Abstract
Placental development initially occurs in a low-oxygen (O2) or hypoxic environment. In this report we show that two hypoxia-inducible factors (HIFs), HIF1α and HIF2α, are essential for determining murine placental cell fates. HIF is a heterodimer composed of HIFα and HIFβ (ARNT) subunits. Placentas from Arnt−/− and Hif1α−/− Hif2α−/− embryos exhibit defective placental vascularization and aberrant cell fate adoption. HIF regulation of Mash2 promotes spongiotrophoblast differentiation, a prerequisite for trophoblast giant cell differentiation. In the absence of Arnt or Hifα, trophoblast stem cells fail to generate these cell types and become labyrinthine trophoblasts instead. Therefore, HIF mediates placental morphogenesis, angiogenesis, and cell fate decisions, demonstrating that O2 tension is a critical regulator of trophoblast lineage determination. This novel genetic approach provides new insights into the role of O2 tension in the development of life-threatening pregnancy-related diseases such as preeclampsia.
Defective placentation is implicated as one of the causes of preeclampsia, a pregnancy-associated disease that affects 4 to 5% of all births and a major cause of maternal and neonatal mortality (49). Women with preeclampsia suffer from edema, hypertension, and proteinuria. Preeclamptic placentas display shallow invasion of cytotrophoblasts (trophectoderm derivatives of the placenta) into the uterus, fewer numbers of invasive cytotrophoblasts, and maternal spiral arteries that have not been converted into enlarged vessels. These pathologies lead to reduced placental blood flow and placental hypoxia (16, 49). Physiological hypoxia may also play an important role in directing placental development. Therefore, delineating the role of O2 tension in trophoblast differentiation during placentation will enable us to better understand and treat pregnancy-related diseases. Furthermore, mouse models of preeclampsia resulting in intrauterine growth retardation will allow us to genetically define pathways that are involved in both placental development and pathologies.
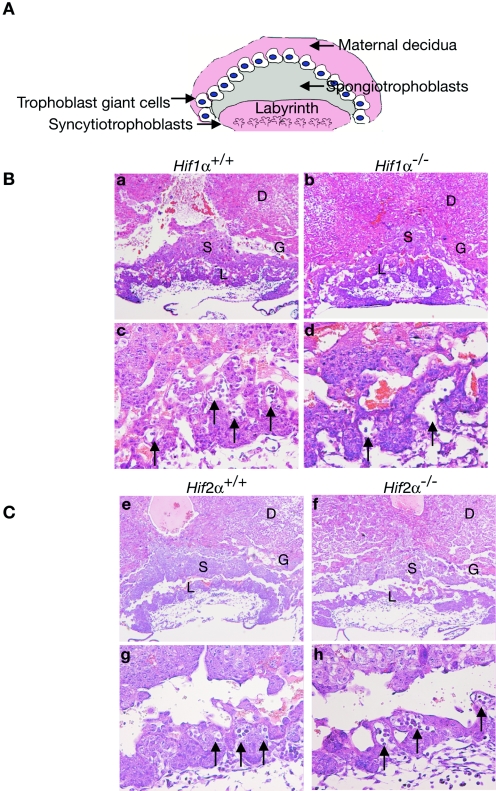
Murine placental development is similar to that of human embryos. The three layers of the chorioallantoic placenta form between embryonic day 9.0 (E9.0) and E10.0. The layer closest to the embryo, the labyrinthine layer, develops when fetal blood vessels from the allantoic mesoderm invade the chorion (Fig. 1A). Fetal blood vessel invasion instructs chorionic trophoblast precursors to differentiate by fusion into syncytiotrophoblasts (38). Labyrinthine fetal and maternal blood vessels become interdigitated in order to facilitate O2, nutrient, and waste exchange (14). Trophoblasts from the polar trophectoderm (in contact with the inner cell mass) give rise to progenitor cells of the ectoplacental cone (EPC). EPC cells differentiate into spongiotrophoblasts forming the supportive middle layer of the placenta (13, 19). Trophoblast giant cells (TGCs) comprise the placental layer that is most invasive into maternal decidua, performing a function similar to that of human invasive cytotrophoblasts (13). TGCs likely emerge from a precursor population in the EPC (e.g., spongiotrophoblasts), are necessary for implantation, promote maternal blood flow, and produce hormones such as placental lactogen 1 and 2 (Pl 1 and Pl 2) (12). In summary, each placental layer plays a necessary function during pregnancy.
FIG. 1.
Vascular defects in Hif1α−/− placentas. (A) Diagram of placental architecture. Trophoblast giant cells (TGCs) are placental lactogen 1+ (Pl 1+), spongiotrophoblasts are Tpbp+, and the syncytiotrophoblasts of the labyrinthine are TFEB+. (B) E9.5 placental sections stained for H&E: Hif1α+/+ (a and c), Hif1α−/− (b and d). (C) E9.5 placental sections stained for H&E: Hif2α+/+ (e and g) and Hif2α−/− (f and h). Hif1α−/− placentas show a decrease in the number of fetal vessels invading from the allantois (arrows in panels e, f, g, and h) into the labyrinthine layer. Original magnification, ×50 (a, b, e, and f) and ×200 (c, d, g, and h). D, maternal deciduum; L, labyrinthine trophoblast; S, spongiotrophoblast; G, TGCs.
The uterine environment profoundly affects placental morphogenesis. In humans the uterine surface experiences low O2 levels (17.9 mm Hg, or 2.5% O2) during early pregnancy; after the placenta has established connections with maternal vasculature, the placenta is relatively O2 rich (60 mm Hg, or about 8.6% O2) (39). Therefore, hypoxia-inducible factors (HIFs) could mediate early placentation. HIF regulates adaptive transcriptional responses to hypoxia by inducing genes involved in glycolysis, red blood cell production, and angiogenesis (42). HIF is a heterodimeric basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor composed of HIFα and HIFβ (ARNT) subunits. ARNT is constitutively transcribed, translated, and detected in the nucleus (1, 24, 50). Two HIFα subunits have been shown to positively regulate the hypoxic response, HIF1α and HIF2α (15, 48, 50). The HIFα subunits are constitutively transcribed and translated; however, they are degraded under normoxia (20% O2) through association with the von Hippel-Lindau E3 ubiquitin ligase (8, 28, 32, 33). Arnt−/− mice die by E10.5 with many cardiovascular defects, including abnormal yolk sac and embryonic blood vessels, decreased hematopoiesis, heart failure, and placental anomalies (2, 3, 27, 31). In addition to HIFα subunits, ARNT associates with multiple bHLH/PAS proteins to regulate responses to a variety of developmental or environmental stimuli. For example, ARNT dimerizes with the aryl hydrocarbon receptor to regulate xenobiotic metabolism (17). Therefore, Arnt−/− placental defects could result from pathways other than those regulated by hypoxia.
Little is known about the role of HIFα subunits during placentation. Both HIF1α and HIF2α are expressed at high levels early in human and mouse gestation (5, 36, and E. Maltepe and M. C. Simon, unpublished observations). However, although abundant in specific cell types, such as fibroblasts and breast epithelial cells, HIF2α is sometimes inactive (20, 34, 43). We show here that placentas from mice lacking both HIF1α and HIF2α exhibit a phenotype identical to that of Arnt−/− placentas, suggesting that O2 levels and O2-sensing pathways are critical regulators of placentation. Arnt−/− and Hif1α−/− Hif2α−/− placentas are avascular and exhibit poor decidual invasion, reduced spongiotrophoblast numbers, expanded TGC numbers, and defective labyrinthine trophoblasts. Therefore, HIF1α and HIF2α perform overlapping functions during placentation. Spongiotrophoblasts are produced in the placenta in a HIF-independent fashion until E8.5. However, maintenance and proliferation of spongiotrophoblasts after E8.5 requires HIF activity. In the absence of HIF, they prematurely differentiate into TGCs. HIF is also essential for spongiotrophoblast and TGC generation in vitro; HIF-deficient trophoblast stem (TS) cells fail to express Mash2 (a transcription factor essential for spongiotrophoblast development) and become syncytiotrophoblasts instead. In aggregate, our results demonstrate that HIF is necessary for the adoption and maintenance of distinct cell fates in vivo and in vitro and suggest that Mash2 is a HIF target gene.
MATERIALS AND METHODS
Generation of mice and trophoblast stem cells.
