Abstract
We identified CENP-50 as a novel kinetochore component. We found that CENP-50 is a constitutive component of the centromere that colocalizes with CENP-A and CENP-H throughout the cell cycle in vertebrate cells. To determine the precise role of CENP-50, we examined its role in centromere function by generating a loss-of-function mutant in the chicken DT40 cell line. The CENP-50 knockout was not lethal; however, the growth rate of cells with this mutation was slower than that of wild-type cells. We observed that the time for CENP-50-deficient cells to complete mitosis was longer than that for wild-type cells. Centromeric localization of CENP-50 was abolished in both CENP-H- and CENP-I-deficient cells. Coimmunoprecipitation experiments revealed that CENP-50 interacted with the CENP-H/CENP-I complex in chicken DT40 cells. We also observed severe mitotic defects in CENP-50-deficient cells with apparent premature sister chromatid separation when the mitotic checkpoint was activated, indicating that CENP-50 is required for recovery from spindle damage.
The centromere plays a fundamental role in accurate chromosome segregation during mitosis and meiosis in eukaryotes. Its functions include sister chromatid adhesion and separation, microtubule attachment, chromosome movement, formation of the heterochromatin structure, and mitotic checkpoint control. Although chromosome segregation errors cause genetic diseases, including some cancers (21), the mechanism by which centromeres interact with microtubules of the spindle apparatus during cell division is not fully understood.
The kinetochore is the specialized structure formed at the centromeres that is responsible for microtubule binding and chromosome movement. Observations of mitotic chromosomes by traditional electron microscopy revealed that the kinetochore of vertebrate cells is a trilaminar, button-like structure on the surface of the centromeric heterochromatin (3, 4, 5, 34). The inner kinetochore plate has an essential role in kinetochore assembly, and the outer kinetochore plate is both a microtubule-binding structure and a mitotic checkpoint structure that includes the Bub and Mad complexes (5, 11). The inner kinetochore contains the centromeric DNA as well as centromere proteins A (CENP-A) and C (CENP-C) (11). CENP-A is a 140-amino-acid centromere-specific protein in which the C-terminal 90 amino acids are 60% identical to those of histone H3 (23, 32, 38). CENP-C, another member of inner kinetochore plate, is essential for mitotic progression and is thought to be necessary for kinetochore assembly (8, 9, 20, 36, 42). Both CENP-A and CENP-C are found only at active centromeres (41, 43). In addition, other proteins thought to be a part of the inner centromere were identified in recent years (11). We previously identified and characterized CENP-H and CENP-I as inner centromere components (10, 28). CENP-H localizes to the centromere throughout the cell cycle, presents at the inner kinetochore plate, and is found only in active centromeres, including neocentromeres (10, 40). Analysis of a conditional knockout of CENP-H in DT40 cells revealed that CENP-H is essential for cell growth and mitotic progression (10). CENP-I is also a constitutive centromere protein that localizes to the centromere throughout the cell cycle (22, 28). A conditional knockout of CENP-I in DT40 cells has been created, and the phenotype of CENP-I knockout cells is similar to that of CENP-H knockout cells. Analyses of both CENP-H and CENP-I knockout cells suggested that CENP-H and CENP-I are mutually interdependent for targeting to the prekinetochore structure and that both are necessary for CENP-C localization to centromeres (28). hMis12 was isolated as a human homolog for Mis12, a centromere component in fission yeast (13). hMis12 localizes to the centromeres during both interphase and mitosis. Furthermore, components of the hMis12 complex were also reported as constitutive centromere proteins (2, 30). Because several centromere components have been identified in recent years, it is possible that there are other unidentified constitutive kinetochore components.
MgcRacGAP is an evolutionarily conserved Rho GTPase-activating protein, and our group and others previously showed that it is involved in the completion of cytokinesis (17, 19, 25). Oceguera-Yanez et al. (31) reported that MgcRacGAP also functions during mitosis through the activation of Cdc42. Consistent with this observation, Obuse et al. (29) found MgcRacGAPin the centromere complex by proteomics analysis. In the present study, we identified a protein that interacts with MgcRacGAP by yeast two-hybrid screening. Because this protein localizes to the centromere throughout the cell cycle, we named it CENP-50 (CENtromere Protein, molecular weight 50 kDa). Recently, CENP-50 was also described as KLIP1/MLF1IP (14, 33). We created a loss-of-function mutant of CENP-50 in the chicken DT40 cell line. CENP-50-deficient cells were viable, but they showed mitotic delay. CENP-50 interacted with the CENP-H/CENP-I complex, and the centromere localization of CENP-50 was abolished in both CENP-H- and CENP-I-deficient cells. Furthermore, we observed that CENP-50-deficient cells showed severe mitotic defects even when the mitotic checkpoint was activated. We observed premature sister chromatid separation in CENP-50-deficient cells, suggesting that CENP-50 is required for maintenance of sister chromatid adhesion.
