Abstract
Pathogenic apicomplexan parasites like Toxoplasma and Plasmodium (malaria) have complex life cycles consisting of multiple stages. The ability to differentiate from one stage to another requires dramatic transcriptional changes, yet there is a paucity of transcription factors in these protozoa. In contrast, we show here that Toxoplasma possesses extensive chromatin remodeling machinery that modulates gene expression relevant to differentiation. We find that, as in other eukaryotes, histone acetylation and arginine methylation are marks of gene activation in Toxoplasma. We have identified mediators of these histone modifications, as well as a histone deacetylase (HDAC), and correlate their presence at target promoters in a stage-specific manner. We purified the first HDAC complex from apicomplexans, which contains novel components in addition to others previously reported in eukaryotes. A Toxoplasma orthologue of the arginine methyltransferase CARM1 appears to work in concert with the acetylase TgGCN5, which exhibits an unusual bias for H3 [K18] in vitro. Inhibition of TgCARM1 induces differentiation, showing that the parasite life cycle can be manipulated by interfering with epigenetic machinery. This may lead to new approaches for therapy against protozoal diseases and highlights Toxoplasma as an informative model to study the evolution of epigenetics in eukaryotic cells.
The phylum Apicomplexa is composed of a number of protozoan parasites of immense clinical and economic importance, including Plasmodium, the causative agent of malaria, and Toxoplasma gondii. Toxoplasma has long been a major medical and veterinary problem, capable of causing abortion or congenital birth defects in both humans and livestock. The advent of AIDS has drawn even more attention to Toxoplasma as a serious opportunistic pathogen.
Toxoplasma is incurable because of its ability to differentiate from the rapidly replicating tachyzoite stage into a latent cyst form (bradyzoite stage) that is impervious to immunity and current drugs. This developmental process, called stage conversion, is triggered by the host immune response. Impairment of the immune response in immunocompromised patients initiates the reconversion of bradyzoite cysts into cytolytic tachyzoites. Stage conversion is associated with dramatic morphological and physiological changes, including alteration of the parasite surface, changes in parasite metabolism, and induction of genes associated with stress response (12, 17, 34, 57). Differentiation is clearly regulated in part at the transcriptional level (37, 40, 49), but the control of gene expression in apicomplexan parasites is profoundly understudied.
Analysis of apicomplexan genome databases for Plasmodium and Cryptosporidium reveals a relatively small number of transcription factors (54). Review of the Toxoplasma genome database reinforces the paucity of conventional transcription factors in these parasites (M. A. Hakimi, unpublished data). The proportion of the transcriptome encoding transcription factors appears to increase as the complexity of the organism increases (3.4% for Saccharomyces cerevisiae, 4.2% for Caenorhabditis elegans, 5.5% for Drosophila melanogaster, 8% for humans) (42). Thus, it has been proposed that the regulation of gene expression in apicomplexan parasites may require the concerted action of a well-developed chromatin-remodeling apparatus and a small set of specific DNA-binding proteins (54).
It is now well accepted that chromatin is dynamic and influences gene expression regulation. Chromatin can be fashioned to be transcriptionally repressive or permissive, dependent in part on posttranslational modifications made to histones, a phenomenon referred to as the histone code (29). In this study, we established chromatin immunoprecipitation (ChIP) techniques for Toxoplasma to investigate histone modifications in the context of stage conversion. For the first time, we show that acetylation and methylation of parasite histones correlate with stage-specific gene expression. In tachyzoites, a novel histone deacetylase (HDAC) corepressor complex (TgCRC) operates at bradyzoite-specific promoters while a TgGCN5 histone acetyltransferase (HAT) (53) operates at tachyzoite-specific promoters. Surprisingly, TgGCN5 exhibits an unexpected substrate preference and exclusively acetylates histone H3 lysine 18 (H3 [K18]). In humans, this modification is mediated by the metazoan HAT CBP/p300 and facilitates recruitment of an arginine methylase, CARM1, to activate transcription (11). Consequently, we characterized a novel CARM1 complex in Toxoplasma that targets H3 [R17] coincident with TgGCN5-mediated stage-specific gene activation. Collectively, the histone modifications associated with differentiation, along with the mediators of those activities reported here, represent a substantial increase in our understanding of the control of gene expression in parasitic protozoa.
MATERIALS AND METHODS
Parasite methods.
The Prugniaud and RH strains of Toxoplasma were grown in human foreskin fibroblasts, transfected, and cloned by limiting dilution as described previously (34). The RH strain lacking HXGPRT was used in selection experiments (14). In vitro bradyzoite induction procedures were performed by the high-pH (8.1) method (58). To determine the ratio of parasites expressing stage-specific antigens within the entire population, the differentiation process was monitored daily by double-immunofluorescence assay with both tachyzoite-specific α-SAG1 and cyst-specific α-CC2 (a kind gift from W. Bohne and U. Gross) antibodies. The ratio of parasites reacting with the antibodies (see Fig. 7B) was determined by counting 50 random fields under a fluorescence microscope.
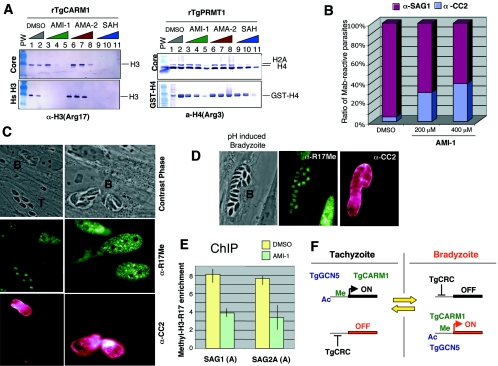
FIG. 7.
AMI-1 inhibits TgCARM1 activity in vitro and induces cyst formation in vivo. (A) In vitro inhibition by AMI-1 and activation by AMA-2 of methylation reactions mediated by rTgCARM1 (0.25 μg) and rTgPRMT1 (0.2 μg). Methylation reactions were performed with histone H3 (1.5 μg), wild-type GST-histone TgH4 (1.5 μg), and free core histone substrates (3 μg) in the presence of dimethyl sulfoxide (DMSO), AMI-1 (50, 200, and 600 μM), AMA-2 (50, 200, and 600 μM), and S-adenosylhomocysteine (SAH; 0.04, 0.16, and 0.32 μM). PW, DNA molecular weight. (B) Estimation of AMI-1-dependent in vitro conversion. The ratio of parasites that were reactive either with tachyzoite-specific α-SAG1 antibody or bradyzoite-specific α-CC2 antibody was determined by double immunofluorescence as described in Materials and Methods. Mab, monoclonal antibody. (C) Phase-contrast microscopy and immunofluorescence assays with AMI-1-treated tachyzoites with α-methyl [R17] H3 and α-CC2 antibodies. The decrease of methylation at arginine 17 localizes within the induced bradyzoites as assessed by α-CC2 immunofluorescence. (D) Immunofluorescence of in vitro-generated bradyzoites induced by an alkaline pH with α-methyl [R17] H3 and α-CC2 antibodies. (E) ChIP experiments with extracellular tachyzoites treated with AMI-1 or dimethyl sulfoxide (control) display the binding of methylated H3 [R17] on TgCARM1 target SAG1 and SAG2A genes (Fig. 6A). The n-fold enrichment was calculated by dividing the α-methyl [R17] H3 intensity signal by the input control signal. The data from five independent experiments were pooled in order to graph the average enrichment. The primers used in the PCR analysis were designed to span the region showing the highest peak of enrichment for each promoter (region A, Fig. 6A). (F) Model of epigenetic control of stage-specific gene expression in T. gondii.
