Abstract
Steroidogenic factor 1 (SF-1) is a nuclear receptor essential for steroidogenic gene expression, but how its activity is regulated is unclear. Here we demonstrate that p300 plays an important role in regulating SF-1 function. SF-1 was acetylated in vitro and in vivo by p300 at the KQQKK motif in the Ftz-F1 (Fushi-tarazu factor 1) box adjacent to its DNA-binding domain. Mutation of the KQQKK motif reduced the DNA-binding activity and p300-dependent activation of SF-1. When stimulated with cyclic AMP (cAMP), adrenocortical Y1 cells expressed more p300, leading to additional SF-1 association with p300 and increased SF-1 acetylation and DNA binding. It also increased SF-1 colocalization with p300 in nuclear foci. Collectively, these results indicate that SF-1 transcriptional activity is regulated by p300 in response to the cAMP signaling pathway by way of increased acetylation, DNA binding, and recruitment to nuclear foci.
Steroidogenic factor 1 (SF-1), also known as Ad4BP (adrenal 4 binding protein), is a member of the nuclear receptor superfamily, designated NR5A1 (39). SF-1 plays a critical role in the development, differentiation, and function of the hypothalamus, pituitary, adrenal glands, and gonads (43). SF-1 controls the expression of a variety of genes, such as steroidogenic genes, Müllerian inhibitory substance, and the α-subunit and β-subunit of gonadotropins (37, 43). SF-1 exerts its transcriptional activity through interaction with numerous proteins, including coactivators, corepressors, and other transcription factors (2, 9-11, 24, 35, 36, 42).
SF-1 is structurally similar to steroid receptors; it contains a zinc finger DNA-binding domain (DBD) and a C-terminal ligand-binding domain but lacks the N-terminal A/B domain (Fig. 1A). Members of the nuclear receptor 5 (NR5) subfamily, including Drosophila melanogaster FTZ-F1 (NR5A3), silkworm BmFTZ-F1, and mammalian LRH-1 (liver receptor homologue 1) (NR5A2) and SF-1 (NR5A1), share a conserved 30-amino-acid (aa) basic region, designated the Ftz-F1 (Fushi-tarazu factor 1) box, adjacent to the C terminus of the DNA-binding domain (52). This box facilitates recognition of the first three bases of the DNA sequence PyCAAGGPyCPu (52). The Ftz-F1 box together with its adjacent proline-rich sequence (aa 78 to 172), called the FP domain, is important for the transactivation function of SF-1 (30). It is also important for nuclear localization as well as interaction with TFIIB and c-Jun (30).
FIG. 1.
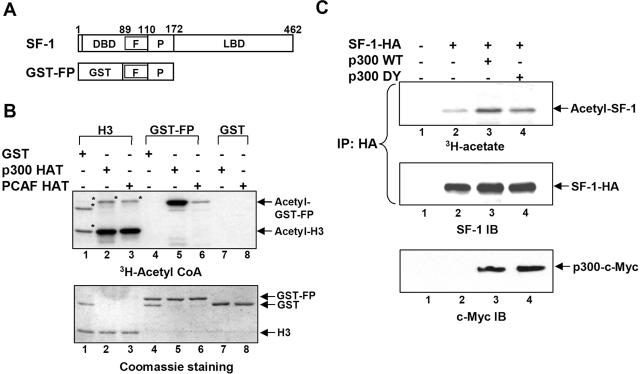
SF-1 is acetylated by p300 in vitro and in vivo. (A) Schematic representation of SF-1 and GST-FP. Residue numbers in SF-1 are indicated. F, Ftz-F1 box; P, proline-rich domain; and LBD, ligand-binding domain. (B) In vitro acetylation of GST-FP. Purified histone H3, GST-FP, or GST was incubated with GST, recombinant p300 HAT, or PCAF HAT in the presence of [3H]acetyl coenzyme A. The acetylated proteins, detected by fluorography, are shown in the top gel; the bottom gel shows proteins stained by Coomassie brilliant blue. The asterisks denote nonspecific signals probably arising from impurities in the commercial histone H3 extract. (C) SF-1 is acetylated by p300 in vivo. Expression plasmid for SF-1-HA was cotransfected with c-Myc-tagged p300 WT or p300 DY plasmid into 293T cells, followed by labeling with [3H]acetate. The acetyl-SF-1-HA proteins (top) were visualized by fluorography after immunoprecipitation (IP) with an anti-HA antibody and gel separation. SF-1-HA and p300 were detected by immunoblotting (IB) with an anti-SF-1 (middle) or anti-c-Myc (bottom) antibody, respectively.
SF-1 is modified by phosphorylation and SUMO conjugation at the hinge domain. The phosphorylation site is mediated by mitogen-activated protein kinase and required for maximal transcriptional activity of SF-1 (17). SUMO conjugation represses SF-1 activity by recruiting transcriptional repressors like DP103 and/or by relocating SF-1 to nuclear speckles (7, 26, 28).
In addition to phosphorylation and SUMO conjugation, SF-1 is also acetylated (25). Two well-characterized histone acetyltransferase (HAT) proteins are in the p300/CBP (CREB-binding protein) family and the PCAF/GCN5 (p300/CBP-associated factor/general control nonderepressed 5) family. These HATs function as coactivators for transcription factors (49), many of which are acetylated, like p53 (15), E2F1 (33), c-Myb (50), EKLF (erythroid Krüppel-like factor) (57), MyoD (44), GATA-1 (4), and androgen receptor (AR) (14). Acetylation modulates the functions of these transcription factors by affecting DNA-binding activity, interaction with other proteins, stability, and nuclear localization. For example, acetylated p53 binds DNA and activates transcription more efficiently than unacetylated p53 (15, 32), probably in a promoter-specific manner (18). SF-1 is acetylated by GCN5 in vitro (25). Acetylation affects the transcriptional activity of SF-1. However, the mechanism of SF-1 activation by acetylation is still unclear.
The subcellular localization of transcription factors is important for gene activation. Activated transcription factors like ligand-induced steroid receptors glucocorticoid receptor (19), AR (51), mineralocorticoid receptor (13), and hypoxia-inducible factor 1 (HIF-1) (46) are concentrated at specific regions of the nuclei. These nuclear clusters partially overlap with activated RNA polymerase II (Pol II) or nascent mRNA. Many proteins in the transcription machinery are also at these foci. These include transcriptional coactivators p300/CBP, SRC-1 (steroid receptor coactivator 1) (48), components of chromatin remodeling complexes (34), and RNA Pol II (46, 53). Thus, nuclear-cluster formation may be a process of gene activation, in which activated transcription factors and coactivators can be recruited to the active transcription sites.
Cyclic AMP (cAMP) is the intracellular molecule that conducts the signal of extracellular tropic hormones to cAMP-dependent protein kinase A (PKA) and the downstream signaling pathway. In adrenocortical cells, activation of the cAMP-PKA pathway increases the expression of several SF-1-regulated steroidogenic genes (5, 20, 29, 41), including Cyp11a1, which encodes cytochrome P450scc, catalyzing the first and rate-limiting step of steroidogenesis. However, the mechanism by which the cAMP-PKA pathway activates SF-1-mediated transcription is still elusive.
Like most acetylated transcription factors, SF-1 is acetylated not just by GCN5; in the current report, we show that p300 can also acetylate and activate SF-1. We demonstrate that p300 acetylates the KQQKK sequence at the Ftz-F1 box of SF-1. This acetylation correlates with DNA binding and p300-potentiated transcriptional activation of SF-1. In addition, SF-1 is also recruited to RNA polymerase II-associated nuclear clusters by p300. Acetylation, association with these nuclear clusters, and DNA binding of SF-1 were increased after cAMP stimulation, which also increases the amount of p300. These results suggest a novel mechanism of the cAMP signaling pathway to stimulate SF-1 activity through the increase of p300 level.
MATERIALS AND METHODS
Cell culture, plasmids, and recombinant proteins.