Mice lacking HIF1α or HIF2α were generated previously (6, 9). Hif1α+/− Hif2α+/− mice were intercrossed to generate Hif1α−/− Hif2α−/− embryos and TS cells. Pregnant females were sacrificed at day 9.5 of gestation to harvest embryos for histology and in situ hybridization or at E3.5 for TS cell generation from preimplantation blastocysts (2, 45). Arnt−/− TS cells were previously described (2). TS cells were differentiated by removing cells from mouse embryo fibroblasts (MEFs), FGF4, and heparin. Unless otherwise indicated, differentiation protocols were performed for 4 days. TS cells were cultured at 3% O2 in an IG750 3 Gas Incubator (Jouan).
Histology and in situ hybridization.
Placentas from E9.5 embryos were fixed in paraformaldehyde for 2 days and subsequently dehydrated in decreasing concentrations of ethanol. Placentas were then paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E) for gross histology. Placental invasion was measured by analyzing H&E sections using National Institutes of Health Image J software. For in situ hybridization, paraffin-embedded sections were deparaffinized and then hybridized to sense and antisense riboprobes as previously described (2).
RNA analysis.
Undifferentiated and differentiated cells were cultured at 20% or 3% O2 for 4 days. Total RNA was extracted with TRIzol reagent (Life Technologies) according to the manufacturer's instructions. The following primers were used for reverse transcription-PCR (RT-PCR): estrogen receptor-related protein β (ERRβ) forward (F), 5′CTACGAGGACTGTACTAGTGG3′; reverse (R), 5′CAAGATGAGAATCTCCATCCAG3′; Fgfr2 F, 5′TCTGGAGATGATGAGGACG3; R, 5′CCAGAGGCAAACTCACCA3′; Tpbp F, 5′-CAGGTACTTGAGACATGACTC-3′; R, 5′-GGCAGAGATTTCTTAGACAATG-3′; Mash2 F, 5′-GAAGGTGCAAACGTCCACTTC-3′; R, 5′-CCTTACTCAGCTTCTTGTTGG-3′; Pl 1 F, 5′-CCACTGAAGACCTGTATACTC-3′; R, 5′-GGACTGCAGTTCTTCGAGTC-3′; Oct4 F, 5′-GGCGTTCTCTTTGGAAAGGTGTTC-3′; R, 5′-CTCGAACCACATCCTTCTCT-3′; and hypoxanthine phosphoribosyltransferase (HPRT) F, 5′-CACAGGACTAGAACACCTGC-3′; R, 5′-GCTGGTGAAAAGGACCTCT-3′.
For Northern blot probes the following fragments were used: a 3′-untranslated region (UTR) of Tpbp as previously described (2), a 3′ UTR of Pl 1 (PCR product from primers above), a 2-kb fragment of the TFEB 3′ UTR, a 300-bp SalI/PstI fragment of the GCM1 coding region (a kind gift from J. Cross), a 1-kb HindIII fragment from the Mash2 coding region, a 800-bp Pst1 fragment from the Proliferin coding region (a kind gift from J. Rossant), a 300-bp fragment from the Hand1 3′ UTR, and a 300-bp EcoRI fragment from β-tubulin.
Q-PCR methods.
Quantitative PCR (Q-PCR) on Hif1α+/+ and Hif1α−/− placental RNA was performed as previously described (7) according to the manufacuturer's protocol (Perkin Elmer) using the Taqman universal PCR master mix kit and the ABI PRISM 79700 sequence detector forward (F), reverse (R), and probes (P), labeled with a fluorescent dye (6-carboxyfluorescein [FAM] or JOE) and a quencher (6-carboxytetramethylrhodamine [TAMRA]). Dyes were attached at the 5′ end, while quenchers were attached to the 3′ end of the probes. Gene expression was standardized for the β-actin gene. Primers used for Q-PCR were the following: β-actin F, 5′-AGAGGGAAATCGTGCGTGAC-3′; R, 5′-CAATAGTGATGACCTGGCCGT-3′; P, 5′ JOE-CACTGCCGCATCCTCTTCCTCCC-TAMRA-3′; Tpbp F, 5′-GGAGTGGCCTCAGCTGTCAT-3′; R, 5′-AACTTCTTTATCCTTCTGCTCTTGCA-3′; P, 5′-FAM-TTCAGCATCCAACTGCGCTTCAGG-3′; Mash2 F, 5′-GTGAAGGTGCAAACGTCCACTT-3′; R, 5′-TCTGGTGCGGCACAGGAA-3′; P, 5′-FAM-CGCACCCGGTTCCTCGCGA-TAMRA-3′.
For Q-PCR on TS cell RNA, we obtained master mix solution and commercially available primer/probe pairs for actin, GCM1, Mash2, and TFEB (Applied Biosciences, Foster City, CA). We made cDNA from 1 μg of sample RNA with random hexamer primer reverse transcription-PCR. cDNA was then diluted 1:10 and assayed in a 20-μl total volume of primer probe pair-Master Mix solution. All reactions were performed in triplicate, and all experiments were performed at least three times in a 7900HT sequence detection system (Applied Biosciences, Foster City, CA). Changes in threshold signal versus actin control (ΔΔcT) calculations were used to calculate relative mRNA levels.
Propidium iodide (PI) incorporation.
TS cells were trypsinized, washed with phosphate-buffered saline (PBS), and fixed at −20°C in 75% ethanol in PBS overnight. Cells were then removed from ethanol by centrifugation, washed with PBS, and resuspended in 10 μg/ml propidium iodide in 1.1% sodium citrate buffer with 1 mg/ml RNase A. Cells were stained in the dark for 1 h, centrifuged, and then resuspended in Isotone buffer. PI incorporation was performed on the FACSCalibur instrument using Cell Quest software.
Phase-contrast microscopy.
For microscopy, TS cells were grown on 60-mm tissue culture-treated plates in the absence of MEFs. Images were acquired using an inverted Nikon Eclipse TE300 microscope using Metamorph software.
RESULTS
Abnormal placentation of Hif1α−/− embryos.
Arnt−/− murine embryos die by E10.5, most likely due to placental failure. We hypothesized that ARNT regulates placentation via dimerization with HIFα subunits. Therefore, we determined what role HIF1α and HIF2α play in placental development. Hif1α+/− males and females were mated, pregnant females dissected, and placentas were isolated at E9.5. Figure 1A illustrates basic murine placental architecture, which is composed of three fetal trophoblast layers: labyrinthine (syncytiotrophoblast), spongiotrophoblast, and TGC. Hif1α−/− placentas displayed a range of phenotypes. Chorioallantoic fusion occurred at the 6- to 7-somite stage (E8.0 to E8.5) in Hif1α+/+ embryos, and embryonic vessels were closely apposed to maternal lacunae by E9.5 (Fig. 1B, panel c). In contrast, the allantois failed to fuse with the chorion in 31% of the Hif1α−/− embryos with more than eight somites. In the 69% of Hif1α−/− embryos where chorioallantoic fusion occurred, vascularization of the chorion was significantly impaired. Hematoxylin and eosin (H&E) staining of these placentas showed that while fetal blood vessels were present, they were fewer in number and appeared dilated compared to wild-type placentas (Fig. 1B, panel d, and Table 1). Quantitative reverse-transcribed-PCR (Q-PCR) analysis of Hif1α−/− placentas revealed that Tpbp, a marker for spongiotrophoblasts, was decreased by more than 50% (190 ± 50 mRNA copies per 103 β-actin mRNA copies in Hif1α−/− versus 420 ± 93 in Hif1α+/+ tissue; n = 6; P < 0.05 at E9.25). In addition, Mash2 mRNA levels were also decreased in the Hif1α−/− placentas (0.15 ± 0.02 in Hif1α−/− versus 0.29 ± 0.02 in Hif1α+/+samples; n = 6; P < 0.05). No change in mRNA expression was observed for a variety of additional genes expressed in the placenta (Hand1, LIMK, placental lactogen 1, adenosine deaminase, matrix metalloproteinase 9, Id2, FLT-1, vascular endothelial growth factor A, FLK-1, and VE-cadherin) in Hif1α−/− placentas compared to the wild type.
TABLE 1.