IFA and Western blot analysis.
Immunofluorescence assays (IFA) were performed as described previously (17). Immunoblotting with alkaline phosphatase was performed as previously described (26).
Antibodies.
Primary antibodies for IFA included a rat monoclonal antibody against the hemagglutinin (HA) epitope tag used at 1:500 (α-HA; Roche Diagnostic), a monoclonal antibody against BSR4 used at 1:300 (p36, Tg4A12; a kind gift from J. F. Dubremetz), a monoclonal antibody against cyst wall antigen used at 1:500 (α-CC2; a kind gift from W. Bohne and U. Gross), and an anti-dimethyl [R17] H3 polyclonal antibody (ab8284; Abcam). An anti-dimethyl [R3] H3 polyclonal antibody (07-213; Upstate) was used for Western blot analysis. A ChIP grade anti-dimethyl [R17] H3 polyclonal antibody (ab8284; Abcam) was used for Western blot analysis and ChIP assays. Additional antibodies used in ChIP assays were polyclonal anti-acetyl [K9-K14] H3 (06-599; Upstate), anti-acetyl [K5-K8-K12-K16] H4 (06-866; Upstate), anti-acetyl [K18] H3 (ab1191; Abcam), α-dimethyl [K4] H3 (07-030; Upstate), α-trimethyl [K4] H3 (ab8580; Abcam), and α-histone H3 (ab12079; Abcam). A rabbit antiserum was raised against recombinant TgCARM1 according to the protocol of the manufacturer (Eurogentec). A 1:3,000 dilution of polyclonal anti-TgCARM1 antibody (α-49) was used to perform Western blot analysis, and 3 μl of immunopurified α-49 was used in each ChIP assay.
ChIP.
ChIP was performed as described previously (26). Briefly, freshly lysed tachyzoites or in vitro bradyzoites were treated for 20 min with 1% formaldehyde at 37°C. The lysate was sonicated under conditions yielding fragments ranging from 400 bp to 800 bp. Samples were purified through PCR Purification Kit columns (QIAGEN) and used as a template in PCRs to detect specific targets. Primer pairs (melting temperature, 55 to 65°C) amplifying 200- to 450-bp fragments were created by using sequences found at ToxoDB.org (see Table S1 in the supplemental material) (33). For each primer pair, the optimal magnesium concentration (1 to 2.5 mM MgCl2) was determined. For individual primer pairs, the annealing temperature and number of cycles were adjusted until no signal was detected for the mock-immunoprecipitated DNA (IgG [immunoglobulin G]), but the amplification on the genomic template was not altered (input). Signals obtained with the antibody-immunoprecipitated DNA under these conditions were considered significant. The amplified DNA was separated on 2% agarose and stained with ethidium bromide.
Affinity chromatographic purification of HA-FLAG-CARM1.
To purify HA-FLAG-TgCARM1, whole-cell extract (WCE) from transgenic parasite clone A3 (5 × 1010 extracellular tachyzoites) was incubated with anti-FLAG M2 affinity gel (Sigma). After extensive washing with buffer A (20 mM tris-HCl [pH 7.9], 0.5 M KCl, 10% glycerol, 1 mM EDTA, 5 mM dithiothreitol [DTT], 0.5% NP-40), the affinity column was eluted with buffer A containing FLAG peptide (500 μg/ml; Sigma) according to the manufacturer's instructions. HA-FLAG-TgCARM1-containing fractions were fractionated on a Superose 6 HR 10/30 (Pharmacia) equilibrated in 0.5 M KCl in buffer A containing 0.1% NP-40, 1 μg/ml aprotinin, leupeptin, and 0.1 mM pepstatin phenylmethylsulfonyl fluoride (PMSF).
Chromatographic purification of TgCRC, a TgHDAC3-containing complex.
The TgHDAC3 complex was purified from 2 g of WCE from transgenic extracellular tachyzoites expressing TgHDAC3-HA-FLAG ectopically. WCE was loaded onto a 250-ml column of phosphocellulose (P11; Whatman) and fractionated stepwise by the indicated KCl concentrations in buffer A (20 mM Tris HCl [pH 7.9], 0.2 mM EDTA, 10 mM β-mercaptoethanol, 10% glycerol, 0.2 mM PMSF). The P11 0.3 M KCl fraction (750 mg) was loaded onto a 45-ml DEAE-Sephacel column (Pharmacia) and eluted with 0.35 M KCl. The 0.35 M KCl elution (420 mg) was incubated with 1 ml of anti-FLAG M2 affinity gel (Sigma) for 2 h at 4°C. Beads were washed with 50 ml of BC500 buffer (20 mM Tris [pH 8], 0.5 M KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 0.5 mM PMSF, 1 μg/ml each aprotinin, leupeptin, and pepstatin) and one time with 10 ml BC100 buffer (20 mM Tris [pH 8], 0.1 M KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 1 μg/ml each aprotinin, leupeptin, and pepstatin). Bound peptides were eluted stepwise with 350 μg/ml of FLAG peptide (Sigma) diluted in BC100 buffer. Eluates enriched in TgHDAC3 were fractionated on a Superose 6 HR 10/30 (Pharmacia) and equilibrated in 0.5 M KCl in buffer A containing 0.1% NP-40 and 1 μg/ml each aprotinin, leupeptin, and pepstatin. Our current purification protocol yields approximately 10 to 30 μg of TgCRC complex from 120 mg of the transgenic parasite clone expressing TgHDAC3-HA-FLAG. This preparation is sufficiently clean that individual peptide bands can be excised and sequenced.
Mass spectrometry peptide sequencing.
Protein bands were excised from colloidal blue-stained gels (Invitrogen), oxidized with 7% H2O2, and subjected to in-gel tryptic digestion. Peptides were extracted with a 5% (vol/vol) formic acid solution and acetonitrile and injected into a CapLC (Waters) nanoLC system that was directly coupled to a QTOF Ultima mass spectrometer (Waters) (18). Tandem mass spectrometry data were acquired and processed automatically by using MassLynx 3.5 software (Waters). Tandem mass spectra were searched against a compiled T. gondii database by using the MASCOT program (Matrix Sciences, London, United Kingdom) available at http://www.osc.edu/hpc/software/apps/mascot.shtml.
Histone methyltransferase assays.