Mouse fibroblast NIH 3T3 and human kidney 293T cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Mouse adrenocortical tumor Y1 cells were maintained in DMEM-F-12 supplemented with 10% fetal bovine serum. The following constructs have previously been described: phscc2.3k-LacZ, pGEX-SF-1-FP (30), pGal4-SF-1 (7), pET-p300 HAT (40), pGEX5X-PCAT-HAT (56), pCMV-p300-myc WT and pCMV-p300-myc DY (23), pcDNA3.1-SF-1-HA (36), and pcDNA3-FLAG-GCN5 (55). Mutated SF-1 constructs (mt1, mt2, dmt, mt3, and mt4 [see Fig. 4]) were generated by PCR-based site-directed mutagenesis in pcDNA3-SF-1-HA. The plasmid pcDNA5-3xFLAG-SF-1 was generated by inserting the coding sequence of SF-1, amplified by PCR from pcDNA3-SF-1-HA, into XhoI/XbaI sites of pcDNA5/TO (Invitrogen). The resulting plasmid was then inserted with the coding sequence of the Flag tag repeating three times at BamHI/XhoI sites. The sequences of constructs were verified by direct sequencing. The recombinant proteins GST-FP, His-p300 HAT, and GST-PCAF HAT (where GST is glutathione S-transferase) were overexpressed in Escherichia coli strain BL21(DE3) pLys and purified as described previously (22). Purified histone H3 protein was purchased from Roche Molecular Biochemicals (Mannheim, Germany).
FIG. 4.
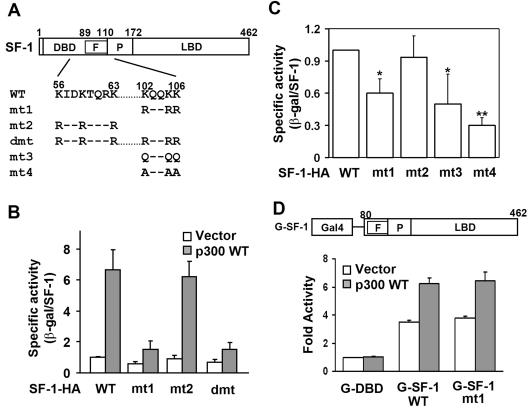
The KQQKK sequence is required for p300-potentiated activation and basal activity of SF-1. (A) Schematic representation of SF-1 constructs. The amino acid sequences of two potential acetylation sites are shown. Lys (K) residues were replaced with Arg (R), Gln (Q), or Ala (A) as indicated. (B) Lys-to-Arg mutation at the KQQKK motif of SF-1 (mt1) diminishes its p300-potentiated transcriptional activity. The specific activities of SF-1 were determined in the presence (p300 WT) or absence (Vector) of p300 in NIH 3T3 cells. (C) Mutation of K to Q or A at the KQQKK motif of SF-1 reduces its basal transcriptional activity. The specific activities of wild-type and mutated SF-1 expressed in NIH 3T3 cells are shown. *, P of <0.05, and **, P of <0.01, compared with wild-type SF-1-HA transfection, by Student's t test. (D) The KQQKK sequence is not required for full activation of SF-1 when linked to the DBD of Gal4 protein. Y1 cells were transfected with the 5×Gal4-tk-Luc reporter gene and expression plasmids for Gal4-DBD (G-DBD), wild-type Gal4-SF-1 (G-SF-1 WT), or mutated Gal4-SF-1 (G-SF-1 mt1). The transactivation activity (n-fold) of each protein relative to that of the G-DBD control is shown as mean ± standard deviation on the y axis. F, Ftz-F1 box; P, proline-rich domain; LBD, ligand-binding domain; WT, wild type; and β-gal, β-galactosidase.
In vitro acetylation assay.
Two micrograms of each substrate (histone H3, GST-SF-1-FP, and GST) was added to HAT buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.1 mM EDTA, and 1 mM dithiothreitol) containing 5 μCi/ml of [3H]acetyl coenzyme A (10 Ci/mmol) with or without 200 ng of purified HAT proteins. Reaction mixtures were incubated at 30°C for 30 min, stopped by adding 1 volume of 2× sample buffer and heating to 100°C for 3 min, and then separated by 12% polyacrylamide gel electrophoresis. The gel was stained with Coomassie brilliant blue, immersed into Amplify solution (Amersham Biosciences) for 20 min, dried, and exposed to X-ray film. Quantitation was performed using Image Gauge version 3.2 software with a FujiFilm LAS-1000plus image reader.
In vivo acetylation assay.
Ten micrograms of each of the expression plasmids for SF-1-HA, wild-type p300 (p300 WT), or HAT-defective p300 (p300 DY) was cotransfected into 293T cells. Forty-six hours after transfection, the medium was replaced with DMEM containing 1 mCi/ml sodium [3H]acetate (5 Ci/mmol) and 100 ng/ml trichostatin A (TSA) for 2 h. Whole-cell extracts were prepared in IPH buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 1× Complete protease inhibitor cocktail) and immunoprecipitated with 2 μg rat anti-hemagglutinin (HA) antibody (clone 3F10; Roche). The immunoprecipitates were separated by 10% polyacrylamide gel electrophoresis followed by fluorography at −70°C for 10 days. The nonradiolabeled parallel group of transfectants was analyzed by immunoblotting with a polyclonal antiserum against full-length SF-1 (7) and visualized by chemiluminescence.
For 3H-incorporation of SF-1 in response to cAMP, Y1 cells were transfected with 3 μg expression plasmids for wild-type or KQQKK mutated 3xFLAG-SF-1. After 24 h, cells were labeled with 1 mCi/ml sodium [3H]acetate in the absence or presence of 1 mM 8-Br-cAMP, a cell-permeable cAMP analog, for 6 h. After immunoprecipitation using 2 μg mouse anti-FLAG antibody (Sigma), the immunoprecipitates were analyzed as described above.
Transcriptional-activity assay.
For transfection into NIH 3T3 cells in 35-mm culture dishes, 200 ng of SF-1-HA plasmid and SF-1-dependent reporter gene phscc2.3k-LacZ were cotransfected with increasing amounts (0.2, 0.5, and 1 μg) of p300 WT or p300 DY as indicated. After 48 h, β-galactosidase activities of whole-cell extracts were assayed as described previously (21). Equal volumes of whole-cell extracts were subjected to 10% polyacrylamide gel electrophoresis and immunoblotted with an SF-1 polyclonal antiserum. The specific activities of SF-1 were calculated from β-galactosidase activities normalized with the levels of transfected SF-1-HA protein.
Stable transfection.
Y1 cells were transfected with pcDNA3-SF-1-HA or pcDNA3-SF-1-HA mt1 1 day before subculture in 100-mm culture dishes supplemented with 500 μg/ml G418 sulfate. Individual clones were generated after 2 weeks of selection. For each construct, 40 to 60 clones were picked, expanded, and tested for SF-1-HA expression by immunoblotting with antibody against the HA tag. Two clones expressing the highest amounts of SF-1-HA were further expanded for subsequent experiments.
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay was performed as described previously, with some modifications (3). Briefly, Y1 SF-1-HA stable clones were grown to 80 to 90% confluence in 100-mm culture dishes treated with or without 1 mM 8-Br-cAMP for 6 h. Cells were cross-linked with 1% formaldehyde for 10 min, and the reaction was stopped with 0.125 M glycine for 5 min at room temperature. Nuclei were collected and sonicated in lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 0.5 mM phenylmethylsulfonyl fluoride, and 1× Complete protease inhibitor cocktail [Roche]). Soluble chromatin was precleared with bovine serum albumin-sheared herring sperm DNA pretreated Staph A cells and immunoprecipitated with antibodies against the HA tag (Santa Cruz), acetylated histone H3 (Upstate), and immunoglobulin G (IgG) control or with no antibody (for input) for 16 h at 4°C. Immunoprecipitates were recovered using Staph A cells. DNA was extracted from immunoprecipitates by phenol-chloroform extraction and ethanol precipitation. Extracted DNA was analyzed by PCR using primers spanning the proximal (nucleotide positions −71 to +209) or distal (+873 to +1141) region of mouse Cyp11a1. Following 30 cycles of amplification, PCR products were run on a 1.5% agarose gel and analyzed by ethidium bromide staining.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays were performed as described previously, with some modifications (16). SF-1-HA containing nuclear extracts was prepared from transfected 293T cells and incubated with 32P-labeled oligonucleotide containing the SF-1-binding sequence on ice for 30 min. Subsequently, samples were separated by 5% native acrylamide gel electrophoresis. Competition for binding was performed by adding a 100-fold excess of unlabeled DNA, and supershift was detected by 1 μg rat anti-HA antibody. The intensity of each band was quantified using an image reader as described above.