Summary of placental phenotypes at E9.5a
| Genotype | No. of fetal vesselsb | Chorioallantoic fusionc | Invasiond | Tpbp+e | PI 1+f |
|---|---|---|---|---|---|
| Arnt+/+ | 12 | + | +++ | +++ | +++ |
| Arnt−/− | 0 | − | − | − | ++++ |
| Hif1α+/+ | >25 | + | +++ | +++ | +++ |
| Hif1α−/−g | 4 | + | ++ | ++ | +++ |
| Hif1α−/−g | 0 | − | + | ++ | +++ |
| Hif2α+/+ | >15 | + | +++ | +++ | +++ |
| Hif2α−/− | 10 | + | +++ | +++ | +++ |
| Hif1α−/− | 0 | − | + | + | ++++ |
| Hif2α+/− | |||||
| Hif1α−/− | 0 | − | − | − | ++++ |
| Hif2α−/− |
Most embryos examined had approximately 20 somites when harvested, except Hif1α−/− Hif2α+/− and Hif1α−/− Hif2α−/− embryos, which had between 6 and 8 somites.
Average number of fetal blood vessels in the labyrinthine layer.
Absence (−) or presence (+) of chorioallantoic fusion.
Extent of maternal tissue invasion: very poor (−), poor (+), good (++), very good (+++). Morphometric analysis revealed that Hif1α−/− Hif2α−/− placentas exhibited a 17% decrease in invasion distance, while Hif1α−/− Hif2α+/− exhibited a 5 to 8% decrease.
Extent of Tpbp expression: little to no expression (−), greatly diminished (+), reduced (++), good (+++).
Extent of PI 1 expression: normal expression (+++), increased expression (++++).
Chorioallantoic fusion occurred in 69% of the Hif1α−/− embryos and was not detected in the remaining 31%.
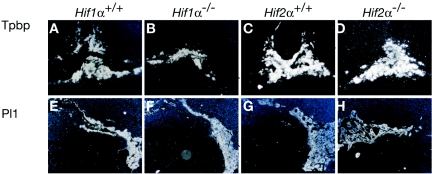
We then evaluated the expression of trophoblast lineage-specific genes in intact placentas. To determine the extent of spongiotrophoblast and TGC development in the absence of HIF1α, in situ hybridization using antisense 35S-labeled riboprobes was performed for Tpbp and the TGC marker placental lactogen 1 (Pl 1). The number of Tpbp+ cells was decreased by approximately 50% in Hif1α−/− placentas (compare Fig. 2A and E to B and F), which is consistent with the Q-PCR data described above. Of note, control sense riboprobes were also used in hybridization assays and showed very little background signal (data not shown). In contrast to Hif1α−/− tissue, Hif2α−/− placentas exhibited normal numbers of fetal blood vessels and proper invasion into maternal tissues (Fig. 1C, panels e, f, g, and h). Additionally, Tpbp+ and Pl 1+ cell numbers in Hif2α−/− placentas were indistinguishable from those of wild-type placentas (Fig. 2C, D, G, and H). Please refer to Table 1 for a summary of all phenotypes. Therefore, placentas isolated from Hif1α−/− mice exhibit less severe defects than those observed in Arnt−/− placentas, while Hif2α−/− placentas appear normal.
FIG. 2.
Expression of cell type-specific markers in trophoblast tissue. E9.5 placentas analyzed by 35S in situ hybridization for Tpbp and Pl 1 in Hif1a+/+ (A and E), Hif1α−/− (B and F), Hif2α+/+ (C and G), and Hif2α−/− (D and H) embryos. Original magnification, ×50.
HIF activity is necessary for placental development in a dose-dependent manner.
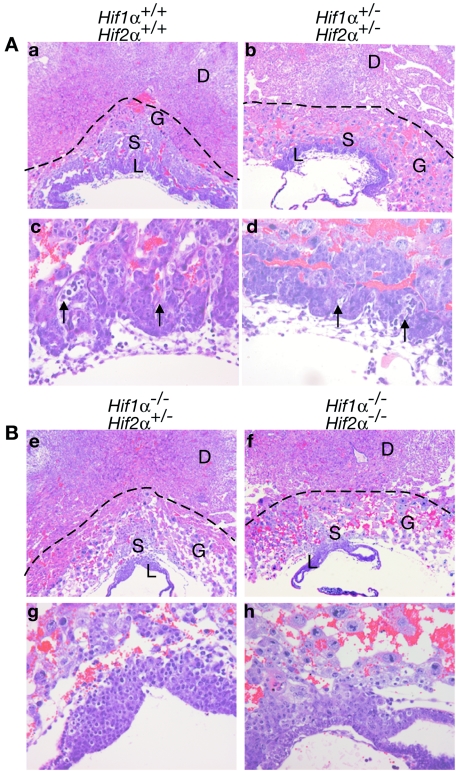
Since neither Hif1α−/− nor Hif2α−/− placentas exhibited phenotypes as severe as Arnt−/− embryos, we evaluated the extent of redundancy between HIF1α and HIF2α during placentation. In crossing Hif1α+/− Hif2α+/− male and female mice, we determined that only 50% of the expected Hif1α+/− Hif2α+/− animals were viable, implying that Hif1α+/− Hif2α+/− mice may exhibit a partially penetrant lethal phenotype. As shown in Table 2, out of 200 live progeny, we obtained only 42 Hif1α+/− Hif2α+/− mice instead of the expected 89 when the Mendelian ratios were modified to account for the embryonic lethality of the following genotypes: Hif1α+/− Hif2α−/−, Hif1α−/− Hif2α+/−, and Hif1α−/− Hif2α−/−. Thus, Hif1α+/− Hif2α+/− progeny were underrepresented. Subsequently, we bred Hif1α+/− Hif2α+/− mice and analyzed 56 E9.5 embryos. All genotypes were present at the appropriate Mendelian ratios. E9.5 Hif1α−/− embryos exhibited defects that have been previously described (23, 40), such as reduced size, open head folds, failure to turn, and abnormal yolk sac vasculature. Of note, Hif1α−/− Hif2α+/− embryos were more severely developmentally delayed than Hif1α−/− embryos, exhibiting no neural tube closure or turning and substantially fewer somites (6 compared to 20 to 23 somites for Hif1α−/− and wild-type embryos). The Hif1α−/− Hif2α−/− embryos exhibited defects similar to those of Hif1α−/− Hif2α+/− embryos but were even smaller (data not shown). Therefore, we analyzed the extent of placental development in Hif1α+/+ Hif2α+/+, Hif1α+/− Hif2α+/−, Hif1α−/− Hif2α+/−, and Hif1α−/− Hif2α−/− embryos at E9.5. Based on H&E staining, Hif1α+/− Hif2α+/− placentas fell into two classes: those with normal invasion of maternal tissue (data not shown) and those that were hypocellular and exhibited shallow invasion (Fig. 3A, panels b and d). Hif1α−/− Hif2α+/− and Hif1α−/− Hif2α−/− placentas displayed a more extensive phenotype, with no allantoic fetal blood vessels and extensive hypocellularity of multiple trophoblast layers (Fig. 3B, panels e, f, g, and h). Invasion of the placenta into maternal decidua was compromised in Hif1α−/− Hif2α+/− placentas and was extremely poor in Hif1α−/− Hif2α−/− embryos (Fig. 3B, panels e, f, g, and h). Morphometric analysis revealed that the Hif1α−/− Hif2α−/− placentas exhibited a 17% decrease in the invasion distance compared to the wild type. Again, please see Table 1 for a summary of these phenotypes. Therefore, HIF1α and HIF2α cooperate in regulating the recruitment of fetal blood vessels into the chorion and placental invasion into maternal decidua as well as establishing overall placental architecture.
TABLE 2.
Hif1α+/− Hif2α+/− × Hif1α+/− Hif2α+/− matings
| Hif1α | Hif2α | No. of live progeny expected | No. of live progeny observeda | No. expected at E9.5c | No. observed at E9.5 |
|---|---|---|---|---|---|
| +/+ | +/+ | 12.5 | 54 | 3.5 | 4 |
| +/+ | −/− | 12.5 | 0 | 3.5 | 3 |
| −/− | +/+ | 12.5 | 0 | 3.5 | 4 |
| −/− | −/− | 12.5 | 0 | 3.5 | 3 |
| +/+ | +/− | 25 | 43 | 7 | 9 |
| +/− | +/+ | 25 | 61 | 7 | 7 |
| +/− | +/− | 50 | 42 | 14 | 18 |
| +/− | −/− | 25 | 0 | 7 | 4 |
| −/− | +/− | 25 | 0 | 7 | 5 |
| Modified ratiosb | |||||
| +/+ | +/+ | 22.25 | 54 | ||
| +/+ | −/− | 0 | 0 | ||
| −/− | +/+ | 0 | 0 | ||
| −/− | −/− | 0 | 0 | ||
| +/+ | +/− | 42.7 | 43 | ||
| +/− | +/+ | 42.7 | 61 | ||
| +/− | +/− | 89 | 42 | ||
| +/− | −/− | 0 | 0 | ||
| −/− | +/− | 0 | 0 |
n = 200 for live progeny.