Recombinant proteins rTgCARM1 (pNS2) and rTgPRMT1 (pMAH2) were purified from bacteria as described previously (26). Protein samples (0.25 μg of rTgCARM1, 0.2 μg of HA-FLAG-TgCARM1 elution, and 0.25 μg of rTgPRMT1) were incubated with recombinant histones or free core histones at 30°C for 60 min in phosphate-buffered saline reaction buffer containing 1 mM DTT and 0.2 μg/μl of S-adenosyl-l-methionine (Sigma) as previously described (48). Assays were performed with 3 μg of free core histones purified from chicken erythrocytes (13-107; Upstate) or from tachyzoites, 1.5 μg of recombinant human histone H3 expressed in Escherichia coli (14-494; Upstate), or 1 μg of purified glutathione S-transferase (GST)-histone TgH4 and 1 μg of mutated GST-histone TgH4 (R3K). The reactions were stopped by the addition of sodium dodecyl sulfate (SDS) sample buffer. Reaction products were then separated on 15% SDS-polyacrylamide gels and transferred onto PVDF membrane. Immunoblot assays were performed by using rabbit antisera as follows: polyclonal antibody anti-dimethyl [R17] H3 (ab8284; Abcam) at 1:1,000 and polyclonal antibody anti-dimethyl [R3] H4 (07-213; Upstate) at 1:1,000. Colorimetric detection with alkaline phosphatase-coupled anti-rabbit IgG (Promega) was performed. Methylation reactions with AMI-1 or AMA-2 were performed as previously described (9).
HAT and HDAC assays.
The HAT assay procedure used was described previously (16). HDAC activity was measured with the colorimetric HDAC activity assay kit (Calbiochem-Novabiochem, Darmstadt, Germany).
RESULTS
Histone acetylation accompanies stage conversion.
Histone acetylation of the promoter precedes the activation of many genes and is thought to establish a chromatin environment suitable for assembly of the transcriptional complex. We established a ChIP (55) assay for Toxoplasma to monitor the status of histone modifications in the promoters of both stage-specific and constitutively expressed genes for tachyzoites and in vitro-generated bradyzoites (Fig. 1A). We designed a series of primer pairs proximal to the putative transcription initiation site of each target gene (Fig. 1B; see Table S1 in the supplemental material). As expected, acetylation of histones H3 and H4 upstream of constitutively expressed housekeeping genes is observed in both tachyzoites and bradyzoites (Fig. 1C). In marked contrast, bradyzoite-specific genes are hypoacetylated in tachyzoite populations and become acetylated under differentiation conditions (Fig. 1C and D). We also noted that tachyzoite-specific genes are hyperacetylated in tachyzoites and acetylation at these promoters is diminished under bradyzoite growth conditions (Fig. 1C). ChIP assays with histone H3 antibody show that the decreased signal from the specific antibodies is due to decreased acetylation, rather than decreased H3 occupancy or accessibility to antibodies. Some residual acetylation of tachyzoite-specific promoters is still evident because in vitro stage conversion is not 100% efficient.
FIG.1.
Histone acetylation is a mark of gene activation during stage conversion. (A) Tachyzoite (T)-to-bradyzoite (B) conversion of Toxoplasma. The Prugniaud strain was induced in vitro with an alkaline pH. The kinetics of stage conversion was monitored by immunofluorescence assay with bradyzoite-specific antibodies (α-CC2 and α-BRS4) and tachyzoite-specific antibody α-SAG1. Note that in vitro conversion to bradyzoites is not 100% efficient, so our converted parasites are labeled B/T to denote parasite populations greatly enriched for bradyzoites (70 to 80%). (B) Stage-specific and constitutively expressed genes examined by ChIP analysis. Genes: BAG, bradyzoite antigen; BSR, bradyzoite-specific recombinant; LDH, lactate dehydrogenase; SAG, surface antigen; SRS, SAG-related sequences; DEOC, deoxyribose-phosphate aldolase; GRA, dense granule protein; eIF4A, eukaryotic translation initiation factor 4A; DHFR, dihydrofolate reductase-thymidylate synthase. (C) ChIP analysis of housekeeping genes (eIF4A and DHFR), tachyzoite-specific genes (SAG1 and SAG2A), and bradyzoite-specific genes (DEOC, LDH2, and BAG1). Tachyzoites (T) and in vitro-generated bradyzoites (B/T) were processed for ChIP analysis with α-acetyl [K9-K14] histone H3 and α-acetyl [K5-K8-K12-K16] histone H4 and α-histone H3 antibodies. A chromatin sample with an irrelevant antibody was used as a negative control (IgG) and genomic DNA as a positive control for PCR (input). The input PCR signal was set at 100%, and the numerical value of the ChIP signal represents the percentage of the input. The amount of immunoprecipitated DNA determined by semiquantitative PCR was normalized to the respective input DNA for each sample (shown in arbitrary units). Each bar is an average of three independent experiments. (D) Long-range ChIP analysis of the BSR4 locus. (E) ChIP analysis of the GRA gene family (GRA1 to GRA9).
Recent evidence indicates that some Toxoplasma genes belonging to gene families are located in close proximity to each other (31). This is the case for the majority of the SRS genes (SAG-related sequences) found throughout the genome in distinct, tandemly arrayed multigene clusters (31). Since transcription of these clusters is coordinated during interconversion, we determined if they exhibit long-range histone acetylation patterns (19). Using ChIP, we scanned a 15-kb stretch of the bradyzoite-specific BRS4 locus for acetylated histone H4 in both tachyzoite and in vitro-induced bradyzoite stages (Fig. 1D). Coincident with bradyzoite conversion, histone acetylation occurs at each gene within the locus. Whereas no significant acetylation is observed for certain regions, some sites displayed great peaks of acetylation (Fig. 1D). These findings demonstrate that long-range histone acetylation patterns spanning many kilobases can be established in Toxoplasma. By contrast, genes from the dense granule family (GRA), which are found in various chromosomal locations, exhibited disparate patterns of acetylation between life cycle stages (Fig. 1E).
Purification of a novel corepressor complex from Toxoplasma.
Histone acetylation activities like those described above are mediated by HATs and HDACs. A GCN5 family HAT has been described in Toxoplasma (27, 53), but no one has characterized an HDAC enzyme in this parasite. Bioinformatic analysis of the Toxoplasma genome (http://ToxoDB.org) (33) reveals a family of six putative HDACs in Toxoplasma, which can be grouped into three classes (I, II, and III) based upon similarity to human homologues (data not shown; 13, 23). We generated stable transgenic parasites (RH strain) expressing recombinant Toxoplasma HDAC homologues dually tagged with HA and FLAG, referred to as TgHDAC1 to -5 and TgSIR2. Whereas some class I- and class II-related enzymes were predominantly cytoplasmic (data not shown), TgHDAC3 (class I) was exclusively localized in the parasite nucleus, being largely excluded from the nucleolus, as observed for acetylated H4 (Fig. 2A). Its nuclear localization prompted us to examine the TgHDAC3 complex in greater detail by utilizing the epitope tag for chromatography purification (Fig. 2B). Analysis by silver staining of Superose 6 gel filtration fractions reveals that TgHDAC3 is embedded in a remarkably robust (it remains stable in 0.5 M NaCl and 0.1% NP-40) and high-molecular-mass complex (>1 MDa, fractions 20 to 22, Fig. 2C). We also observed a peak of immunoreactivity of the monomeric form (fractions 30 to 32; Fig. 2C). Colloidal blue staining of trichloroacetic acid-precipitated fraction 20 displays multiple bands, whose intensity suggests a stoichiometric complex (named TgCRC for T. gondii corepressor complex). Peptide sequences obtained from excised bands were compared to predicted proteins in the Toxoplasma sequence database (Fig. 2D; see Table S2 in the supplemental material).