Immunostaining and confocal microscopy.
Y1 cells were subcultured on coverslips at a density of 1 × 104 cells/well in 6-well plates. Before immunostaining, cells were treated with 1 mM 8-Br-cAMP for 6 h or transfected with the expression plasmids as indicated for 24 h by using Lipofectamine Plus according to the manufacturer's protocol. Treated or transfected cells were fixed in methanol at −20°C (or in 4% paraformaldehyde for green fluorescent protein fusion proteins) for 3 min, permeabilized with 0.2% Triton X-100-phosphate-buffered saline, and blocked in 2% blocking reagent (Roche). Subsequently, the cells were immunostained with antibody against SF-1, p300 (clone RW128, no cross-reaction with CBP; Upstate), RNA Pol II (C21, recognizing both hyper- and hypophosphorylated Pol II; Santa Cruz), c-Myc, or FLAG as indicated in blocking buffer at 4°C overnight. The cells were washed with 0.2% Triton X-100-phosphate-buffered saline and stained with Alexa 488- or Alexa 546-conjugated secondary antibodies (Molecular Probes) in blocking buffer at room temperature for 1 h. After three extensive washes, the coverslips were mounted on glass slides in 50% glycerol-phosphate-buffered saline. Fluorescent cells were examined with a Zeiss LSM 510 confocal microscope.
Measurement of colocalization.
The percentage of protein colocalization was determined by using LSM software release 3.2 (Carl Zeiss). Briefly, single-cell images were obtained from dual-immunolabeled Y1 cells treated with or without 1 mM 8-Br-cAMP by using confocal laser scanning microscopy. Optical sections from the middles of cells were used for the generation of scatter plots. The thresholds for both red and green fluorescent signals were determined after reducing the backgrounds of cell images to the lowest setting, and the same condition was applied to all images. Eighteen cells were randomly selected and analyzed in four separate immunostaining experiments in the absence or presence of 1 mM 8-Br-cAMP treatment. The percentage of SF-1 or Pol II colocalization with p300 was calculated by dividing pixels that were colocalized with p300 in area 3 by the total pixels in areas 2 and 3, with error bars representing standard deviations.
Coimmunoprecipitation assay.
Y1 cells were grown in 10-cm-diameter plates for 24 h and transfected with 3 μg expression plasmids for 3xFLAG-SF-1. After 24 h, cells were treated with or without 1 mM 8-Br-cAMP for 6 h. Whole-cell extracts were prepared in IPH buffer containing 240 mM NaCl. Equal amounts of whole-cell extracts were used for coimmunoprecipitation with 2 μg anti-p300 antibody (N-15; Santa Cruz) for 1 h at 4°C. The immunoprecipitates and 10% input of whole-cell extracts were separated by 7% polyacrylamide gel electrophoresis, followed by immunoblotting with antibodies against p300, SF-1, or Pol II, and visualized by chemiluminescence.
RESULTS
SF-1 is acetylated by p300 in vitro and in vivo.
SF-1 is a zinc finger protein. Many zinc finger proteins, like GATA-1, EKLF, and AR, are acetylated around the zinc finger region (4, 14, 57). Given that SF-1-mediated transcription can be potentiated by p300/CBP (36), we reasoned SF-1 might be acetylated by these HATs as well. To test this hypothesis, the FP domain of SF-1, which contains the Ftz-F1 box and the Pro-rich sequence, was fused with GST to be a substrate for the in vitro acetylation assay (Fig. 1A). The acetylation enzymes tested were the HAT domains from p300 (aa 1195 to 1673) and PCAF (aa 352 to 832). As shown in Fig. 1B, the HAT domains of p300 and PCAF acetylated their known substrate, histone H3 (lanes 2 and 3), but not GST (lanes 7 and 8), showing that both HATs were active and substrate specific. GST-FP was acetylated efficiently by p300 but poorly by PCAF (Fig. 1B, lanes 5 and 6). This result suggests that the FP domain of SF-1 is preferentially acetylated by p300 rather than by PCAF.
To examine whether SF-1 is acetylated by p300 in vivo, we expressed both SF-1-HA and p300 in 293T cells and incubated the cells with [3H]acetate before immunoprecipitating SF-1-HA. In the absence of overexpressed p300, the [3H]acetyl group was incorporated into SF-1, indicating that SF-1 was acetylated at a basal level (Fig. 1C, top, lane 2). This basal level of acetylation was enhanced by p300 WT, as revealed by the increased amount of acetyl-SF-1 (Fig. 1C, lane 3). Thus, p300 can acetylate SF-1 in vivo. A HAT-defective p300 (p300 DY), however, could not acetylate SF-1 to the same extent as p300 WT could (Fig. 1C, lane 4 versus lane 3), implying the importance of the HAT activity of p300 in SF-1 acetylation. Our control experiment showed that equivalent amounts of SF-1-HA were expressed and immunoprecipitated from each lane (Fig. 1C, middle), indicating that the increased level of acetyl-SF-1 was due to increased acetylation, not due to an increased amount of SF-1. These results indicate that p300 transfers acetyl groups to SF-1 in vivo and in vitro and that the acetylation site lies in the FP domain of SF-1.
Mutation of the KQQKK sequence in the Ftz-F1 box abolishes p300-promoted acetylation of SF-1.
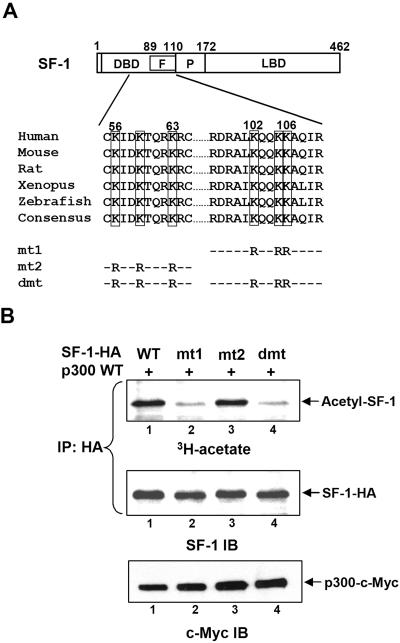
In order to map the p300 acetylation site of SF-1, we searched and identified two potential acetylation sequences in SF-1: one in the middle of the DNA-binding domain, similar to acetylation sequence KXXKXXXK in histone H3, and the other at the C terminus of the Ftz-F1 box, similar to p53 acetylation sequence KXKK (15). Both sequences are conserved in SF-1 proteins from several species (Fig. 2A). To examine whether these two sites could be acetylated, we replaced the Lys residues of both potential acetylation sites with Arg to form mt1, mt2, and dmt (Fig. 2A). In vivo acetylation assays demonstrated that the acetylation level of SF-1 mt1, in which the KQQKK motif in the Ftz-F1 box was mutated, was much lower than that of the wild-type SF-1 even in the presence of p300 (Fig. 2B, lane 2 versus lane 1). The lack of p300-specific acetylation in SF-1 mt1 indicated that p300 specifically acetylated this KQQKK sequence. The acetylation level of SF-1 mt2 was unchanged (Fig. 2B, lane 3), thus indicating that the Lys residues in KXXKXXXK are not p300 substrates. SF-1 dmt, mutated at both sites, also exhibited a low acetylation level (Fig. 2B, lane 4). Collectively, these results indicate that the KQQKK motif in the Ftz-F1 box of SF-1 is acetylated by p300.
FIG. 2.
The KQQKK sequence at the Ftz-F1 box of SF-1 is acetylated by p300 in vivo. (A) Potential acetylation sites of SF-1. The amino acid sequences of two potential acetylation sites of SF-1 are shown, and the conserved Lys (K) residues are marked. In mutated SF-1 (mt1, mt2, and dmt), the K residues were replaced with Arg (R). F, Ftz-F1 box; P, proline-rich domain; and LBD, ligand-binding domain. (B) Mutation of KQQKK sequence abolishes p300-enhanced acetylation of SF-1. The incorporation of [3H]acetate into wild-type (WT) and mutated SF-1 following expression and immunoprecipitation (IP) of SF-1-HA are shown; the amounts of immunoprecipitated SF-1 and p300 expression were determined by SF-1 and c-Myc immunoblotting (IB), respectively.