Mendelian ratios were modified to account for the embryonic lethality of five of the genotypes.
n = 56 for timed matings at embryonic day 9.5.
FIG. 3.
Vascular and architectural defects in Hif1α+/− Hif2α+/−, Hif1α−/− Hif2α+/−, and Hif1α−/− Hif2α−/− placentas. The dotted lines indicate the extent of desidual invasion for each placental section. (A) E9.5 placental sections stained for H&E: Hif1α+/+ Hif2α+/+ (a and c) Hif1α+/− Hif2α+/− and (b and d). (B) E9.5 placental sections stained for H&E: Hif1α−/− Hif2α+/− (e and g) and Hif1α−/− Hif2α−/− (f and h). Arrows denote fetal blood vessels. No fetal vessels (g and h) were detected in the labyrinthine layer of Hif1α−/− Hif2α+/− and Hif1α−/− Hif2α−/− placentas. Original magnification, ×50 (a, b, e, and f) and ×200 (c, d, g, and h). D, maternal deciduum; L, labyrinthine trophoblast; S, spongiotrophoblast; G, TGCs.
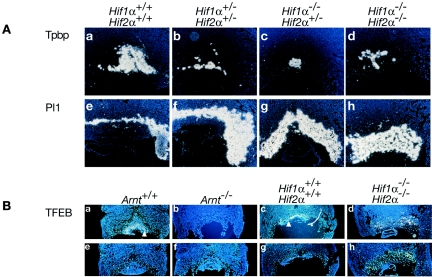
We next determined the extent of spongiotrophoblast and TGC layer differentiation for HIFα-deficient mice. In situ hybridization for Tpbp expression in Hif1α+/− Hif2α+/− embryos showed that poorly invasive placentas also exhibited decreased Tpbp+ cell numbers and expanded Pl 1+ cell numbers, while normally invasive Hif1α+/− Hif2α+/− placentas demonstrated wild-type numbers of Tpbp+ and Pl 1+ cells (Fig. 4A, panels b and f). Hif1α−/− Hif2α+/− placentas exhibited a dramatic decrease in Tpbp+ cells (Fig. 4A, panel c) and increase in Pl 1+ cells (Fig. 4A, panel g). Hif1α+/− Hif2α+/− embryos also exhibited fewer Tpbp+ spongiotrophoblasts and more Pl 1+ TGCs (Fig. 4A, panels d and h, and Table 1). Additionally, the placental architecture was aberrant in Hif1α−/− Hif2α−/− placentas, and some trophoblasts appeared to express both Tpbp and Pl 1 (Fig. 4, panels d and h). These phenotypes are similar to those noted for Arnt−/− placentas (2). Furthermore, the correlation between genotype and the severity of placental phenotype suggests that the dosage of HIF activity in the placenta is critical to this developmental program.
FIG. 4.
Expression of cell type-specific markers in trophoblast tissue. (A) E9.5 placentas analyzed by 35S in situ hybridization for Tpbp and Pl 1 in Hif1α+/+ Hif2α+/+ (a and e), Hif1α+/− Hif2α+/− (b and f), Hif1α−/− Hif2α+/− (c and g), and Hif1α−/− Hif2α−/− (d and h) embryos. Original magnification, ×50. (B) E9.5 placentas analyzed by 35S in situ hybridization for expression of TFEB in Arnt+/+ (a and e), Arnt−/− (b and f) Hif1α+/+ Hif2α+/+ (c and g), and Hif1α−/− Hif2α−/− (d and h) embryos. Antisense probes are shown in a, b, c, and d, and sense probes are shown in e, f, g, and h. Original magnification, ×50. Arrowheads indicate cells expressing TFEB.
Having shown that HIF is essential for proper fetal blood vessel invasion of the labyrinthine layer, we next determined if HIF also plays a role in labyrinthine trophoblast differentiation. We performed in situ hybridization for TFEB, a transcription factor exclusively expressed in the syncytiotrophoblasts making up the bulk of the labyrinthine layer (44). Figure 4B, panels a to d, present in situ hybridization with antisense TFEB probes, whereas panels e to h depict hybridization with sense probes. Both antisense and control sense hybridizations are shown, because antisense probe staining is specific but weak. TFEB expression was detected in labyrinthine trophoblasts in wild-type placentas (Fig. 4B, panels a and c). In contrast, Arnt−/− and Hif1α−/− Hif2α−/− placentas produced no TFEB+ cells (Fig. 4B, panels b and d). These data suggest that HIF-deficient placentas do not generate functional syncytiotrophoblasts in vivo. Therefore, HIF1α and HIF2α work in concert via dimerization with ARNT to direct proper development of all three placental cell layers.
Trophoblast stem cells lacking Arnt or Hifα do not generate spongiotrophoblasts or trophoblast giant cells.
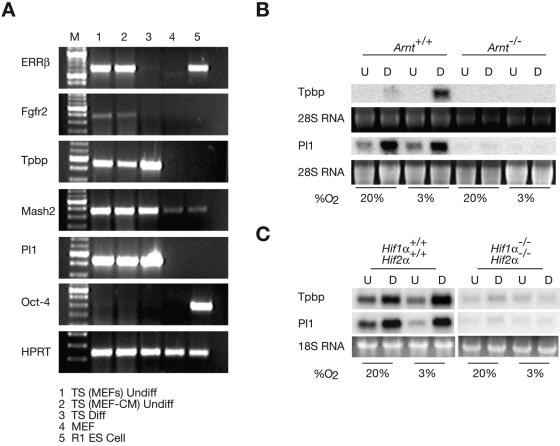
To further dissect the role of hypoxia in placental cell fate specification, we utilized trophoblast stem (TS) cell technology (2, 45). We obtained primary TS cell cultures by harvesting and disaggregating blastocysts. TS cells derived from the trophectoderm of disaggregated E3.5 blastocysts can be cultured in vitro and differentiated into the various syncytiotrophoblast, spongiotrophoblast, and TGC lineages. TS cells represent a primary cell type that is immortal and capable of recapitulating many aspects of placental development. Each of the wild-type lines were derived from different blastocysts from the various crosses. Therefore, the various wild-type lines exhibit slight differences likely resulting from variation in deriving the lines and genetic background. The use of TS cells with targeted mutations allowed us to study the role of hypoxia and HIFα and HIFβ (ARNT) subunits in trophoblast differentiation in a way not possible in vivo. In addition to the previously described Arnt+/+ and Arnt−/− TS cells, we generated 16 independent TS cell lines and several lines of extraembryonic endoderm or “XEN” cells (29). The following TS-cell lines were obtained, and there was no evidence of skewing from the expected Mendelian ratios: one Hif1α+/+ Hif2α+/+, two Hif1α+/+ Hif2α+/−, one Hif1α+/− Hif2α+/+, three Hif1α+/− Hif2α+/−, one Hif1α+/− Hif2α−/−, three Hif1α−/− Hif2α+/+, four Hif1α−/− Hif2α+/−, and one Hif1α−/− Hif2α−/−. TS cells were characterized by semiquantitative RT-PCR for the expression of differentiation and trophoblast-specific transcripts, and the results for a wild-type TS line are presented in Fig. 5A. Embryonic stem (ES) cells and mouse embryo fibroblasts (MEFs) were used as controls. As expected (45), undifferentiated TS cells (cultured on MEFs or in MEF-conditioned media), and ES cells expressed estrogen receptor-related protein β (ERRβ) and FGFR2 (Fig. 5A, lanes 1, 2, and 5). Expression of ERRβ and FGFR2 was extinguished upon TS differentiation for 4 days (lane 3). Northern blot analysis also confirmed that ERRβ mRNA decreased upon differentiating the Arnt+/+, Arnt−/−, Hif1α+/+ Hif2α+/+, and Hif1α−/− Hif2α−/− TS cells (data not shown). All TS cells expressed the trophoblast lineage-specific genes Tpbp, Mash2, and Pl 1 (45), as they undergo a small degree of precocious differentiation and the RT-PCR assays for their expression are highly sensitive (Fig. 5A, lanes 1 to 3). Importantly, the inner-cell-mass-specific transcript Oct-4 (45) was only detected in ES cells (Fig. 5A lane 5), while equal levels of hypoxanthine phosphoribosyltransferase (HPRT) appeared in undifferentiated TS cells, differentiated TS cells, ES cells, and feeder MEFs (Fig. 5A).