FIG. 2.
Chromatographic purification of TgCRC, a TgHDAC3-containing complex. (A) Recombinant protein in transgenic parasites expressing TgHDAC3-HA-FLAG was detected by IFA with HA antibody and compared to nuclear localization of acetylated histone H4 (α-AcH4). (B) Purification scheme of the TgHDAC3-HA-FLAG complex. WCE from tachyzoites was fractionated by chromatography as described in Materials and Methods. The 0.35 M KCl elution of DEAE-Sephacel was purified with an anti-FLAG M2 affinity column. The bound proteins were further analyzed by Superose 6 gel filtration. The horizontal and diagonal lines indicate stepwise and gradient elutions, respectively. Molar concentrations are given. FT, flowthrough. (C) Silver staining and Western blotting (α-HA) of Superose 6 fractions (15 μl). Molecular weight markers are indicated on the left. (D) Superose 6 fraction 20 was trichloroacetic acid precipitated, separated by SDS-polyacrylamide gel electrophoresis (4% to 12%), and visualized by colloidal blue staining. Molecular weight markers are indicated on the left. The proteins identified by mass spectrometry sequencing are indicated. Polypeptides marked with asterisks are contaminants. (E) Structural features of TgCRC-100/TgTBL1, TgCRC-230, and TgCRC-350. aa, amino acids.
We identified the TgCRC-62 and -100 bands as TgHDAC3 (accession no. DQ004745) and TgTBL1 (T. gondii transducin beta-like protein 1; accession no. DQ004746), respectively. Interestingly, TgTBL1 contains one LisH and six WD40 domains most similar to its human counterpart (Fig. 2E). TBL1 was originally identified as deleted in patients with sensorineural deafness associated with an X-linked syndrome of ocular albinism (3). HDAC3 is also found associated with TBL1 in human N-CoR (nuclear receptor corepressor)- and SMRT (silencing mediator of retinoid and thyroid hormone receptors)-containing complexes (24, 28, 36, 63, 64). However, no peptide sequences corresponding to parasite homologues of N-CoR or SMRT proteins were purified with TgHDAC3, consistent with their absence from the Toxoplasma genome database. Nonetheless, TgCRC exhibits other features of HDAC3-containing complexes besides the TBL1 association. For instance, the actin-binding protein IR10 was reported as a specific component of the N-CoR complex, suggesting a functional connection with actin (63). Interestingly, we identified TgCRC-45 as actin (accession no. AAC13766). TgHDAC3 also forms a stable and stoichiometric association with two HSP70-like proteins, TgCRC-75 (TgHSP70a; accession no. AAF23321), and TgCRC-80 (TgHSP70b; accession no. AAC72001), as previously reported in human cells (25, 63). We also established TgCRC-64, -66, and -68 bands as subunits of a Toxoplasma TCP-1 ring complex, TRiC (see Table S2 in the supplemental material) (50). TRiC stably associates with human HDAC3 (25) and the yeast HDAC HOS2 (46).
TgCRC-120 was identified as a novel parasite HSP90-like protein (accession no. DQ004747) distinct from the one previously described as being secreted into the parasitophorous vacuole (accession no. AY292370) (1). Whereas in human cells the HDAC3-Hsp90 association was described as transient (25), we observed a stable and robust interaction between TgHDAC3 and TgCRC-120 (Fig. 2D). However, treatment of transgenic parasites with the Hsp90 inhibitor geldanamycin does not prevent nuclear localization of TgHDAC3-HA-FLAG, as shown in human cells (data not shown; 25).
Convincing data suggest that human HDAC3 interacts with SMRT only after priming by cellular chaperones, including the TCP-1 ring complex, HSP70, and HSP90 (25). It has also been shown that HDAC3 requires SMRT for HDAC enzyme activity (25, 28, 63, 64). While no clear N-CoR- or SMRT-related homologues exist in Toxoplasma, two new polypeptides were discovered that correspond to functionally uncharacterized genes (TgCRC-350 and TgCRC-230, Fig. 2E). Analysis of a number of preparations indicates that TgCRC-350 is a substoichiometric component of this complex whereas TgCRC-230 is sensitive to breakdown and abundant, with more than 30 tryptic peptides detected (Fig. 2D). Both TgCRC-230 and TgCRC-350 display a bipartite nuclear targeting sequence, as predicted by PSORT (Fig. 2E). SMART analysis reveals that TgCRC-230 possesses six KELCH repeats and a HATPase domain (Fig. 2E). The roles of the TgCRC-350 and TgCRC-230 proteins remain to be elucidated, but it is tempting to speculate that they could be activating factors for TgHDAC3 that can potentiate transcriptional repression in a manner similar to that reported for SMRT or N-CoR (24, 28, 36, 63, 64).
To rigorously demonstrate that these proteins are components of the TgCRC, we employed a HA-FLAG-tagged version of TgTBL1 as bait to reisolate it. This protein ectopically expressed in tachyzoites localizes exclusively in the nucleus and is also excluded from the nucleolus, as observed previously for TgHDAC3 (Fig. 3A). No stable transgenic parasites could be obtained, suggesting that an additional copy of TgTBL1 is not tolerated. However, silver staining analysis of the affinity eluates from transiently transfected tachyzoites confirms that many of the polypeptides are members of the TgCRC. The differences include the lack of the TgCRC-230 subunit and the presence of two novel uncharacterized subunits named p50 and p130 (Fig. 3B and C).
FIG.3.
Repurification of TgCRC with TgTBL1 as bait and determination of enzymatic activity. (A) Transiently expressed recombinant protein HA-FLAG-TgTBL1 was detected by IFA with an HA antibody and compared to nuclear localization of acetylated histone H4 (α-AcH4). (B) Purification scheme of TgTBL1-associated proteins. FT, flowthrough. (C) Silver staining of affinity-purified TgTBL1-containing complex compared to TgCRC (Superose 6, fraction 20). The proteins analyzed are indicated on the right. Western blot analysis using HA antibody detects tagged TgTBL1. WCE from untransfected wild-type RH tachyzoites was processed similarly as a control (mock). Molecular weight markers are indicated on the left. (D) HDAC activity assays of FLAG affinity-purified TgTBL1 or TgHDAC3. TSA, trichostatin A; APAH, aroyl-pyrrole-hydroxyamides.
HDAC activities of the TgCRC purified by virtue of HA-FLAG-TBL1 or TgHDAC3-HA-FLAG were determined and found to be susceptible to known HDAC inhibitors (Fig. 3D). The purified complexes also displayed HDAC activity on core histones (data not shown).
Differential association of TgGCN5 and TgHDAC3 at stage-specific promoters.