The HAT activity of p300 is required for full activation of SF-1.
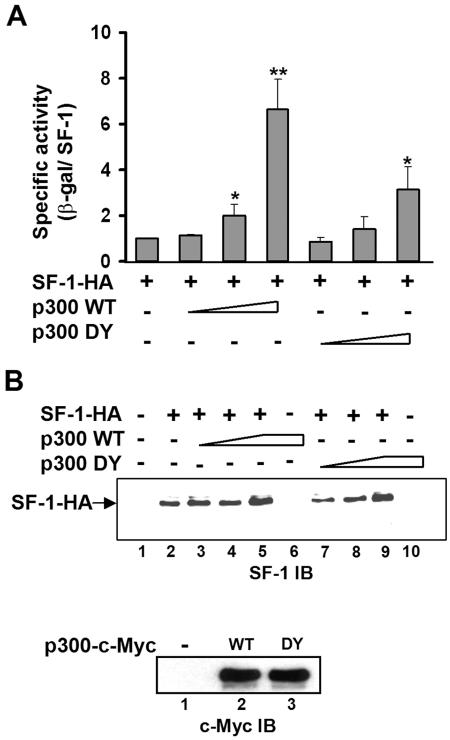
Since SF-1 is acetylated by p300, we next investigated whether this acetylation regulates its activity. SF-1 activity was measured by its ability to activate an SF-1-dependent reporter gene, phscc-2.3K-LacZ, which contains a 2.3-kb CYP11A1 promoter linked to a reporter. NIH 3T3 cells were chosen because they express less endogenous p300. The specific activity of SF-1 was calculated from the reporter gene activity normalized against the protein level of SF-1, obtained after immunoblotting of whole-cell lysate (Fig. 3B). As shown in Fig. 3A, the specific activity of SF-1 was enhanced by p300 WT in a dose-dependent manner. Although expressed at a level comparable to that of p300 WT, the HAT-defective p300 (p300 DY) could not potentiate SF-1 activity as efficiently. Since p300 DY was also less effective in acetylating SF-1 (Fig. 1C), the ability of p300 to acetylate SF-1 parallels that of p300 to potentiate SF-1. These observations indicate that the HAT activity of p300 is required for full activation of SF-1.
FIG. 3.
p300 HAT activity is required for full activation of SF-1. Expression plasmids for SF-1-HA (0.2 μg) and increasing amounts of p300 WT (0.1, 0.5, and 1 μg) or p300 DY were cotransfected with 0.3 μg SF-1-dependent reporter gene phscc-2.3K-LacZ into NIH 3T3 cells. (A) The specific activity of each transfectant consisted of the β-galactosidase (β-gal) activity normalized with the SF-1 expression level. The specific activity of SF-1-HA alone was set to 1. All values represent the results of at least three separate transfection experiments, with error bars representing standard deviations. *, P of <0.05, and **, P of <0.01, compared with wild-type SF-1-HA transfection only, by Student's t test. (B) Immunoblotting (IB) for expression of SF-1-HA (top) and c-Myc-tagged p300 (bottom; transfection with 1μg plasmid) of cell lysate from panel A. WT, wild type. Triangles and trapezoids denote increasing amounts of p300.
Acetylation-deficient SF-1 cannot be fully activated by p300.
We next examined the role of p300 acetylation in transcription by comparing the transcriptional activities of wild-type and acetylation-deficient SF-1 in the presence or absence of p300. p300 enhanced the activities of wild-type SF-1 and mt2 but not of mt1 and dmt (Fig. 4A and B). Since mt1 and dmt were not acetylated by p300, the lack of p300 acetylation correlates with the loss of p300-enhanced transcriptional activity. This result indicates the importance of the KQQKK motif in p300-potentiated transcription activity of SF-1.
It has been proposed that acetylation of histone H3 neutralizes the positive charge of Lys, leading to a disruption of charge interactions between histone H3 and DNA, chromatin remodeling, and transcription activation (8). In order to assess whether SF-1 acetylation follows a similar mechanism, we replaced Lys of the KQQKK motif with neutral amino acids Gln and Ala to form SF-1 mt3 and mt4, respectively (Fig. 4A). It was expected that higher transcriptional activities would have been seen for mt3 and mt4 had charge neutralization had an effect on transcriptional activity. On the contrary, the basal transcriptional activities of mt3 and mt4, similar to those for mt1, were significantly lower than that of wild-type SF-1 (Fig. 4C). SF-1 mt2, whose mutation site is not in the KQQKK motif, had activity similar to that of wild-type SF-1. Therefore, simple charge neutralization cannot explain the function of acetylation in regulating SF-1 activity.
Since the Ftz-F1 box helps specific DNA recognition through the DBD (52), we next examined whether abolishment of acetylation by mutation would affect SF-1 function if the ability to bind DNA was retained. The DBD of SF-1 was replaced by the Gal4 DBD, which would bind the Gal4 recognition site, and the function of the resultant G-SF-1, which contains either the wild-type or mutated Ftz-F1 box, was tested. Both wild-type and mutated G-SF-1 could activate the reporter gene through the Gal4 recognition sequence, and the presence of p300 could potentiate both activities (Fig. 4D). This result indicates that acetylation-deficient SF-1 can still activate transcription as long as it can bind DNA through other DBDs. Thus, the acetylation of the Ftz-F1 box probably affects only the DNA-binding ability of SF-1.
Mutation of the KQQKK motif in SF-1 results in reduced DNA binding.
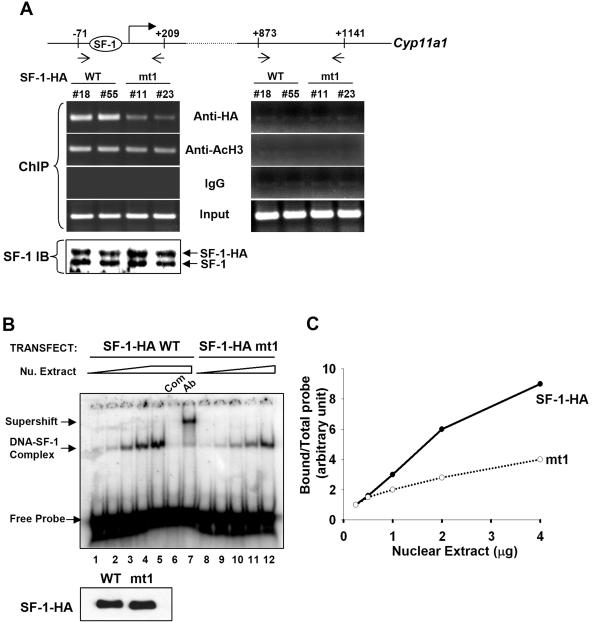
We next examined whether acetylation of the Ftz-F1 box modulates the DNA-binding activity of SF-1. To compare DNA-binding activities of wild-type and acetylation-deficient SF-1 in vivo, we established mouse adrenocortical tumor Y1 cell clones that overexpress similar amounts of wild-type and mt1 SF-1-HA proteins (Fig. 5A, bottom). Two clones expressing wild-type SF-1-HA (no. 18 and no. 55) and two clones expressing mt1 (no. 11 and no. 23) were tested. After ChIP using antibody against the HA tag, SF-1-HA bound to the region of nucleotide positions −71 to +209 of Cyp11a1, which contains an SF-1-binding site. SF-1-HA mt1, however, bound to this region with a reduced efficiency (Fig. 5A, left). As a control, acetylated histone H3 from all four clones of cells bound to this region of DNA equally well, and IgG did not pull down chromatin. No proteins bound to intron 1 of Cyp11a1 (+873 to +1141), indicating the specificity of the assay (Fig. 5A, right). These results show that mutation of the acetylation site reduces the DNA-binding ability of SF-1 in vivo.
FIG. 5.
