar]
- 49.Svitkin, Y. V., V. A. Ginevskaya, T. Y. Ugarova, and V. I. Agol. 1978. A cell-free model of the encephalomyocarditis virus-induced inhibition of host cell protein synthesis. Virology 87:199-203. [DOI] [PubMed] [Google Scholar]
- 50.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svitkin, Y. V., H. Hahn, A.-C. Gingras, A. C. Palmenberg, and N. Sonenberg. 1998. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 72:5811-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interaction with eIF4G stimulates picornavirus IRES-dependent translation. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 53.Svitkin, Y. V., L. P. Ovchinnikov, G. Dreyfuss, and N. Sonenberg. 1996. General RNA binding proteins render translation cap dependent. EMBO J. 15:7147-7155. [PMC free article] [PubMed] [Google Scholar]
- 54.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svitkin, Y. V., and N. Sonenberg. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 257:155-170. [DOI] [PubMed] [Google Scholar]
- 57.Thach, R. E. 1992. Cap recap: the involvement of eIF-4F in regulating gene expression. Cell 68:177-180. [DOI] [PubMed] [Google Scholar]
- 58.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.