We hypothesized that during the tachyzoite stage HATs would be present at tachyzoite-specific promoters while HDACs would be at bradyzoite-specific promoters. We performed ChIP assays on stable transgenic parasite clones expressing either TgGCN5 or TgHDAC3 as a FLAG-tagged fusion protein (Fig. 4) (5). ChIP assays using anti-FLAG show that TgHDAC3 is recruited upstream of the bradyzoite-specific SAG2C and BAG1 genes in tachyzoites (Fig. 4). Importantly, as shown by ChIP assays using antibodies to acetylated H3 and H4, hypoacetylation is observed at the same loci (data not shown). In contrast, TgGCN5 is found in association with tachyzoite-specific promoters (SAG2A and an unknown expressed sequence tag [EST], Fig. 4). Therefore, the differential association of TgGCN5 and TgHDAC3 at stage-specific promoters contributes to gene regulation via acetylation of histone tails. Other acetylases should be involved, since hyperacetylation of H3 [K9-K14] and H4 [K5-K8-K12-K16] is detected at these loci (data not shown).
FIG. 4.
ChIP analysis of TgHDAC3 and TgGCN5 binding sites. Tachyzoites from stable transgenic parasite clones expressing either FLAG-TgGCN5 or TgHDAC3-HA-FLAG were processed for ChIP with αα-FLAG antibody. Stage-specific promoters that were tested included SAG2A and an unknown EST (tachyzoite specific), as well as BAG1 and SAG2C (bradyzoite specific). A chromatin sample with an irrelevant antibody was used as a negative control (IgG) and genomic DNA as a positive control for PCR (input).
Arginine methylation of Toxoplasma histones.
In mammalian cells, methylation of histone arginine residues by CARM1 (coactivator-associated arginine methyltransferase 1) and PRMT1 (protein arginine methyltransferase 1) occurs at promoters during transcriptional activation in response to hormone induction (4, 38, 51, 56) or in association with p53 (2). No evidence exists that arginine methylation affects gene expression in protozoa. We detected methylation of H3 [R17] within tachyzoite nuclei by using immunofluorescent staining (Fig. 5A). Bioinformatic analysis of the Toxoplasma genome sequence identifies five putative arginine methyltransferases with a conserved catalytic core. Since CARM1 and PRMT1 are the methyltransferases predominantly involved in gene regulation in human cells, we focused on the putative orthologues TgCARM1 (accession no. AY820755) and TgPRMT1 (accession no. AY820756) (Fig. 5B).
FIG.5.
Arginine methylation of Toxoplasma histones. (A) Immunofluorescence colocalization of methylated [R17] H3 and Hoechst 33258 within the nuclei of intracellular tachyzoites. (B) Evolutionary relationships between arginine methyltransferases of T. gondii TgCARM1 and TgPRMT1, Plasmodium falciparum PfCARM1 (accession no. CAD51260) and PfPRMT1 (accession no. AAN36855), Cryptosporidium parvum CpCARM1 (genomic annotation), Arabidopsis thaliana AtCARM1 (accession no. AAF26997), D. melanogaster DmCARM1 (CARMER, accession no. AAF54471) and DmPRMT1 (accession no. AAM11369), C. elegans CePRMT1 (accession no. T26447), S. cerevisiae ScHMT1 (accession no. NP_009590), and Homo sapiens HsCARM1 (accession no. AAH46240) and HsPRMT1 (accession no. AAF62895). The unrooted phylogenetic tree was inferred from the core domain alignment. (C) Recombinant histidine-tagged proteins (rTgCARM1 and rTgPRMT1) were expressed in E. coli and purified by Ni-nitrilotriacetic acid affinity chromatography. Methylation of H3 [R17] is mediated in vitro by rTgCARM1 (0.25 μg). The assays with recombinant H3 (1.5 μg) and free core histones (3 μg) were performed by immunoblotting with α-methyl [R17] H3 antibody. Methylation of H4 [R3] is mediated in vitro by rTgPRMT1 (2.5 μg). The assays with wild-type GST-histone TgH4 (1.5 μg), mutant GST-histone TgH4 (R3K, 1.5 μg), and free core histones (3 μg) were performed by immunoblotting with α-methyl [R3] H4 antibody. (D) Immunofluorescence assay with α-HA of transgenic parasite clone A3, engineered to ectopically express HA-FLAG-TgCARM1. (E) WCE of 5 × 1010 tachyzoites expressing HA-FLAG-TgCARM1 was fractionated with an anti-FLAG M2 affinity column. Bound proteins were further analyzed by chromatography on a Superose 6 column. Silver staining analysis of the anti-FLAG affinity eluates corresponding to fractions 17 to 41 was done. Western blotting was performed with the antibodies indicated on the left. W.T, wild type. (F) Methylation of nucleosomal histones (3 μg) by rTgCARM1 (0.25 μg) supplemented with 5 μg of Toxoplasma protein extract enriched in ATP-dependent nucleosome disruption activity. The concentration of ATP was 1.0 mM where added. The reaction mixtures were resolved by SDS-polyacrylamide gel electrophoresis, and [3H]histone H3 was excised for counting in scintillation buffer.
In vitro histone methylation assays show that recombinant TgCARM1 methylates H3 [R17] while TgPRMT1 methylates H4 [R3], consistent with their respective human counterparts (Fig. 5C). Moreover, mutation of arginine 3 of H4 abolishes the enzymatic activity of TgPRMT1 (Fig. 5C). Ectopically expressed HA-FLAG-TgCARM1 is able to localize to the parasite nucleus, consistent with a role in transcriptional regulation (Fig. 5D). Affinity-purified HA-FLAG-TgCARM1 from the transgenic parasite clone reveals a dimer of tagged and endogenous TgCARM1 capable of methylating histone H3 [R17] in free core histones (Fig. 5E). The apparent molecular mass is 600 kDa, suggesting that TgCARM1 exists as a hexamer, as reported previously for HMT1 and PRMT3 (59, 65).
Human CARM1 acquires the ability to covalently modify nucleosomal histones when complexed with SWI/SNF ATPases (60). We examined this in Toxoplasma since SWI/SNF proteins have been identified (52) by mixing recombinant TgCARM1 with parasite extract enriched in ATP-dependent nucleosome disruption activity. In the presence of ATP, this mixture was capable of methylating nucleosomal histones (Fig. 5F), suggesting that TgCARM1 likely acts in concert with SWI/SNF chromatin remodelers in Toxoplasma in vivo.
Concurrent histone methylation and acetylation on stage-specific promoters.
We generated a polyclonal antibody against TgCARM1 (α-49) for use in ChIP experiments to identify target genes of TgCARM1. ChIP analysis of tachyzoites reveals that TgCARM1 is associated with the promoters of tachyzoite-specific genes SAG1 and SAG2A and the unknown EST proximal to the BAG1 locus (Fig. 6A). After induction of stage conversion, TgCARM1 was found in association with the bradyzoite-specific SAG2C promoter (Fig. 6A). Importantly, methylated arginine 17 and acetylated lysine 18 of histone H3 were consistently present at the same loci (Fig. 6A). Cross talk between these histone modifications was previously reported in human cells, where CBP/p300 acetylates H3 [K18] to facilitate CARM1 recruitment and subsequent H3 [R17] methylation (11). Apicomplexans do not possess a CBP/p300 homologue, but purified FLAG-TgGCN5 exhibits an unexpected bias to acetylate only H3 [K18] (Fig. 6B). This is intriguing because the preferred substrate for GCN5 in other species is H3 [K14]. These studies suggest that TgGCN5 has evolved an unusual substrate preference and may contribute to combinatorial histone modifications to affect gene expression in Toxoplasma. However, it should be noted that yeast GCN5 exhibits an increased ability to acetylate multiple H3 lysines when partnered with additional proteins found in native GCN5 complexes (22).