Mutation of the acetylation site in the Ftz-F1 box of SF-1 results in decreased binding to DNA. (A) ChIP. Soluble chromatin was prepared from stable Y1 cell clones expressing SF-1-HA (clones no. 18 and no. 55) and SF-1-HA mt1 (clones no. 11 and no. 23) and immunoprecipitated with antibodies against HA, acetyl-H3 (AcH3), or IgG control. DNA in the extracts was further analyzed by PCR using primers that cover the regions of nucleotide positions −71 to +209 or +873 to +1141 of the Cyp11a1 gene, as indicated. Protein expression levels of SF-1 in stable clones were determined by immunoblotting (IB) with antiserum against SF-1 (bottom). (B) Wild-type SF-1-HA (SF-1-HA WT) or mutated SF-1-HA (SF-1-HA mt1) was coexpressed with p300 in 293T cells prior to DNA binding and electrophoretic mobility shift assays. The DNA-SF-1 complex was formed with increasing amounts of nuclear (Nu.) extract (0.25, 0.5, 1, 2, and 4 μg). Com, competition with 100× unlabeled oligonucleotide; Ab, supershift with anti-HA antibody. The immunoblot with anti-HA antibody showing both WT and mt1 expressed at the same level is shown at bottom. The triangle and trapezoid denote increasing amounts of nuclear extracts. (C) The relative binding abilities of SF-1-HA WT and SF-1-HA mt1 are presented as the ratios of bound probe/total probe versus the amounts of nuclear extract.
To further confirm that the acetylation site is important for DNA binding, electrophoretic mobility shift assays were performed with nuclear extract from 293T cells cotransfected with p300 and SF-1-HA wt or mt1 as a source of acetylated or unacetylated SF-1. As shown in Fig. 5B, the DNA-SF-1 complex was increased when the amount of nuclear extract was increased (lanes 1 to 5). This DNA-SF-1 complex was specific, as it disappeared when competed with 100-fold of unlabeled oligonucleotide (Fig. 5B, lane 6) or supershifted with anti-HA antibody (Fig. 5B, lane 7). Although expressed at the same level as wild-type SF-1 (Fig. 5B, bottom), SF-1 mt1 bound to DNA less efficiently (Fig. 5B, lanes 8 to 12). After quantitation of the protein-DNA complexes, we found that SF-1 mt1 bound to DNA at a reduced efficiency (Fig. 5C). These results suggest that mutation of the KQQKK sequence of SF-1 results in reduced DNA-binding activity.
p300 recruits SF-1 to nuclear foci.
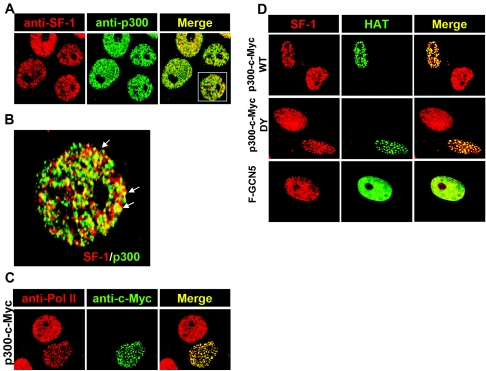
Previously, we reported that conjugation of SF-1 by SUMO changes its nuclear distribution (7). It is interesting to ask whether p300 acetylation would also modulate the distribution of SF-1. We performed double-color immunostaining with antibodies against SF-1 and p300 in Y1 cells. SF-1 and p300 were each located in discrete nuclear domains of the nucleus (Fig. 6A); some of these signals overlap when examined in the magnified image (Fig. 6B). p300 was located in transcriptionally active loci, as shown by colocalization of p300-c-Myc with Pol II (Fig. 6C). Therefore, SF-1 was partly activated in the basal condition. When Y1 cells were transfected with p300-c-Myc, the spots of SF-1 and p300-c-Myc colocalization were increased (Fig. 6D), indicating that SF-1 could be recruited by p300 to these transcriptionally active loci.
FIG. 6.
p300 recruits SF-1 to p300-RNA Pol II loci. (A) SF-1 partially colocalizes with p300 in the basal condition. Y1 cells grown on coverslips were fixed and labeled with antibodies against SF-1 (red) and p300 (green). Immunofluorescence staining was analyzed by confocal laser scanning microscopy. The area marked by a rectangle is enlarged in panel B. The arrows indicate yellow colors as a result of colocalization of SF-1 and p300. (C) p300 foci contain RNA Pol II. Expression plasmid for c-Myc-tagged p300 was transfected into Y1 cells. Fixed cells were labeled with antibodies against RNA Pol II (red) and c-Myc (green). An optical section of immunofluorescence staining is shown. (D) The HAT activity of p300 is not required to recruit SF-1 to p300-foci. Y1 cells were transfected with expression plasmid for c-Myc-tagged p300 WT (top), p300 DY (middle), or FLAG-tagged GCN5 (F-GCN5) (bottom). Fixed cells were labeled with antibodies against SF-1 (red), c-Myc (green), or FLAG (green). Optical sections of immunofluorescence staining are shown.
As described above, p300 DY that lost acetyltransferase activity could still partially activate SF-1 activity (Fig. 3). We then examined whether p300 DY retained the ability to recruit SF-1 to p300 loci, and we found that it could (Fig. 6D). In line with this observation, acetylation-deficient SF-1, mt1, was also localized to these foci in the presence of p300 (data not shown). Thus, HAT activity of p300 and acetylation of SF-1 are not required for SF-1/p300 colocalization. We also checked whether the other HAT enzyme, GCN5, could target SF-1 to discrete nuclear loci. Both Flag-GCN5 and SF-1 signals were uniformly distributed in the nucleus (Fig. 6D), suggesting that p300 but not GCN5 can direct SF-1 to discrete foci in the nucleus.
Cyclic AMP increases the colocalization of SF-1 and p300.
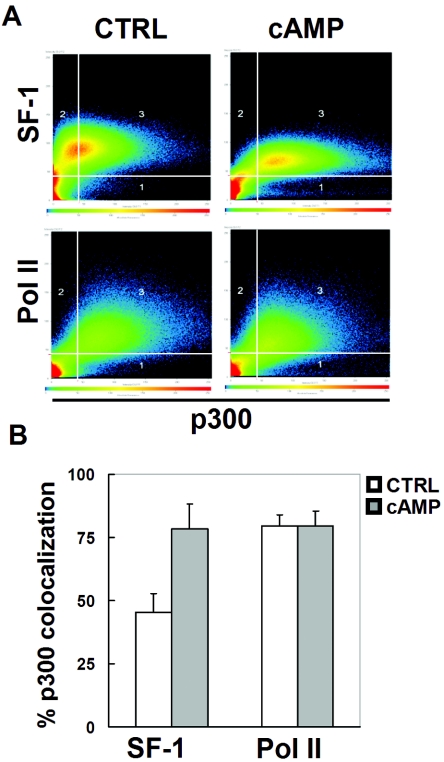
The activity of SF-1 to stimulate steroidogenic genes in Y1 cells is modulated by the cAMP signaling pathway. To determine how cAMP modulates SF-1 activity, we examined the distribution of SF-1 and its pattern of colocalization with p300 in the nucleus following cAMP stimulation (Fig. 7). In the absence of 8-Br-cAMP, many of the SF-1-positive signals were devoid of p300 signals (Fig. 7A). In the presence of 8-Br-cAMP, most of the SF-1-positive signals were also high in p300 signals. Quantification showed that the pixels of SF-1 and p300 colocalization increased from 47% to 78% after cAMP stimulation (Fig. 7B). As a control, cAMP stimulation does not affect the colocalization of p300 and Pol II. This result suggests that cAMP might target SF-1 to p300-positive foci.
FIG. 7.
cAMP increases colocalization of SF-1 and p300. (A) Y1 cells were treated without (control [CTRL]) or with 1 mM 8-Br-cAMP for 6 h. Fixed cells were labeled with antibodies against p300 and SF-1 (top) or RNA Pol II (bottom). After confocal laser scanning microscopy, the scatter diagrams for each staining were generated and are shown. The thresholds for scoring p300, SF-1, or Pol II staining as positive are shown as vertical or horizontal lines in each panel. Area 3 in each scatter plot represents pixels showing strong staining for both p300 and SF-1 (Pol II). (B) Measurement of p300 colocalization with SF-1 or Pol II. Eighteen cells were randomly selected and analyzed in four separate immunostaining experiments in the absence (CTRL) or presence of 1 mM 8-Br-cAMP treatment. The percentages of SF-1 or Pol II colocalization with p300, calculated by computer software, are presented.