FIG. 5.
Generation and differentiation of trophoblast stem (TS) cells. (A) Wild-type undifferentiated TS cells on MEFs or in MEF-conditioned media (lanes 1 and 2), differentiated TS cells (lane 3), MEFs alone, and R1 ES cells were analyzed by RT-PCR for ERRβ, FGFR2, Tpbp, Mash2, Pl 1, Oct-4, and HPRT. (B) Arnt+/+ and Arnt−/− TS cells were differentiated and examined by Northern blot assay for expression of Tpbp and Pl 1 under 20% O2 and 3% O2. U, undifferentiated (undiff) cells; D, differentiated (diff) cells. (C) Hif1α+/+ Hif2α+/+ and Hif1α−/− Hif2α−/− TS cells were differentiated and examined by Northern blot for Tpbp and Pl 1 expression.
To quantitatively assess TS-cell differentiation, Northern blot analysis was performed for Tpbp and Pl 1 mRNA. Undifferentiated Arnt+/+ TS cells did not express Tpbp by this assay. However, Tpbp mRNA levels increased upon differentiation at 20% O2 for 4 days and were further enhanced by 3% O2, suggesting that hypoxia promotes spongiotrophoblast formation (2). TS cells were cultured at 3% O2 to mimic physiological O2 levels TS cells encounter during early placentation (see Introduction). Intriguingly, Tpbp was not detected in Arnt−/− TS cells regardless of differentiation or pO2 (reference 2 and Fig. 5B). Arnt+/+ TS cells exhibited increased expression of the giant cell marker Pl 1 under normoxic and hypoxic differentiating conditions. In contrast to what was observed in vivo, Arnt−/− TS cells did not express Pl 1 under any condition (Fig. 5B).
We then examined HIFα-deficient TS cells for their ability to differentiate into distinct trophoblast lineages. Hif1α+/+ Hif2α+/+ TS cells exhibited elevated levels of Tpbp mRNA upon differentiation, which was further enhanced at 3% O2 (Fig. 5C). Pl 1 mRNA increased under differentiating conditions in Hif1α+/+ Hif2α+/+ TS cells independent of pO2 (Fig. 5C). Interestingly, Hif1α−/− Hif2α−/− TS cells expressed little Tpbp or Pl 1 (Fig. 5C), as noted for Arnt−/− TS cells. In aggregate, TS cells lacking Arnt or both Hif1α and Hif2α are unable to differentiate into spongiotrophoblasts or TGCs. Therefore, Arnt−/− and Hif1α−/− Hif2α−/− TS cells represent a precursor population of the placenta with severe developmental defects. Of note, HIF-deficient TS cells display phenotypes in both normoxic and hypoxic culture conditions. This is likely due to the presence of HIFα/ARNT dimers at 20% O2 in these cells, as previously noted for ES cells (11, 23). The in vitro results clearly are in contrast to in vivo findings and will be discussed below.
HIF-deficient TS cells differentiate into syncytiotrophoblasts.
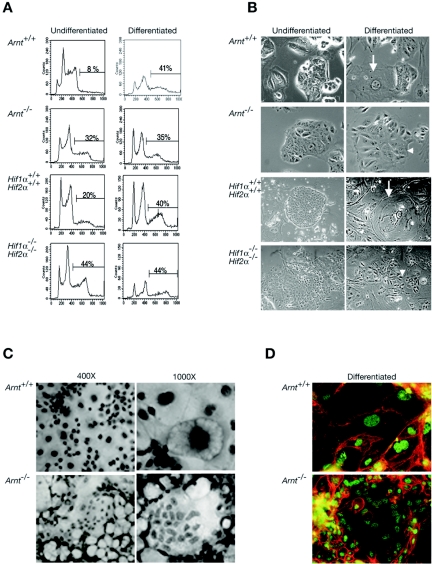
Arnt−/− and Hif1α−/− Hif2α−/− TS cells appear to undergo some differentiation events as ERRβ expression was decreased upon withdrawal of FGF4 (Fig. 5A and data not shown). Therefore, we determined if the HIF-deficient TS cells acquired TGC features or an alternate cell fate. We analyzed TS differentiation by both DNA content and morphology. TGCs are polyploid cells that endoreduplicate their genome. We stained TS cells with propidium iodide (PI) and subsequently performed flow cytometry analysis. Eight percent of the Arnt+/+ TS cells exhibited genomes of more than 4 N after 4 days in undifferentiating conditions, which increased to 40% upon differentiation (Fig. 6A). In contrast, Arnt−/− TS-cell cultures exhibited 35% polyploid cells under both undifferentiating and differentiating conditions (Fig. 6A). Twenty percent of the undifferentiated Hif1α+/+ Hif2α+/+ TS cells were more than 4 N, increasing to 40% upon 4 days of differentiation. Hif1α−/− Hif2α−/− TS cultures contained 44% polyploid cells prior to and after differentiation (Fig. 6A). Therefore, ARNT and HIFα deficient TS cells precociously begin a polyploid cell differentiation program.
FIG. 6.
HIF-deficient TS cells differentiate into polyploid cells. (A) DNA content of undifferentiated and differentiated TS cells. Arnt+/+, Arnt−/−, Hif1α+/+ Hif2α+/+, and Hif1α−/− Hif2α−/− TS cells were analyzed for DNA content by PI incorporation and flow cytometry. Percentage of cells with >4 N DNA content is indicated. (B) Morphology of undifferentiated and differentiated TS cells. TS cells were photographed using a phase-contrast microscope (×100 magnification). Undifferentiated Arnt+/+, Arnt−/−, Hif1α+/+ Hif2α+/+, and Hif1α−/− Hif2α−/− TS cells form epithelial-like colonies. Differentiated Arnt+/+ and Hif1α+/+ Hif2α+/+ TS cells exhibited a giant-cell-like morphology with large nuclei and cytoplasm. In contrast, differentiated Arnt−/− and Hif1α−/− Hif2α−/− TS cells form sheets of cells with many nuclei and undefined borders. Arrows indicate giant cells in wild-type cultures. Arrowheads indicate differentiated HIF-deficient cells. Original magnification, ×1,000. (C) Arnt+/+ and Arnt−/− TS cells were grown for 4 days on plastic and stained by H&E (magnification, ×400 and ×1,000). (D) Arnt+/+ and Arnt−/− TS cells were grown on coverslips during a 4-day differentiation period and then immunostained for β-catenin (red) and HDAC1 (green). Original magnification, ×1,000.
Undifferentiated and differentiated TS cells were then photographed by phase-contrast microscopy. Undifferentiated Arnt+/+ and Hif1α+/+ Hif2α+/+ TS cells formed epithelial sheets characteristic of these cells (Fig. 6B), and cell size dramatically increased upon differentiation. Differentiated cells contained large nuclei and cytoplasm, perinuclear deposits, and decreased expression of E-cadherin (Fig. 6B and data not shown); therefore, they appeared to develop into TGCs (45). Arnt−/− and Hif1α−/− Hif2α−/− TS cells also formed epithelial sheets in the undifferentiated state (Fig. 6B). Importantly, although undifferentiated Arnt−/− and Hif1α−/− Hif2α−/− TS cells exhibited higher DNA content (>4 N), they appeared morphologically indistinguishable from wild-type TS cells. After 4 days of differentiation, both Arnt−/− and Hif1α−/− Hif2α−/− TS cells formed large multinuclear sheets of cells, which were quite distinct from TGCs (Fig. 6B). Differentiated HIF-deficient cells never acquired large nuclei, based on 4′, 6′-diamidino-2-phenylindole staining (data not shown) and phase-contrast microscopy (Fig. 6B). Instead, they grew as sheets of cells with poorly defined borders and elongated cytoplasm (Fig. 6B). This is similar to what has been reported for TS cells that differentiated into syncytiotrophoblasts (21). Furthermore, differentiated Arnt−/− TS cells stained with H&E clearly formed multinucleated syncytia (Fig. 6C). As shown in Fig. 6D, immunocytochemistry using antibodies to β-catenin (which reveals the plasma membrane) and histone deacetylase 1 (HDAC1; delineating the nucleus) confirms the photomicroscopic data in Fig. 6B and C, indicating that differentiated Arnt−/− TS cells become multinucleate after 4 days. We concluded that Arnt−/− and Hif1α−/− Hif2α−/− TS cells differentiated into polyploid nongiant cells that are likely to be syncytiotrophoblasts.