FIG. 6.
Concurrent histone acetylation and methylation in the epigenetic control of stage conversion. (A) ChIP analysis of tachyzoite-specific genes (unknown tachyzoite EST, SAG1, and SAG2A) and the bradyzoite-specific gene SAG2C. Tachyzoites (T) and in vitro-generated bradyzoites (Bradyzoite/Tachyzoite) were processed for ChIP with TgCARM1 antibody (α-49), α-methyl [R17] H3, α-acetyl [K18] H3, or α-histone H3 antibodies. A chromatin sample with an irrelevant antibody was used as a negative control (IgG) and genomic DNA as a positive control for PCR (input). (B) Yeast (Sc) and Toxoplasma (Tg) GCN5 acetylation of recombinant histone H3 (1 μg) in vitro. Substrate specificity was monitored by immunoblotting HAT assay reaction mixtures with antibodies to specific acetylated (Ac) lysine residues.
We also observed accumulation of multiple methyl groups at H3 [K4] (α-K4-2ME and -3ME [data not shown]). Dimethylation occurs at both inactive and active euchromatic Toxoplasma genes, whereas trimethylation is present exclusively at active genes, as reported in yeast (data not shown; 47). Trimethylation is significantly enriched in the neighborhood of tachyzoite-specific genes during that life cycle stage and enhanced for the bradyzoite gene SAG2C following differentiation. Therefore, the presence of a trimethylated lysine 4 defines an active state of gene expression in Toxoplasma, much like arginine 17 methylation and K18 acetylation.
Inhibiting TgCARM1 induces cyst formation.
Multiple attempts to generate a knockout of the TgCARM1 gene failed, suggesting that the gene is essential in tachyzoites (data not shown). This is consistent with the observation that mice null for CARM1 die perinatally and are small (61). We next attempted to express a dominant-negative form of TgCARM1 by mutating the conserved catalytic residues to abolish enzyme activity (VMDΔAAA) (7). Surprisingly, HA-FLAG-tagged TgCARM1VMDΔAAA is exclusively cytoplasmic (data not shown). Unfortunately, no stable parasites could be obtained expressing this mutant protein. Even when transiently expressed, HA-FLAG-TgCARM1VMDΔAAA significantly reduces parasite division (data not shown).
We then explored pharmacological options to study TgCARM1 in greater detail. AMI-1 is a small molecule that specifically inhibits arginine N-methyltransferase activity (9). AMI-1 does not compete for the AdoMet binding site and inhibits arginine but not lysine methyltransferases in vitro (9). AMA-2 is an activator of Hmt1p (9). We investigated the effect of AMI-1 and AMA-2 in methylation reactions performed with recombinant TgCARM1 and TgPRMT1 (Fig. 7A). AMA-2 stimulates the methylation activity of TgCARM1 up to 10-fold, whereas it has no effect on TgPRMT1; AMI-1 efficiently inhibits TgCARM1-mediated methylation of H3 (Fig. 7A). In contrast, AMI-1 exhibits a minimal effect on TgPRMT1 activity whereas S-adenosylhomocysteine totally abrogates both TgCARM1 and TgPRMT1 activities at 0.04 μM and 0.40 μM, respectively (Fig. 7A).
Specific inhibition by AMI-1 of TgCARM1-mediated methylation prompted us to test its effect on Toxoplasma. Treatment of intracellular parasites had no observable effect on cell division or cyst formation (data not shown). However, pretreatment of extracellular tachyzoites with AMI-1 prior to infection induced cyst formation following two to three rounds of intracellular division (Fig. 7B). Interestingly, AMI-1-induced stage conversion correlates with a significant decrease in global H3 [R17] methylation (Fig. 7C). We also used ChIP assays to confirm that AMI-1 treatment triggers an up to 50% decrease in methylated H3 [R17] in promoters of the tachyzoite-specific genes SAG1 and SAG2A (Fig. 7E). These observations imply that methylated H3 [R17] might be a hallmark of the tachyzoite stage; however, in vitro-generated bradyzoites also exhibit methylation of H3 [R17] (Fig. 7D), showing clearly that TgCARM1 is active in both stages. One explanation for the effect of AMI-1 is that by inhibiting TgCARM1 activity, unknown “differentiation checkpoint genes” may have been turned off. Down-regulation of these genes would therefore initiate differentiation of tachyzoites to bradyzoites. Alternatively, arginine methyltransferases have been shown to target nonhistone proteins, and inhibiting those modifications may be responsible for the effects of AMI-1.
DISCUSSION
Regulation of gene expression is vastly understudied in pathogenic protozoan parasites, where it is likely to be of critical importance given their complex multiple life cycle stages. Recent completion of several apicomplexan genomes reveals a glaring absence of conventional transcription factors; however, it is becoming apparent that an abundance of chromatin remodeling machinery is present. Insight into the epigenetics and how transcription is regulated in these ancient eukaryotes has not only important evolutionary implications but also the potential to reveal novel ways to therapeutically intervene to treat protozoal diseases. Indeed, antigenic variation in Trypanosoma and Plasmodium involves epigenetics, and disruption of the SIR2 HDAC in the latter leads to a loss of control over var gene expression (15, 20, 45). Further, the antiprotozoal agent apicidin targets a parasite HDAC (10). In this study, we were unable to disrupt the TgCARM1 loci, implying that the gene product is essential for parasite welfare. Hence, we sought to explore epigenetics as a means of controlling stage-specific gene expression in the medically relevant apicomplexan Toxoplasma. This is the first study to demonstrate the use of histone modifications for life cycle stage conversion and elucidates novel parasite-specific HDAC and CARM1 complexes mediating these activities in the context of a previously described GCN5 HAT.
Histone modifications and life cycle stage conversion.
Having made strong correlations between histone acetylation status and stage-specific gene expression in Toxoplasma (Fig. 1), we have made our next objective to identify the chromatin remodeling complexes responsible for mediating these activities. A GCN5 HAT has been described previously (53), and here we characterize TgCRC, a multisubunit TgHDAC3-containing complex found in association with transcriptionally inactive promoters in Toxoplasma. While Toxoplasma lacks unambiguous homologues to N-CoR and SMRT, the TgCRC is a likely evolutionary precursor to the complexes containing NcoR/SMRT in higher eukaryotes. While some components, such as TBL1, HSP70, and the TCP-1 ring complex, are conserved between TgCRC and N-CoR/SMRT complexes, novel parasite-specific proteins (TgCRC-230 and -350) highlight the uniqueness of the Toxoplasma machinery, whose physiological and phylogenetic properties warrant further investigation. Of particular note is that TgCRC-350 is a substoichiometric component that hence may not always be associated with the complex, suggesting that it may function as the DNA-binding component. Consistent with a crucial role for TgCRC in apicomplexans in general, HDAC3 (30) and TgTBL1 (accession no. Q8IEK5) orthologs are present in Plasmodium, as are possible orthologs to TgCRC-230 (accession no. AAN37262) and -350 (accession no. CAD49240).