Cyclic AMP increases the acetylation and DNA-binding activity of SF-1.
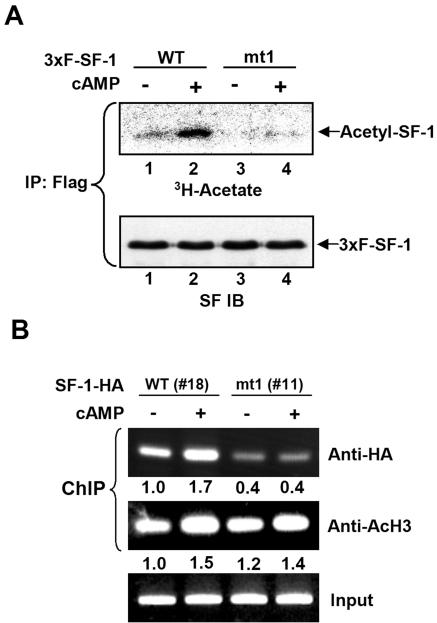
Based on the result that cAMP stimulation increases the colocalization of SF-1 and p300 in Y1 cells, we next examined whether cAMP enhances the acetylation of SF-1. As shown in Fig. 8A, a basal level of acetyl-SF-1 was detected in Y1 cells expressing FLAG-tagged wild-type 3xF-SF-1, and the acetylation signal was increased after cAMP stimulation. In Y1 cells expressing the mutated SF-1 mt1, very little basal acetylation was detected, and 8-Br-cAMP did not enhance its acetylation. This result suggests cAMP stimulates the acetylation of the KQQKK sequence of SF-1. To further investigate the function of cAMP stimulation in DNA binding in vivo, we performed a ChIP assay. Cyclic AMP also increased the DNA-binding activity of wild-type SF-1-HA in vivo (Fig. 8B) but had no effect on the binding of mutated SF-1-HA mt1 to DNA. Collectively, these results suggest that cAMP induced the increase of SF-1 acetylation and its binding to DNA and that the KQQKK sequence is the target of this cAMP-induced acetylation.
FIG. 8.
cAMP increases the acetylation and DNA-binding activity of SF-1 in vivo. (A) Mutation of KQQKK sequence abolishes cAMP-induced acetylation of SF-1. The expression plasmids for wild-type (WT) 3xFLAG-SF-1 (3×F-SF-1) or mutated mt1 were transfected into Y1 cells. After 24 h, cells were labeled with [3H]acetate in the presence (+) or absence (−) of 1 mM 8-Br-cAMP for 6 h. The results for acetyl-3xFLAG-SF-1 (top) following immunoprecipitation (IP) with anti-FLAG antibody are shown; the amounts of immunoprecipitated 3xFLAG-SF-1 were determined by SF-1 immunoblotting (IB) (bottom). (B) cAMP increases the DNA-binding activity of WT but not acetylation-deficient SF-1. Stable Y1 cells expressing SF-1-HA (clone no. 18) and SF-1-HA mt1 (clone no. 11) were treated with (+) or without (−) 1 mM 8-Br-cAMP for 6 h. ChIP assays were performed with antibodies against HA or acetyl-H3 (AcH3). The results of PCR amplification using primers that cover the region of nucleotide positions −71 to +209 of the Cyp11a1 gene are shown. Numbers below each lane are quantitations of the band intensities.
Cyclic AMP increases the interaction between SF-1 and p300.
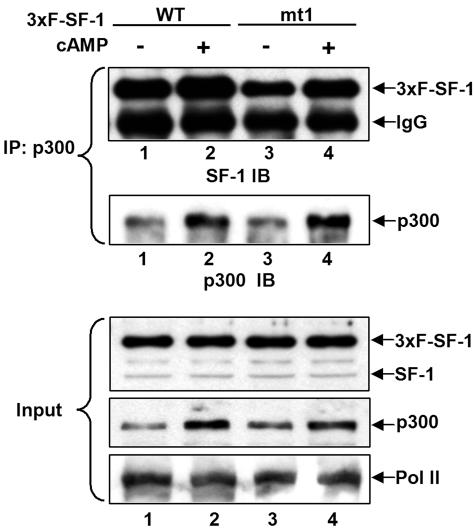
To analyze the interaction of SF-1 and p300 after cAMP stimulation, 3xF-SF-1 expressed from Y1 cells was coimmunoprecipitated with an anti-p300 antibody. As shown in Fig. 9, both wild-type and mutated 3xF-SF-1 mt1 were efficiently coprecipitated with endogenous p300 (lanes 1 and 3, top), and more SF-1 proteins were coprecipitated after cAMP stimulation (lanes 2 and 4 versus lanes 1 and 3, respectively). The amount of p300 was increased by cAMP stimulation (Fig. 9, lanes 2 and 4), while the protein level of Pol II was unchanged. Thus, cAMP increases the amount of p300 and also enhances the binding of SF-1 to p300.
FIG. 9.
cAMP increases SF-1/p300 association in Y1 cells. The expression plasmids for wild-type (WT) 3xFLAG-SF-1 (3xF-SF-1) or mutated mt1 were transfected into Y1 cells. After 24 h, cells were treated with (+) or without (−) 1 mM 8-Br-cAMP for 6 h. Results of immunoblotting (IB) of SF-1 and p300 recovered from immunoprecipitates (IP) of cell lysates using anti-p300 antibody are shown. Immunoblotting of SF-1, p300, and Pol II in 10% of input cell lysate (Input) are shown at the bottom.
DISCUSSION
In this report, we demonstrate that SF-1 is modified and regulated by p300 through acetylation and localization to specific nuclear foci. Mutation of the KQQKK acetylation sequence in the Ftz-F1 box results in decreased DNA binding as well as reduction of basal and p300-potentiated transcriptional activity of SF-1. In addition, SF-1 acetylation, its localization to p300-positive nuclear foci, and its binding to DNA can be enhanced by cAMP signaling through an increased amount of p300. These results indicate a novel mechanism for SF-1 activation in response to cAMP stimulation.
Acetylation modulates the function of the Ftz-F1 box.
We show that SF-1 is acetylated at the KQQKK motif of the Ftz-F1 box by p300. In addition, mutation of the acetylation sequence results in a reduction of the DNA-binding activity of SF-1; therefore, acetylation of the Ftz-F1 box correlates with increased DNA binding.
How acetylation affects DNA binding is still not fully understood. Our data show that this is unrelated to charge neutralization (Fig. 4C). It appears that the site of acetylation in relation to the DNA-binding domain may affect the ability of proteins to bind DNA. Acetylation of DNA-binding domains of YY1 (56) and HMGI(Y) (38) decreases DNA binding; on the contrary, acetylation near the DNA-binding domain generally increases the DNA-binding activity of transcription factors such as p53 (15, 38), E2F1 (33), GATA-1 (4), and EKLF (57). Acetylation of the Ftz-F1 box near the core DNA-binding sequence of SF-1 also increases its DNA-binding activity. One possible mechanism for the increased DNA-binding activity is that acetylation modifies SF-1 conformation to favor DNA binding, as has been hypothesized for p53 before (15). Further investigation should be conducted to uncover the underlying mechanism.
Acetylation of SF-1.
We show that SF-1 is acetylated at the KQQKK motif by p300. Aside from acetylation catalyzed by p300, a basal level of acetylation was observed for wild-type and mutated SF-1 (Fig. 1C and Fig. 2B). This implies that SF-1 has multiple acetylation sites for different HATs, such as CBP/p300 and GCN5/PCAF. Indeed, SF-1 is acetylated by GCN5 (25). GCN5, however, probably did not acetylate the FP domain of SF-1 since its homologue, PCAF, acetylates FP poorly in vitro (Fig. 1B).