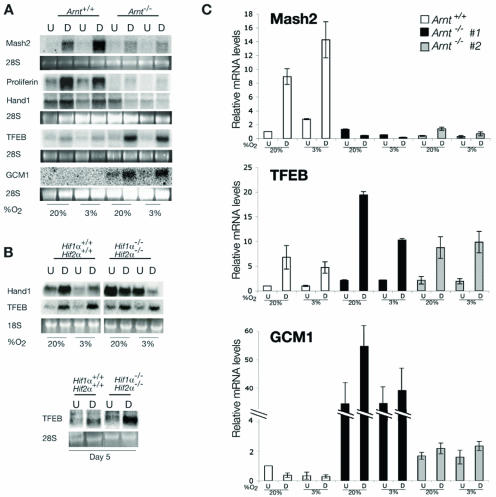
Syncytiotrophoblasts and TGCs are polyploid cells generated via distinct mechanisms (13). Labyrinthine syncytiotrophoblasts form by cell fusion, while TGCs become polyploid through endoreduplication without intervening mitoses (13). To further demonstrate that differentiated mutant TS cells produce syncytiotrophoblasts, we analyzed TS cells for additional lineage-specific markers. We examined the expression of Mash2, a bHLH transcription factor essential for spongiotrophoblast formation (25, 26). Undifferentiated Arnt+/+ and Hif1α+/+ Hif2α+/+ TS cells expressed low Mash2 mRNA levels, which increased upon differentiation for 4 days (Fig. 7A and data not shown). Importantly, differentiation at 3% O2 enhanced Mash2 expression and diminished Proliferin (a TGC marker) and Hand1 expression, suggesting that low O2 maintains the spongiotrophoblast population at the expense of TGCs. In contrast, Mash2 levels were low in Arnt−/− TS under both differentiating and undifferentiating conditions and were not changed by hypoxia. As stated previously, HIF-deficient placentas also expressed lower levels of Mash2 based on quantitative RT-PCR assay (50% less than wild-type placentas), demonstrating that Mash2 mRNA is decreased in vivo in the absence of HIF. Proliferin expression was nearly undetectable in Arnt−/− and Hif1α−/− Hif2α−/− TS cells, and Hand1, a bHLH factor critical for secondary TGC formation (21, 37), was not induced by differentiation (Fig. 7A and B). These data suggest that HIF activity is necessary to generate spongiotrophoblasts and TGCs in vitro. Since HIF-deficient TS cells become multinucleated cells upon differentiation, we examined Arnt−/− TS cells for expression of the syncyciotrophoblast factors TFEB and GCM1. Arnt+/+ TS cells expressed TFEB and low levels of GCM1 (Fig. 7A). However, Arnt−/− TS cells exhibited elevated levels of TFEB and GCM1 upon differentiation. After 4 days of differentiation, TFEB expression was induced equally well in Hif1α+/+ Hif2α+/+ and Hif1α−/− Hif2α−/− cells (Fig. 7B). However, by 5 days of differentiation, TFEB was expressed at higher levels in the differentiated Hif1α−/− Hif2α−/− TS cells than in the wild type (Fig. 7B).
FIG. 7.
HIF-deficient trophoblasts adopt aberrant cell fates in vitro. (A) Arnt+/+ and Arnt−/− TS cells were differentiated under 20% and 3% O2 and examined by Northern blot for expression of a spongiotrophoblast marker (Mash2), TGC-specific genes (Proliferin and Hand1), and labyrinthine trophoblast markers (TFEB and GCM1). (B) Hif1α+/+ Hif2α+/+ and Hif1α−/− Hif2α−/− TS cells were differentiated for 4 days under 20% and 3% O2 and examined by Northern blot for expression of TFEB and Hand1. The lower panel shows TFEB expression after 5 days of differentiation for Hif1α+/+ Hif2α+/+ and Hif1α−/− Hif2α−/− TS cells. (C) Quantitative real-time PCR (Q-PCR) for the spongiotrophoblast gene Mash2 and the syncytiotrophoblast genes TFEB and GCM1. A second Arnt−/− TS line (no. 2) was added to these experiments to demonstrate that these phenotypes are due to HIF deficiency and are associated with multiple independent lines. U, undifferentiated cells; D, differentiated cells.
Q-PCR analysis of multiple Arnt−/− and Hifα−/− TS-cell lines confirmed the Northern assays. As shown in Fig. 7C, two independently derived Arnt−/− TS lines exhibited 9- to 18-fold less Mash2 mRNA than the wild type independent of partial O2 pressure. Of note, Arnt+/+ TS cells exhibited a 9-fold increase in Mash2 mRNA levels, and these levels were further increased to 14-fold at 3% O2 (Fig. 7C). Q-PCR analyses of differentiated Hif1α−/− Hif2α−/− TS cells demonstrated that they also exhibited less Mash2 mRNA (twofold) than wild-type controls. Consistent with the Northern data (Fig. 7A), Arnt+/+ TS cells exhibited higher levels of TFEB upon differentiation. Of note, both Arnt−/− TS-cell lines exhibited higher levels of TFEB than wild-type cells (Fig. 7C). GCM1 mRNA was substantially induced in Arnt−/− TS line no. 1, while Arnt−/− line no. 2 exhibited a twofold increase relative to the wild type. These data in conjunction with the immunocytochemistry analysis displayed in Fig. 6D strongly indicate that Arnt−/− and Hif1α−/− Hif2α−/− TS cells become syncytia when differentiated. We concluded that TS cells lacking either ARNT or HIFα subunits do not form spongiotrophoblasts or TGCs but instead differentiate into syncytiotrophoblasts. The implications of these findings are shown in Fig. 8 and are discussed below.
FIG. 8.
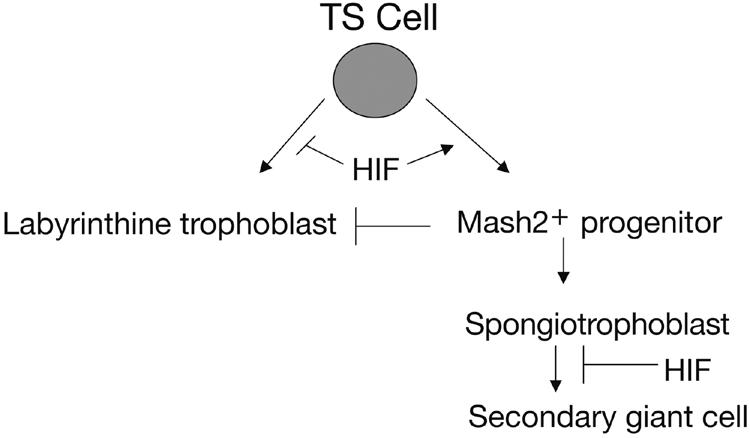
Model for the role of HIF in trophoblast cell fate decisions. Expression of the HIF complex promotes formation of a precursor trophoblast population that expresses Mash2. Mash2 expression acts to repress syncytiotrophoblast differentiation. Mash2+ cells become spongiotrophoblasts that are maintained by hypoxic expression of HIF. Spongiotrophoblasts are then able to terminally differentiate into secondary TGCs as O2 levels increase. This occurs naturally as cells encounter the maternal spiral arteries. HIF negatively regulates labyrinthine trophoblast formation by inducing Mash2 and preferentially promoting the TGC pathway.
DISCUSSION
The ability to sense and then respond to O2 availability is critical for multiple developmental processes. Therefore, we evaluated a possible role for HIF and O2 levels during placentation. Placentas from mice lacking both Hif1α and Hif2α contained no fetal blood vessels and exhibited expanded TGC numbers, reduced spongiotrophoblast numbers, and aberrant placental architecture. Hif1α−/− Hif2α−/− placentas are also poorly invasive, based on morphometric analyses in vivo and poor in vitro invasion reported for mutant TS cells (11). Since the defects in Hif1α−/− placentas are exacerbated by further deleting either one or two copies of Hif2α, we established that HIF2α plays an important role in placental cell fate adoption and that Hif1α and Hif2α are somewhat redundant in this tissue. Importantly, since the Hif1α−/− Hif2α−/− placentas phenocopy the Arnt deficiency, O2 levels and O2 sensing are essential in patterning placental layers. Therefore, hypoxia stimulates the HIF complex through both HIF1α/ARNT and HIF2α/ARNT dimers, allowing HIF to activate genes important for proper placentation.