MATERIALS AND METHODS
Molecular biology, cell culture, and transfection.
All plasmids were constructed by standard methods. mRFP plasmids (1) were kind gifts from R. Y. Tsien. For the disruption constructs, a histidinol or puromycin resistance cassette under control of the β-actin promoter was inserted between the two arms. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad).
DT40 cells were cultured and transfected as described previously (8, 12). All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, penicillin, and streptomycin. Conditional loss-of-function mutants for CENP-H (10), CENP-I (28), Nuf2 (18), and Hec1 (18) were cultured as described previously. To suppress expression of the tetracycline-responsive transgenes, tetracycline (Tet) (Sigma) was added to the culture medium at a final concentration of 2 μg/ml.
Immunocytochemistry and FISH.
Immunofluorescent staining of whole cells was performed as described previously (9, 12). Cells were collected onto slides with a cytocentrifuge and fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, permeabilized in 0.5% NP-40 in PBS for 15 min at room temperature, rinsed three times in 0.5% bovine serum albumin, and incubated for 1 h at 37°C with primary antibody. Binding of primary antibody was then detected with Cy3- or fluorescein isothiocyanate-conjugated goat antirabbit immunoglobulin G (Jackson ImmunoResearch) diluted to an appropriate concentration in PBS-0.5% bovine serum albumin. Chromosomes and nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) at 0.2 μg/ml in Vectashield Antifade (Vector Labs). Fluorescent in situ hybridization (FISH) experiments with chicken painting probes were carried out as described previously (9). All immunofluorescence and FISH images were collected with a cooled charge-coupled-device camera (Cool Snap HQ; Photometrics Image Point) mounted on an Olympus IX71 inverted microscope with a ×60 objective lens (PlanApo 60X; numerical aperture [NA] for the lense = 1.40) together with a filter wheel. Images were analyzed with IPLab software (Signal Analytics).
Immunoprecipitation and Western blot analysis.
The nuclear fraction of DT40 cells was collected by centrifugation at low speed and suspended in buffer A (20 mM HEPES [pH 8.0], 150 mM KCl, 1 mM dithiothreitol). The chromatin fraction was then collected. Micrococcal nuclease was added to the samples and incubated at 4°C for 1 h. KCl (final concentration, 300 mM) and NP-40 (final concentration, 0.1%) were added to soluble samples of the chromatin fraction. M2 FLAG beads (Sigma) were used for immunoprecipitation. After immunoprecipitation, the beads were washed three times in buffer B (20 mM HEPES, KOH [pH 8.0], 300 mM KCl, 1 mM dithiothreitol, 0.1% NP-40) and collected by centrifugation. Pelleted beads were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels. Proteins were transferred to Hybond P membranes (Amersham). Blots were blocked with 5% skim milk and then incubated with anti-rabbit polyclonal anti-CENP-50 (1:2,000), anti-CENP-H (10) (1:2,000), CENP-I (28) (1:2,000), and anti-Hec1 (18) (1:2,000) antibodies. Horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:15,000; Jackson ImmunoResearch) was used to detect bound primary antibody. Blots were developed with the ECL-Plus kit (Amersham), and protein bands were visualized with a STORM imager (Molecular Dynamics).
Fluorescence microscopy in living cells.
Fluorescently stained living cells were observed with an Olympus inverted microscope (IX70) with an oil immersion objective lens (PlanApo ×60; NA for the lens = 1.40). The DeltaVision microscope system used in this study was purchased from Applied Precision, Inc. For temperature control during microscopic observations, the system was assembled in a custom-made, temperature-controlled room (15, 16).
Photobleaching.
Inverse fluorescence recovery after photobleaching (iFRAP) experiments were performed with a confocal microscope (LSM510Meta; Carl Zeiss) with a ×63 objective lens (PlanApo 63X; NA for the lens = 1.40) in an air chamber at 37°C. The entire area except a small centromeric region of an individual cell expressing the green fluorescent protein (GFP) fusion protein was photobleached with the 488-nm laser line (100% transmission, 5 scans). Cells were monitored in z-series (six stacks with 1.0-μm intervals) at 5-s intervals for 25 s before bleaching and for the first 30 s after bleaching and then monitored at 15-s intervals for 30 min (1.0% laser transmission; zoom, 5.0; scan speed, 12; 256 by 256 pixels). For quantification, z-series images were subjected to maximum projection with LSM image browser (Carl Zeiss), and the fluorescence intensities of the unbleached area and background were measured with MetaMorph (Universal Imaging Corporation) software. The net intensity was obtained by subtracting the background intensity. The average signal intensity was obtained from the analysis of at least 10 live cells. For fluorescence loss in photobleaching (FLIP) analysis, five z-series images were collected as fluorescence recovery after photobleaching before repeating the bleaching of a small area in a nucleus and the image acquisition.