In our study (Fig. 1C), we observed decreased but not absent acetylation after transfection of p300 DY, which lacks acetyltransferase activity (23). This is probably because p300 DY can still recruit SF-1 to nuclear foci (Fig. 6D), where p300 might function as a scaffold to attract other HATs, like PCAF/GCN5 (6), which can in turn acetylate SF-1. Current investigations demonstrate that SF-1 can be acetylated by both p300 (Fig. 1) and PCAF/GCN5 (25) and that acetylation by both HAT proteins correlates with transcriptional activation. This phenomenon is similar to that of p53, which can be modified by p300 (15) and PCAF (32).
Jacob et al. reported that TSA, an inhibitor of histone deacetylase, induced the export of SF-1 from the nucleus to the cytoplasm (25). We also examined the effect of TSA on SF-1 in Y1 cells but obtained different results. We found that TSA downregulated SF-1 expression in Y1 cells. This has been confirmed by promoter analysis, Western blot analysis, and immunofluorescence detection (our unpublished data). We suspect that Jacob et al. also failed to detect reasonable SF-1 expression after TSA treatment and that what they observed is probably overexposure of residual amounts of partially degraded SF-1 located in both nucleus and cytoplasm.
Acetylation of steroid receptors.
In addition to SF-1, other steroid receptors, such as AR and estrogen receptor (ER), are also substrates for acetylation (14, 54). The acetylation sequences of SF-1, AR, and ER are similar, either KXKK or KXXKK, but different HATs acetylate these sites with different efficiencies: ER is selectively acetylated by p300, while AR is efficiently acetylated by both p300 and PCAF. Our data show that KQQKK of SF-1 is preferentially acetylated by p300 rather than by PCAF in vitro. The mechanism governing substrate specificity of HATs is still unclear.
Mutation of the acetylation site of AR reduces its ligand-dependent transactivation function by lowering its ligand sensitivity (14). In contrast, mutation of the acetylation site of ER increases its ligand sensitivity (54). Unlike these two steroid receptors, mutation of the acetylation site does not affect the transactivation function of SF-1 (Fig. 4D). We show that KQQKK acetylation correlates with increased DNA-binding activity of SF-1 (Fig. 5). Thus, although these steroid receptors can be similarly acetylated at similar sites, the effects of their acetylation are vastly different. What controls this apparent functional difference remains to be further investigated.
SF-1 localization to p300 foci in the nucleus.
The cell nucleus is compartmentalized, and various nuclear foci that exert different biological actions have been described previously. Activated transcription factors have been found in specific nuclear foci (13, 19, 47), and p300/CBP are colocalized with RNA Pol II in transcriptionally active domains (46, 53). We found SF-1 in discrete nuclear sites by immunostaining with two different anti-SF-1 antibodies (Fig. 6A; also data not shown). This observation is different from the observation of even distribution of exogenous SF-1 (12; also data not shown). The reason for the diffuse distribution of exogenous SF-1 is probably due to the effect of overexpression.
SF-1 constitutively activates basal expression of steroidogenic genes, and SF-1 activity can be potentiated by p300. This is consistent with the finding that SF-1 is partially localized to p300 nuclear foci in Y1 cells, and SF-1 localization to p300 nuclear foci can be enhanced by overexpression of p300. These results suggest that p300 potentiates the activity of SF-1 by two means: acetylation and recruitment to specific nuclear foci.
To examine whether SF-1 localization to p300 nuclear foci requires acetylation, we examined the location of the acetylation-deficient SF-1, mt1. SF-1 mt1 can interact with p300 (Fig. 9) and be recruited to specific nuclear foci together with p300 (data not shown). Therefore, acetylation is not required for recruitment to p300 foci. Consistent with this observation, the HAT-defective p300 (p300 DY) also has the ability to form nuclear foci and to recruit SF-1 into them. It appears that p300 first recruits SF-1 to specific nuclear foci and it then transfers acetyl groups to SF-1, resulting in increased SF-1 binding to DNA and final gene activation. This mode of action seems different from that of GCN5, which does not recruit SF-1 to specific nuclear foci (Fig. 6).
In addition to examining acetylation-deficient SF-1, we examined other mutated SF-1, including those defective in DNA binding (G35E) (1), ligand binding (V349W) (31), intrastructure interaction (R314M) (27), and AF-2 domain (30); all of these mutants were still localized to p300 nuclear foci (data not shown). This observation suggests that these mutations may not abolish the interaction of SF-1 with p300; thus, these mutated SF-1 can be enriched in nuclear foci. These results are compatible with the recent observation of HIF-1 being recruited to nuclear foci by CBP (45). Only the transcriptionally inert form of HIF-1, whose CBP-interacting domains were completely disrupted, cannot interact with CBP and cannot be recruited to nuclear foci (45). It is believed that p300/CBP foci may also serve as storage, supply sites, or sites of transcriptional initiation complex regeneration (53). Based on these observations, we propose that SF-1 localization to p300 foci may be prior to its binding to cognate DNA and interacting with other coactivators, such as SRC-1. However, the mechanism of recruiting transcription factors to p300/CBP foci remains unclear and should be further investigated.
It is well known that activation of the cAMP-dependent PKA signaling pathway can enhance SF-1-mediated transcription of many steroidogenic genes (5, 20, 29, 41). Activation of the PKA signaling pathway reorganizes SF-1/green fluorescent protein fusion protein from a diffuse distribution pattern to specific foci (12). In the current report, we further found that colocalization of endogenous SF-1 with p300 was increased by the addition of 8-Br-cAMP, whereas RNA polymerase II is associated with p300 even in the absence of cAMP stimulation. We found that cAMP increases the amount of p300 in Y1 cells, leading to increased interaction of SF-1 and p300 and their colocalization in specific nuclear foci. Furthermore, cAMP enhances the acetylation and DNA-binding ability of SF-1 (Fig. 8). These observations provide a direct link between the cAMP signaling pathway and p300-mediated acetylation and stimulation of SF-1. Collectively, our results suggest that SF-1 localization to p300 foci and acetylation by p300 might be a novel regulatory mechanism underlying the activation of the intracellular cAMP signaling pathway.
Acknowledgments
We thank Tso-Pang Yao, Yoshihiro Nakatani, Edward Seto, Shigeaki Kato, and Dean Hum for pCMV-p300, pET-p300 HAT, pGEX5X-PCAF-HAT, pcDNA3-FLAG-GCN5, and pcDNA3.1-SF-1-HA plasmids, respectively. We also thank Ken Deen for editing the manuscript.
This work was supported by grant NSC 91-2311-B-001-085 from the National Science Council and by grant AS02IMB4PP from Academia Sinica, Republic of China.