Recent studies suggest that HIF2α is nonfunctional in some cell types (20, 34). Hif1α−/− and Arnt−/− mice exhibit angiogenic defects in the embryo and yolk sac vasculature (2, 10, 23, 27, 31, 40). However, three independent lines of HIF2α-deficient mice display a wide array of phenotypes, most notably reduced circulating catecholamine levels, lung remodeling defects, and heart failure (9, 35, 47). We now show that HIF2α is important for both recruiting blood vessels into the chorion and regulating placental cell fate adoption. Therefore, HIF2α is clearly important for trophoblast differentiation and function. While Arnt−/− and Hif1α−/− Hif2α−/− embryos as well as their corresponding placentas are defective, we believe that many of the placental phenotypes are primary, because many of the in vivo defects can be recapitulated in vitro by TS cells (see below). Tetraploid chimeras were generated with Arnt−/− ES cells (2). The Arnt−/− embryos, when supplied with wild-type placentas, had normal labyrinthine and spongiotrophoblast layers but only survived one more day due to cardiac failure.
To further delineate the role of HIF and O2 tension in cell fate decisions, we generated TS cells lacking components of the HIF pathway. Arnt−/− and Hif1α−/− Hif2α−/− TS cells are unable to generate spongiotrophoblasts or TGCs but instead differentiate into syncytiotrophoblasts. We observed a number of HIF-mediated phenotypes in TS cells grown at 20% O2. Wild-type TS cells (but not Arnt−/− TS cells) exhibit HIF DNA binding activity and target gene expression at 20% O2, as did ES cells (11). Therefore, TS cells represent a situation where some HIF activity is present at normoxic O2 levels. Interestingly, differentiation at 3% O2 preferentially induces Tpbp and Mash2 expression, suggesting that spongiotrophoblast differentiation, proliferation, or maintenance is enhanced by hypoxia. HIF-deficient placentas failed to exhibit TFEB+ cells, while the number of TFEB+ syncytiotrophoblasts was increased during the differentiation of HIF-deficient TS cells. How can we explain the discrepancy of the in vivo and in vitro results? Syncytiotrophoblast differentiation occurs after E9.0, when fetal blood vessels are beginning to invade the chorionic plate (12). Placentas lacking the DnaJ-related cochaperone Mrj exhibit no chorioallantoic fusion, and the chorionic trophoblasts never form a labyrinth (22). Additionally, Mrj−/− placentas have greatly reduced GCM1, suggesting that chorioallantoic fusion is an essential prerequisite for syncytiotrophoblast differentiation in vivo (22). Since mice lacking Arnt or Hifα do not exhibit chorioallantoic fusion, labyrinthine differentiation may not occur properly. Our data suggest that chorioallantoic fusion is not essential for syncytiotrophoblast differentiation in vitro. Therefore, if the cells cannot adopt the spongiotrophoblast-TGC fate, they become synciotrophoblasts as a default lineage. Of note, the placental phenotypes seen in vivo were identical for mice deficient in Arnt or Hif1α and Hif2α; likewise, the TS cells lacking ARNT of HIFα subunits exhibited remarkably similar phenotypes. As Hif1α−/− Hif2a−/− E3.5 blastocysts are relatively rare (1 in 16), we established only one Hif1α−/− Hif2α−/− TS line. However, two independently derived Arnt−/− TS lines and one Hif1α−/− Hifα−/− line display concordant defects on all assays reported here and by Cowden Dahl et al. (11).
Our data indicate that the TS cells we generated are capable of two distinct differentiation programs: the TGC pathway and the syncytiotrophoblast pathway (Fig. 8). Cells that differentiate towards secondary TGCs transit through a Mash2+ spongiotrophoblast. Yan et al. proposed the existence of Mash2+ progenitors based on evidence that Mash2 mRNA expression is induced 2 days earlier than Tpbp in TS cells during differentiation (51). However, our data suggest that TS cells must transit though a Mash2+/Tpbp+ progenitor to become TGCs (Fig. 8). Because the Arnt−/− and Hif1α−/− Hif2α−/− TS cells are unable to adopt the spongiotrophoblast/TGC pathway, they aberrantly differentiate into labyrinthine trophoblasts. TS cells isolated at E3.5 recapitulate some, but not all, of the in vivo phenotypes of HIF-deficient placentas. The reason for this is that there are likely to be different sources of murine TS cells in vivo which may exhibit distinct gene expression profiles (30, 46). TS cells isolated at E3.5 are bipotential, and distinct TS populations may give rise to the expanded number of TGCs in HIF-deficient placentas in vivo.
The bHLH protein Mash2 is essential for establishing specific trophoblast lineages (12). Mash2 expression is restricted to the EPC and spongiotrophoblast lineage (18, 19). Since Mash2 expression is necessary to generate spongiotrophoblasts, the failure to induce Mash2 expression in Arnt−/− TS cells likely explains why both spongiotrophoblasts and TGCs were not produced in vitro. HIF may directly regulate Mash2, since it contains a putative hypoxia-responsive element (HRE) in its promoter at −214 from the transcriptional start site. Since Mash2 expression is decreased in Hif1α−/− placentas, we propose that HIF activates a cohort of genes, including Mash2 (Fig. 8); Mash2 in turn initiates a program that represses syncytiotrophoblast formation and promotes the spongiotrophoblast differentiation pathway. At E9.5, HIF induces genes in vivo necessary to maintain the number of spongiotrophoblasts, some of which subsequently differentiate into TGCs. HIF appears to negatively regulate TGC differentiation (Fig. 8) as most spongiotrophoblasts have differentiated into TGCs by E9.5 in mutant placentas (Fig. 4A and reference 2). Interestingly, Mash2−/− placentas (like Arnt−/− and Hif1α−/− Hif2α−/− placentas) exhibit a reduced labyrinthine layer, no spongiotrophoblasts, and expanded TGC layers (18, 19). The genetic data suggest that HIF and Mash2 may be part of the same pathway during placental morphogenesis. Dysregulation of this pathway likely contributes to aspects of the clinical syndrome preeclampsia.
The placenta, where O2 tension is tightly regulated, requires a precise amount of HIF activity between E8 and E10. O2 is an essential regulator of placental development, and understanding the pathways regulated by O2 will provide insight into the causation and treatment of preeclampsia. Little is known about what occurs during preeclampsia at the molecular level. In order to rationally design therapeutics, it will be important to define the molecular pathways involved in placentation. Our study defines a pathway in placentation that is imperative for sensing a natural O2 gradient. Additional studies using microarray analysis of TS cells lacking HIF activity could potentially be used to identify hypoxic and nonhypoxic genes involved in placental cell fate decisions. These genes are likely to be important for placentation and could be misexpressed in preeclampsia. One HIF target, transforming growth factor β3 (TGFβ3), is clearly dysregulated during preeclampsia (41). TGFβ3 has been shown to be overexpressed in preeclamptic placentas, and inhibition of TGFβ3 with blocking antibodies restores cytotrophoblast invasion and migration (4, 5). Once additional candidate genes are identified as being misexpressed during preeclampsia, wild-type TS cells could subsequently be used to assay pharmacological compounds that potentially alter the expression of such genes in preeclampsia. Therefore, elucidating the basis for HIF regulation of placental development will likely be important to understanding the etiology of and identifying new treatment options for preeclampsia.
Acknowledgments
We thank Susan Fisher, Tilo Kunath, Janet Rossant, and James Cross for reagents and helpful discussions. We also thank Frank Winslow and Anja Runge for assistance with mouse husbandry.
This work was supported by an NIH NRSA predoctoral fellowship award (no. F31 HL10378) (K.D.C.D.), NIH R01 grant (no. 66331), and the Abramson Family Cancer Research Institute (K.D.C.D. and M.C.S.). M.C.S. is an Investigator for the Howard Hughes Medical Institute.