RESULTS
Identification of CENP-50/KLIP1/ML1IP as a constitutive kinetochore component.
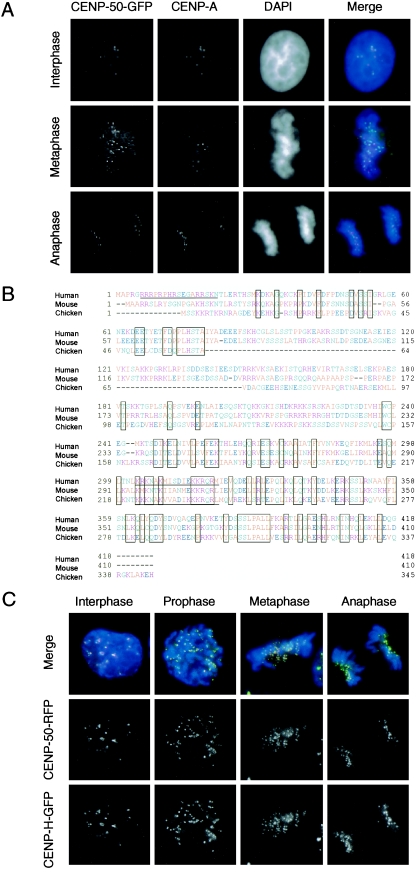
MgcRacGAP is required for the completion of cytokinesis (17, 25) and also functions during mitosis through the activation of Cdc42 (31). To understand the various functions of MgcRacGAP, we have attempted to identify proteins that interact with MgcRacGAP by yeast two-hybrid screening. Among several isolated proteins, we focused on an interesting protein with a molecular weight of 50 kDa, which we named centromere protein p50 (CENP-50) because it localizes to the centromere throughout the cell cycle (see below). The amino acid sequence of this protein was also reported as KLIP1/MLF1IP (14, 33); the authors reported that KLIP1/MLF1IP was distributed throughout the nucleus. We expressed CENP-50 as a GFP fusion protein in HeLa cells and observed discrete signals in interphase nuclei and on mitotic chromosomes, suggesting that CENP-50 is localized at the centromere throughout the cell cycle and is not distributed diffusely throughout the nucleus (Fig. 1). To confirm the centromeric localization of CENP-50, we stained cells expressing this protein as a GFP fusion protein with an anti-CENP-A antibody. We observed perfect colocalization of CENP-50 with CENP-A throughout the cell cycle. We also investigated the subcellular localization of the chicken homolog of this protein in DT40 cells. We isolated a chicken cDNA, compared the amino acid sequence of chicken CENP-50 with that of the human and mouse homologs, and found that these proteins had similar structures (Fig. 1B). The sequence information of chicken CENP-50 was deposited in the DDBJ/EMBL/NCBI database with accession number AB233421. We then tagged the C terminus of chicken CENP-50 with red fluorescent protein (RFP) and introduced the fusion construct into DT40 cells expressing CENP-H fused with GFP (CENP-H-GFP), which was used as a centromere marker because it localizes to the centromere throughout the cell cycle. Subcellular localization of chicken CENP-50-RFP and CENP-H-GFP was observed by fluorescence microscopy. CENP-50-RFP signals were perfectly colocalized with CENP-H-GFP signals as discrete signals throughout the cell cycle in chicken DT40 cells (Fig. 1C). On the basis of the localization data for human and chicken cells, we concluded that CENP-50 is a constitutive centromere component.
FIG.1.
CENP-50 localizes to centromeres throughout the cell cycle. (A) Localization of human CENP-50-GFP in different stages of the cell cycle in HeLa cells. Cells were stained with anti-CENP-A antibody. Nuclei and chromosomes were counterstained with DAPI. (B) Comparison of amino acid sequences of human, mouse, and chicken CENP-50 proteins. Amino acids conserved in all three species are boxed. Putative nuclear localization signals are underlined. (C) Localization of chicken CENP-50-RFP at progressive stages of the cell cycle in DT40 cells that express chicken CENP-H-GFP. Perfect colocalization of CENP-50 with CENP-H was observed.
CENP50 is stably associated with centromeres.