REFERENCES
- 1.Achermann, J. C., M. Ito, P. C. Hindmarsh, and J. L. Jameson. 1999. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat. Genet. 22:125-126. [DOI] [PubMed] [Google Scholar]
- 2.Borud, B., T. Hoang, M. Bakke, A. L. Jacob, J. Lund, and G. Mellgren. 2002. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol. Endocrinol. 16:757-773. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 5.Carlone, D. L., and J. S. Richards. 1997. Functional interactions, phosphorylation, and levels of 3′,5′-cyclic adenosine monophosphate-regulatory element binding protein and steroidogenic factor-1 mediate hormone-regulated and constitutive expression of aromatase in gonadal cells. Mol. Endocrinol. 11:292-304. [DOI] [PubMed] [Google Scholar]
- 6.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W.-Y., W.-C. Lee, N.-C. Hsu, F. Huang, and B.-C. Chung. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279:38730-38735. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, W. L., S. D. Briggs, and C. D. Allis. 2000. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12:326-333. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, P. A., C. Dorn, Y. Sadovsky, and J. Milbrandt. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 18:2949-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, P. A., J. A. Polish, G. Ganpule, and Y. Sadovsky. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol. Endocrinol. 11:1626-1635. [DOI] [PubMed] [Google Scholar]
- 11.De Santa Barbara, P., N. Bonneaud, B. Boizet, M. Desclozeaux, B. Moniot, P. Sudbeck, G. Scherer, F. Poulat, and P. Berta. 1998. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell. Biol. 18:6653-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, W., T. Yanase, Y. Wu, H. Kawate, M. Saitoh, K. Oba, M. Nomura, T. Okabe, K. Goto, J. Yanagisawa, S. Kato, R. Takayanagi, and H. Nawata. 2004. Protein kinase A potentiates adrenal 4 binding protein/steroidogenic factor 1 transactivation by reintegrating the subcellular dynamic interactions of the nuclear receptor with its cofactors, general control nonderepressed-5/transformation/transcription domain-associated protein, and suppressor, dosage-sensitive sex reversal-1: a laser confocal imaging study in living KGN cells. Mol. Endocrinol. 18:127-141. [DOI] [PubMed] [Google Scholar]
- 13.Fejes-Toth, G., D. Pearce, and A. Naray-Fejes-Toth. 1998. Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc. Natl. Acad. Sci. USA 95:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, M., C. Wang, J. Wang, X. Zhang, T. Sakamaki, Y. G. Yeung, C. Chang, T. Hopp, S. A. W. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Jänne, and R. G. Pestell. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 16.Guo, I.-C., H.-M. Tsai, and B.-C. Chung. 1994. Actions of two different cAMP-responsive sequences and an enhancer of the human CYP11A1 (P450scc) gene in adrenal Y1 and placental JEG-3 cells. J. Biol. Chem. 269:6362-6369. [PubMed] [Google Scholar]
- 17.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, C.-H., M. D. T. Chang, K.-Y. Tai, Y.-T. Yang, P.-S. Wang, C.-J. Chen, Y.-H. Wang, S.-C. Lee, C.-W. Wu, and L.-J. Juan. 2004. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 23:2269-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Htun, H., J. Barsony, I. Renyi, D. L. Gould, and G. L. Hager. 1996. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc. Natl. Acad. Sci. USA 93:4845-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, M. C., E. F. Chiang, S. K. Tong, W. Lai, N. C. Hsu, L. C. Wang, and B. C. Chung. 2001. Regulation of steroidogenesis in transgenic mice and zebrafish. Mol. Cell. Endocrinol. 171:9-14. [DOI] [PubMed] [Google Scholar]
- 21.Hu, M.-C., S.-J. Chou, Y.-Y. Huang, N.-C. Hsu, H. Li, and B.-C. Chung. 1999. Tissue-specific, hormonal, and developmental regulation of SCC-LacZ expression in transgenic mice leads to adrenocortical zone characterization. Endocrinology 140:5609-5618. [DOI] [PubMed] [Google Scholar]
- 22.Hu, M. C., and B. C. Chung. 1990. Expression of human 21-hydroxylase (P450c21) in bacterial and mammalian cells: a system to characterize normal and mutant enzymes. Mol. Endocrinol. 4:893-898. [DOI] [PubMed] [Google Scholar]
- 23.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, M., R. Yu, and J. L. Jameson. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 17:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob, A. L., J. Lund, P. Martinez, and L. Hedin. 2001. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J. Biol. Chem. 276:37659-37664. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu, T., H. Mizusaki, T. Mukai, H. Ogawa, D. Baba, M. Shirakawa, S. Hatakeyama, K. I. Nakayama, H. Yamamoto, A. Kikuchi, and K. Morohashi. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18:2451-2462. [DOI] [PubMed] [Google Scholar]
- 27.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343-355. [DOI] [PubMed] [Google Scholar]
- 28.Lee, M. B., L. A. Lebedeva, M. Suzawa, S. A. Wadekar, M. Desclozeaux, and H. A. Ingraham. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 25:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L. A., Y. C. Chang, C. J. Wang, F. Y. Tsai, S. B. Jong, and B. C. Chung. 2004. Steroidogenic factor 1 differentially regulates basal and inducible steroidogenic gene expression and steroid synthesis in human adrenocortical H295R cells. J. Steroid Biochem. Mol. Biol. 91:11-20. [DOI] [PubMed] [Google Scholar]
- 30.Li, L.-A., E. F.-L. Chiang, J.-C. Chen, N.-C. Hsu, Y.-J. Chen, and B.-C. Chung. 1999. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol. Endocrinol. 13:1588-1598. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., M. Choi, G. Cavey, J. Daugherty, K. Suino, A. Kovach, N. C. Bingham, S. A. Kliewer, and H. E. Xu. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17:491-502. [DOI] [PubMed] [Google Scholar]
- 32.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuda, K., I. Ochiai, M. Nishi, and M. Kawata. 2002. Colocalization and ligand-dependent discrete distribution of the estrogen receptor (ER)alpha and ERbeta. Mol. Endocrinol. 16:2215-2230. [DOI] [PubMed] [Google Scholar]
- 35.Mizusaki, H., K. Kawabe, T. Mukai, E. Ariyoshi, M. Kasahara, H. Yoshioka, A. Swain, and K.-I. Morohashi. 2003. Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by Wnt4 in the female developing gonad. Mol. Endocrinol. 17:507-519. [DOI] [PubMed] [Google Scholar]
- 36.Monte, D., F. DeWitte, and D. W. Hum. 1998. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem. 273:4585-4591. [DOI] [PubMed] [Google Scholar]
- 37.Morohashi, K.-I., S.-I. Honda, Y. Inomata, H. Handa, and T. Omura. 1992. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol. Chem. 267:17913-17919. [PubMed] [Google Scholar]
- 38.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 39.Nuclear Receptors Nomenclature Committee. 1999. A unified nomenclature system for the nuclear receptor superfamily. Cell 97:161-163. [DOI] [PubMed] [Google Scholar]
- 40.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 41.Omura, T., and K. Morohashi. 1995. Gene regulation of steroidogenesis. J Steroid Biochem. Mol. Biol. 53:19-25. [DOI] [PubMed] [Google Scholar]
- 42.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 43.Parker, K. L., and B. P. Schimmer. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 18:361-377. [DOI] [PubMed] [Google Scholar]
- 44.Polesskaya, A., A. Duquet, I. Naguibneva, C. Weise, A. Vervisch, E. Bengal, F. Hucho, P. Robin, and A. Harel-Bellan. 2000. CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem. 275:34359-34364. [DOI] [PubMed] [Google Scholar]
- 45.Ruas, J. L., L. Poellinger, and T. Pereira. 2002. Functional analysis of hypoxia-inducible factor-1 alpha-mediated transactivation. Identification of amino acid residues critical for transcriptional activation and/or interaction with CREB-binding protein. J. Biol. Chem. 277:38723-38730. [DOI] [PubMed] [Google Scholar]
- 46.Ruas, J. L., L. Poellinger, and T. Pereira. 2005. Role of CBP in regulating HIF-1-mediated activation of transcription. J. Cell Sci. 118:301-311. [DOI] [PubMed] [Google Scholar]
- 47.Stenoien, D. L., M. G. Mancini, K. Patel, E. A. Allegretto, C. L. Smith, and M. A. Mancini. 2000. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol. Endocrinol. 14:518-534. [DOI] [PubMed] [Google Scholar]
- 48.Stenoien, D. L., A. C. Nye, M. G. Mancini, K. Patel, M. Dutertre, B. W. O'Malley, C. L. Smith, A. S. Belmont, and M. A. Mancini. 2001. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α-coactivator complexes in living cells. Mol. Cell. Biol. 21:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomita, A., M. Towatari, S. Tsuzuki, F. Hayakawa, H. Kosugi, K. Tamai, T. Miyazaki, T. Kinoshita, and H. Saito. 2000. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene 19:444-451. [DOI] [PubMed] [Google Scholar]
- 51.Tomura, A., K. Goto, H. Morinaga, M. Nomura, T. Okabe, T. Yanase, R. Takayanagi, and H. Nawata. 2001. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J. Biol. Chem. 276:28395-28401. [DOI] [PubMed] [Google Scholar]
- 52.Ueda, H., G.-C. Sun, T. Murata, and S. Hirose. 1992. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol. Cell. Biol. 12:5667-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, C., M. Fu, R. H. Angeletti, L. Siconolfi-Baez, A. T. Reutens, C. Albanese, M. P. Lisanti, B. S. Katzenellenbogen, S. Kato, T. Hopp, S. A. W. Fuqua, G. N. Lopez, P. J. Kushner, and R. G. Pestell. 2001. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276:18375-18383. [DOI] [PubMed] [Google Scholar]
- 55.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9:553-562. [DOI] [PubMed] [Google Scholar]
- 56.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, W., and J. J. Bieker. 1998. Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA 95:9855-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]