REFERENCES
- 1.Abbott, B. D., and M. R. Probst. 1995. Developmental expression of two members of a new class of transcription factors. II. Expression of aryl hydrocarbon receptor nuclear translocator in the C57BL/6N mouse embryo. Dev. Dyn. 204:144-155. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, D. M., M. Gertsenstein, A. Nagy, M. C. Simon, and E. Maltepe. 2000. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 14:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adelman, D. M., E. Maltepe, and M. C. Simon. 1999. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes Dev. 13:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caniggia, I., S. Grisaru-Gravnosky, M. Kuliszewsky, M. Post, and S. J. Lye. 1999. Inhibition of TGF-beta 3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J. Clin. Investig. 103:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caniggia, I., H. Mostachfi, J. Winter, M. Gassmann, S. J. Lye, M. Kuliszewski, and M. Post. 2000. Hypoxia-inducible factor-1 mediates the biological effects of oxygen on human trophoblast differentiation through TGFbeta(3). J. Clin. Investig. 105:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet, P., Y. S. Ng, D. Nuyens, G. Theilmeier, K. Brusselmans, I. Cornelissen, E. Ehler, V. V. Kakkar, I. Stalmans, V. Mattot, J. C. Perriard, M. Dewerchin, W. Flameng, A. Nagy, F. Lupu, L. Moons, D. Collen, P. A. D'Amore, and D. T. Shima. 1999. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nat. Med. 5:495-502. [DOI] [PubMed] [Google Scholar]
- 8.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 9.Compernolle, V., K. Brusselmans, T. Acker, P. Hoet, M. Tjwa, H. Beck, S. Plaisance, Y. Dor, E. Keshet, F. Lupu, B. Nemery, M. Dewerchin, P. Van Veldhoven, K. Plate, L. Moons, D. Collen, and P. Carmeliet. 2002. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 8:702-710. [DOI] [PubMed] [Google Scholar]
- 10.Compernolle, V., K. Brusselmans, D. Franco, A. Moorman, M. Dewerchin, D. Collen, and P. Carmeliet. 2003. Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia-inducible factor-1alpha. Cardiovasc. Res. 60:569-579. [DOI] [PubMed] [Google Scholar]
- 11.Cowden Dahl, K. D., S. E. Robertson, V. M. Weaver, and M. C. Simon. 2005. Hypoxia-inducible factor regulates alphavbeta3 integrin cell surface expression. Mol. Biol. Cell 16:1901-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, J. C. 2000. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin. Cell Dev. Biol. 11:105-113. [DOI] [PubMed] [Google Scholar]
- 13.Cross, J. C., L. Anson-Cartwright, and I. C. Scott. 2002. Transcription factors underlying the development and endocrine functions of the placenta. Recent Prog. Horm. Res. 57:221-234. [DOI] [PubMed] [Google Scholar]
- 14.Cross, J. C., Z. Werb, and S. J. Fisher. 1994. Implantation and the placenta: key pieces of the development puzzle. Science 266:1508-1518. [DOI] [PubMed] [Google Scholar]
- 15.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman-Wohl, D., and S. Yagel. 2002. Regulation of trophoblast invasion: from normal implantation to pre-eclampsia. Mol. Cell Endocrinol. 187:233-238. [DOI] [PubMed] [Google Scholar]
- 17.Gu, Y. Z., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 18.Guillemot, F., T. Caspary, S. M. Tilghman, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, D. J. Anderson, A. L. Joyner, J. Rossant, and A. Nagy. 1995. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9:235-242. [DOI] [PubMed] [Google Scholar]
- 19.Guillemot, F., A. Nagy, A. Auerbach, J. Rossant, and A. L. Joyner. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371:333-336. [DOI] [PubMed] [Google Scholar]
- 20.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, M., N. Dobric, I. C. Scott, L. Su, M. Starovic, B. St.-Pierre, S. E. Egan, J. C. Kingdom, and J. C. Cross. 2004. The Hand1, Stra13 and Gcm1 transcription factors override FGF signaling to promote terminal differentiation of trophoblast stem cells. Dev. Biol. 271:26-37. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, P. J., B. J. Swanson, M. A. Haendel, G. E. Lyons, and J. C. Cross. 1999. Mrj encodes a DnaJ-related co-chaperone that is essential for murine placental development. Development 126:1247-1258. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain, S., E. Maltepe, M. M. Lu, C. Simon, and C. A. Bradfield. 1998. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech. Dev. 73:117-123. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, B., A. Kamat, and C. R. Mendelson. 2000. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2). Mol. Endocrinol. 14:1661-1673. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, B., and C. R. Mendelson. 2003. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol. Cell. Biol. 23:6117-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak, K. R., B. Abbott, and O. Hankinson. 1997. ARNT-deficient mice and placental differentiation. Dev. Biol. 191:297-305. [DOI] [PubMed] [Google Scholar]
- 28.Krieg, M., R. Haas, H. Brauch, T. Acker, I. Flamme, and K. H. Plate. 2000. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene 19:5435-5443. [DOI] [PubMed] [Google Scholar]
- 29.Kunath, T., D. Arnaud, G. D. Uy, I. Okamoto, C. Chureau, Y. Yamanaka, E. Heard, R. L. Gardner, P. Avner, and J. Rossant. 2005. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132:1649-1661. [DOI] [PubMed] [Google Scholar]
- 30.Kunath, T., D. Strumpf, and J. Rossant. 2004. Early trophoblast determination and stem cell maintenance in the mouse-a review. Placenta 25(Suppl A):S32-S38. [DOI] [PubMed] [Google Scholar]
- 31.Maltepe, E., J. V. Schmidt, D. Baunoch, C. A. Bradfield, and M. C. Simon. 1997. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature 386:403-407. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 33.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 34.Park, S. K., A. M. Dadak, V. H. Haase, L. Fontana, A. J. Giaccia, and R. S. Johnson. 2003. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1α (HIF-1α): role of cytoplasmic trapping of HIF-2α. Mol. Cell. Biol. 23:4959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng, J., L. Zhang, L. Drysdale, and G. H. Fong. 2000. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. USA 97:8386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajakumar, A., and K. P. Conrad. 2000. Expression, ontogeny, and regulation of hypoxia-inducible transcription factors in the human placenta. Biol. Reprod. 63:559-569. [DOI] [PubMed] [Google Scholar]
- 37.Riley, P., L. Anson-Cartwright, and J. C. Cross. 1998. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 18:271-275. [DOI] [PubMed] [Google Scholar]
- 38.Rinkenberger, J., and Z. Werb. 2000. The labyrinthine placenta. Nat. Genet. 25:248-250. [DOI] [PubMed] [Google Scholar]
- 39.Rodesch, F., P. Simon, C. Donner, and E. Jauniaux. 1992. Oxygen measurements in endometrial and trophoblastic tissues during early pregnancy. Obstet. Gynecol. 80:283-285. [PubMed] [Google Scholar]
- 40.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaffer, L., A. Scheid, P. Spielmann, C. Breymann, R. Zimmermann, M. Meuli, M. Gassmann, H. H. Marti, and R. H. Wenger. 2003. Oxygen-regulated expression of TGF-beta 3, a growth factor involved in trophoblast differentiation. Placenta 24:941-950. [DOI] [PubMed] [Google Scholar]
- 42.Semenza, G. L. 2000. Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit. Rev. Biochem. Mol. Biol. 35:71-103. [DOI] [PubMed] [Google Scholar]
- 43.Sowter, H. M., R. R. Raval, J. W. Moore, P. J. Ratcliffe, and A. L. Harris. 2003. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 63:6130-6134. [PubMed] [Google Scholar]
- 44.Steingrimsson, E., L. Tessarollo, S. W. Reid, N. A. Jenkins, and N. G. Copeland. 1998. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 125:4607-4616. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, S., T. Kunath, A. K. Hadjantonakis, A. Nagy, and J. Rossant. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072-2075. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, T. S., T. Kunath, W. L. Kimber, S. A. Jaradat, C. A. Stagg, M. Usuda, T. Yokota, H. Niwa, J. Rossant, and M. S. Ko. 2002. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 12:1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian, H., R. E. Hammer, A. M. Matsumoto, D. W. Russell, and S. L. McKnight. 1998. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 12:3320-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 49.VanWijk, M. J., K. Kublickiene, K. Boer, and E. VanBavel. 2000. Vascular function in preeclampsia. Cardiovasc. Res. 47:38-48. [DOI] [PubMed] [Google Scholar]
- 50.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan, J., S. Tanaka, M. Oda, T. Makino, J. Ohgane, and K. Shiota. 2001. Retinoic acid promotes differentiation of trophoblast stem cells to a giant cell fate. Dev. Biol. 235:422-432. [DOI] [PubMed] [Google Scholar]