We also show here for the first time that arginine methylation, linked to transcriptional activation in other organisms, is a key histone modification in Toxoplasma that also correlates with parasite differentiation. This is in agreement with other observations showing that arginine methylation functions in differentiation (8). We noted that TgCARM1 acquires the ability to methylate nucleosomes in the presence of ATP and parasite fractions enriched for SWI/SNF activities (Fig. 5F). It is tempting to speculate that it interacts with TgSRCAP, a SWI/SNF ATPase noted to increase in expression during bradyzoite differentiation (52) and whose human homologue does interact with CARM1 (43). Finally, it would be of interest to study if TgCARM1 methylates nonhistone substrates as a means of triggering differentiation, as noted in thymocytes (32).
Evolution of histone modifications.
It was surprising to discover that Toxoplasma possesses a sophisticated capacity to modify histones, rivaling the system observed in higher eukaryotic cells. For instance, a more extensive repertoire of arginine methylation machinery is present in apicomplexans compared to yeast and C. elegans, which each possess only one PRMT-related enzyme (Fig. 5B). This observation is consistent with the idea that the lack of many known transcription factors in apicomplexans may be compensated for at an epigenetic level. Even more surprising is the parasite's employment of H3 [K18] acetylation and H3 [R17] methylation to activate transcription in a stage-specific manner. This combination has particular relevance as it has only been reported to occur in humans, whereby CBP/p300 recruits CARM1 by virtue of H3 [K18] acetylation (11). While there is no CBP/p300 in apicomplexans, we made the remarkable discovery that recombinant TgGCN5 exquisitely targets H3 [K18] in in vitro HAT assays. As observed in yeast, TgGCN5 may display greater H3 acetylation capacity in vivo (22). Nevertheless, this concurrent acetylation-methylation observed in Toxoplasma may represent an example of convergent evolution.
Model for epigenetic involvement in parasite differentiation.
Our model thus far for the epigenetic regulation of stage conversion in Toxoplasma is shown in Fig. 7F. As noted, key signatures of Toxoplasma gene activation are acetylation of H3 [K18] and methylation of H3 [R17], which we have shown to be mediated by TgGCN5 and TgCARM1, respectively. We have also shown that Toxoplasma can silence genes via deacetylases, such as the TgHDAC3-containing TgCRC. There are undoubtedly other histone modifications employed by Toxoplasma to modulate gene expression, and our establishment of a working ChIP protocol for this organism will be invaluable in teasing them apart. The parasite-specific subunits contained within these chromatin-remodeling complexes are some of the most compelling components worthy of further investigation. These or other identified remodeling components (44) are likely to be master regulators or members of the stress-signaling pathway triggering bradyzoite differentiation. Either will be critical clues illuminating the molecular mechanisms driving parasite differentiation. Finally, our report demonstrates that besides its importance in pathogenesis, stage conversion in Toxoplasma can serve as a model system to study epigenetic mechanisms of eukaryotic cell differentiation.
Supplementary Material
Acknowledgments
Thanks go to J. F. Dubremetz, U. W. Bohne, and U. Gross for antibodies and D. S. Roos for the λZAPII library. We thank J. Gagnon for reviewing the manuscript. Preliminary genomic and/or cDNA sequence data were accessed via http://ToxoDB.org (33).
W. J. Sullivan is supported by an R01 grant from the National Institutes of Health (GM065051). N. Saksouk is supported by a fellowship from CNRSL (Lebanon). M. A. Hakimi is supported by grants from CNRS (ATIP program), the Fondation pour la Recherche Médicale (FRM), Emergence (Rhône-Alpes), and INSERM. Genomic data were provided by the Institute for Genomic Research (supported by NIH grant AI05093) and by the Sanger Center (Wellcome Trust). EST sequences were generated by Washington University (NIH grant 1R01AI045806-01A1).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, H. J., S. Kim, and H. W. Nam. 2003. Molecular cloning of the 82-kDa heat shock protein (HSP90) of Toxoplasma gondii associated with the entry into and growth in host cells. Biochem. Biophys. Res. Commun. 311:654-659. [DOI] [PubMed] [Google Scholar]
- 2.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 3.Bassi, M. T., R. S. Ramesar, B. Caciotti, I. M. Winship, A. De Grandi, M. Riboni, P. L. Townes, P. Beighton, A. Ballabio, and G. Borsani. 1999. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am. J. Hum. Genet. 64:1604-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, U. M., S. Daujat, S. J. Nielsen, K. Nightingale, and T. Kouzarides. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatti, M. M., and W. J. Sullivan, Jr. 2005. Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J. Biol. Chem. 280:5902-5908. [DOI] [PubMed] [Google Scholar]
- 6.Bohne, W., U. Gross, D. J. Ferguson, and J. Heesemann. 1995. Cloning and characterization of a bradyzoite-specifically expressed gene (hsp30/bag1) of Toxoplasma gondii, related to genes encoding small heat-shock proteins of plants. Mol. Microbiol. 16:1221-1230. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. L., K. A. Loffler, D. Chen, M. R. Stallcup, and G. E. Muscat. 2002. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 277:4324-4333. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, D., N. Yadav, R. W. King, M. S. Swanson, E. J. Weinstein, and M. T. Bedford. 2004. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 279:23892-23899. [DOI] [PubMed] [Google Scholar]
- 10.Darkin-Rattray, S. J., A. M. Gurnett, R. W. Myers, P. M. Dulski, T; M. Crumley, J. J. Allocco, C. Cannova, P. T. Meinke, S. L. Colletti, M. A. Bednarek, S. B. Singh, M. A. Goetz, A. W. Dombrowski, J. D. Polishook, and D. M. Schmatz. 1996. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. USA 93:13143-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daujat, S., U. M. Bauer, V. Shah, B. Turner, S. Berger, and T. Kouzarides. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12:2090-2097. [DOI] [PubMed] [Google Scholar]
- 12.Denton, H., C. W. Roberts, J. Alexander, K. W. Thong, and G. H. Coombs. 1996. Enzymes of energy metabolism in the bradyzoites and tachyzoites of Toxoplasma gondii. FEMS Microbiol. Lett. 137:103-108. [DOI] [PubMed] [Google Scholar]
- 13.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donald, R. G., D. Carter, B. Ullman, and D. S. Roos. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribo-syltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010-14019. [DOI] [PubMed] [Google Scholar]
- 15.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13-24. [DOI] [PubMed] [Google Scholar]
- 16.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson, D. J. 2004. Use of molecular and ultrastructural markers to evaluate stage conversion of Toxoplasma gondii in both the intermediate and definitive host. Int. J. Parasitol. 34:347-360. [DOI] [PubMed] [Google Scholar]
- 18.Ferro, M., D. Salvi, S. Brugiere, S. Miras, S. Kowalski, M. Louwagie, J. Garin, J. Joyard, and N. Rolland. 2003. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2:325-345. [DOI] [PubMed] [Google Scholar]
- 19.Forsberg, E. C., and E. H. Bresnick. 2001. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23:820-830. [DOI] [PubMed] [Google Scholar]
- 20.Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel- Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25-36. [DOI] [PubMed] [Google Scholar]
- 21.Gastens, M. H., and H. G. Fischer. 2002. Toxoplasma gondii eukaryotic translation initiation factor 4A associated with tachyzoite virulence is down-regulated in the bradyzoite stage. Int. J. Parasitol. 32:1225-1234. [DOI] [PubMed] [Google Scholar]
- 22.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 23.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 24.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther, M. G., J. Yu, G. D. Kao, T. J. Yen, and M. A. Lazar. 2002. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 16:3130-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 27.Hettmann, C., and D. Soldati. 1999. Cloning and analysis of a Toxoplasma gondii histone acetyltransferase: a novel chromatin remodelling factor in apicomplexan parasites. Nucleic Acids Res. 27:4344-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 30.Joshi, M. B., D. T. Lin, P. H. Chiang, N. D. Goldman, H. Fujioka, M. Aikawa, and C. Syin. 1999. Molecular cloning and nuclear localization of a histone deacetylase homologue in Plasmodium falciparum. Mol. Biochem. Parasitol. 99:11-19. [DOI] [PubMed] [Google Scholar]
- 31.Jung, C., C. Y. Lee, and M. E. Grigg. 2004. The SRS superfamily of Toxoplasma surface proteins. Int. J. Parasitol. 34:285-296. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J., J. Lee, N. Yadav, Q. Wu, C. Carter, S. Richard, E. Richie, and M. T. Bedford. 2004. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J. Biol. Chem. 279:25339-25344. [DOI] [PubMed] [Google Scholar]
- 33.Kissinger, J. C., B. Gajria, L. Li, I. T. Paulsen, and D. S. Roos. 2003. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 31:234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoll, L. J., and J. C. Boothroyd. 1998. Isolation of developmentally regulated genes from Toxoplasma gondii by a gene trap with the positive and negative selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase. Mol. Cell. Biol. 18:807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lekutis, C., D. J. Ferguson, and J. C. Boothroyd. 2000. Toxoplasma gondii: identification of a developmentally regulated family of genes related to SAG2. Exp. Parasitol. 96:89-96. [DOI] [PubMed] [Google Scholar]
- 36.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons, R. E., R. McLeod, and C. W. Roberts. 2002. Toxoplasma gondii tachyzoite-bradyzoite interconversion. Trends Parasitol. 18:198-201. [DOI] [PubMed] [Google Scholar]
- 38.Ma, H., C. T. Baumann, H. Li, B. D. Strahl, R. Rice, M. A. Jelinek, D. W. Aswad, C. D. Allis, G. L. Hager, and M. R. Stallcup. 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 11:1981-1985. [DOI] [PubMed] [Google Scholar]
- 39.Manger, I. D., A. B. Hehl, and J. C. Boothroyd. 1998. The surface of Toxoplasma tachyzoites is dominated by a family of glycosylphosphatidylinositol-anchored antigens related to SAG1. Infect. Immun. 66:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matrajt, M., R. G. Donald, U. Singh, and D. S. Roos. 2002. Identification and characterization of differentiation mutants in the protozoan parasite Toxoplasma gondii. Mol. Microbiol. 44:735-747. [DOI] [PubMed] [Google Scholar]
- 41.Matrajt, M., C. D. Platt, A. D. Sagar, A. Lindsay, C. Moulton, and D. S. Roos. 2004. Transcript initiation, polyadenylation, and functional promoter mapping for the dihydrofolate reductase-thymidylate synthase gene of Toxoplasma gondii. Mol. Biochem. Parasitol. 137:229-238. [DOI] [PubMed] [Google Scholar]
- 42.Messina, D. N., J. Glasscock, W. Gish, and M. Lovett. 2004. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 14:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monroy, M. A., N. M. Schott, L. Cox, J. D. Chen, M. Ruh, and J. C. Chrivia. 2003. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor-mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 17:2519-2528. [DOI] [PubMed] [Google Scholar]
- 44.Nallani, K. C., and W. J. Sullivan, Jr. 2005. Identification of proteins interacting with Toxoplasma SRCAP by yeast two-hybrid screening. Parasitol. Res. 95:236-242. [DOI] [PubMed] [Google Scholar]
- 45.Navarro, M., G. A. Cross, and E. Wirtz. 1999. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 18:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, R., and A. J. Bannister. 2002. Protein N-methyltransferase assays in the study of gene transcription. Methods 26:226-232. [DOI] [PubMed] [Google Scholar]
- 49.Singh, U., J. L. Brewer, and J. C. Boothroyd. 2002. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol. Microbiol. 44:721-733. [DOI] [PubMed] [Google Scholar]
- 50.Spiess, C., A. S. Meyer, S. Reissmann, and J. Frydman. 2004. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell. Biol. 14:598-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan, W. J., Jr., M. A. Monroy, W. Bohne, K. C. Nallani, J. Chrivia, P. Yaciuk, C. K. Smith II, and S. F. Queener. 2003. Molecular cloning and characterization of an SRCAP chromatin remodeling homologue in Toxoplasma gondii. Parasitol. Res. 90:1-8. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan, W. J., Jr., and C. K. Smith II. 2000. Cloning and characterization of a novel histone acetyltransferase homologue from the protozoan parasite Toxoplasma gondii reveals a distinct GCN5 family member. Gene 242:193-200. [DOI] [PubMed] [Google Scholar]
- 54.Templeton, T. J., L. M. Iyer, V. Anantharaman, S. Enomoto, J. E. Abrahante, G. M. Subramanian, S. L. Hoffman, M. S. Abrahamsen, and L. Aravind. 2004. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 14:1686-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogelauer, M., J. Wu, N. Suka, and M. Grunsteins. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 56.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 57.Weiss, L. M., and K. Kim. 2000. The development and biology of bradyzoites of Toxoplasma gondii. Front. Biosci. 5:D391-D405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss, L. M., D. Laplace, P. M. Takvorian, H. B. Tanowitz, A. Cali, and M. Wittner. 1995. A cell culture system for study of the development of Toxoplasma gondii bradyzoites. J. Eukaryot. Microbiol. 42:150-157. [DOI] [PubMed] [Google Scholar]
- 59.Weiss, V. H., A. E. McBride, M. A. Soriano, D. J. Filman, P. A. Silver, and J. M. Hogle. 2000. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 7:1165-1171. [DOI] [PubMed] [Google Scholar]
- 60.Xu, W., H. Cho, S. Kadam, E. M. Banayo, S. Anderson III, J. R. Yates, B. M. Emerson, and R. M. Evans. 2004. A methylation-mediator complex in hormone signaling. Genes Dev. 18:144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yadav, N., J. Lee, J. Kim, J. Shen, M. C. Hu, C. M. Aldaz, and M. T. Bedford. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 100:6464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, S., and S. F. Parmley. 1997. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene 184:1-12. [DOI] [PubMed] [Google Scholar]
- 63.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, X., L. Zhou, and X. Cheng. 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.