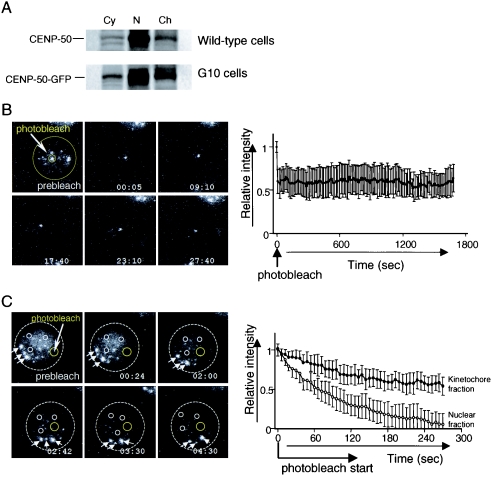
Whereas CENP-50 was localized to the centromere, it was also present in the soluble nuclear fraction (Fig. 2A). This is in contrast to CENP-H and CENP-I, most of each of which was present in the chromatin fraction (data not shown). Because CENP-50 is localized constitutively to the centromere, we hypothesized that CENP-50 moves rapidly into the kinetochore from the nuclear soluble fraction. To gain insight into the dynamics of CENP-50 in living cells, we used the G10 cell line, which was created by introduction of chicken CENP-50-GFP under control of a cytomegalovirus promoter into chicken DT40 cells with a conditional knockout of CENP-50 under the control of a Tet-responsive promoter. Creation of a conditional knockout of CENP-50 is described in the next section. In G10 cells, expression of CENP-50 is replaced with that of CENP-50-GFP after Tet addition. After addition of Tet, CENP-50-GFP was detected in both the soluble nuclear fraction and kinetochores, which was identical to the distribution observed in wild-type cells (Fig. 2A). We used iFRAP to study the kinetics of GFP-tagged CENP-50 in living cells (Fig. 2B). With iFRAP, an entire chromosome or nucleus, with the exception of the small region of interest containing the CENP-50-GFP signal, is photobleached with a pulsed laser. The loss of fluorescent signal is then monitored by time-lapse microscopy. iFRAP, as opposed to fluorescence recovery after photobleaching, is the method of choice for such experiments because it provides relatively direct evidence of a protein's time of residence in the chromosome or nucleus, and the measurement is independent of the size of these structures (6, 7, 24). We first performed iFRAP analysis to examine the stability of the CENP-50-GFP association with centromeres and observed little loss of fluorescence intensity of CENP-50-GFP during the 30-min observation period (Fig. 2B) (see Movie S1 in the supplemental material). This observation indicates that CENP-50-GFP associates stably with the centromere, like CENP-C and CENP-H (24, 37). In contrast, CENP-50-GFP in the nuclear fraction except for the centromere region moved rapidly (data not shown). To characterize the differences in the mobility of CENP-50 in the nuclear and centromere fractions, we used FLIP analysis (Fig. 2C) (see Movie S2 in the supplemental material). In FLIP experiments, imaging and bleaching of a small area of the nucleus were repeated, and the fluorescence intensity in the unbleached area (both the soluble nuclear fraction and the centromere region) was measured. If CENP-50-GFP is mobile, the fluorescence intensity in unbleached areas will decrease because the labeled molecules can diffuse into the bleached area. We observed that the fluorescence of CENP-50-GFP in the nucleoplasm was lost rapidly, whereas the fluorescence of CENP-50-GFP in the kinetochore was relatively stable (Fig. 2C). The initial loss in fluorescence of the kinetochore likely reflects the loss of a high level of background due to the mobile (nucleoplasmic) CENP-50-GFP, because the fluorescent intensity in the kinetochore also dropped to the same level (∼60%) in the iFRAP experiments (Fig. 2B). These results from photobleaching studies suggest that CENP-50 can move rapidly in the nucleus, but once it is assembled into the kinetochore complex, the association is stable, similar to that of CENP-H and CENP-I.
FIG. 2.
CENP-50 stably associates with the centromere. (A) Cytoplasmic, nuclear soluble, and chromatin fractions were prepared from wild-type and G10 cells (in which the expression of CENP-50 is replaced with that of CENP-50-GFP) and separated by SDS-PAGE. Western blotting analysis was performed with anti-CENP-50 antibody. The banding pattern of wild-type cells is similar to that of G10 cells, which were used for photobleaching experiments. (B) Images of iFRAP experiments and quantification of iFRAP kinetics. With the exception of a small region of the centromere (small circle), the entire G10 cell (big circle) expressing CENP-50-GFP was photobleached with a laser, and images were collected by fluorescence time-lapse microscopy for 30 min. Images before (prebleach) and after bleaching of a typical cell are shown. The average of the relative intensity is plotted with the standard deviation. The fluorescence intensity of unbleached regions for CENP-50-GFP was essentially unchanged. (C) FLIP analysis. A small region of nucleus (yellow circle) was photobleached repeatedly. The fluorescence intensity in the nucleoplasm (small white circle) and in the centromere region (arrows) was measured. The average relative intensity is plotted with standard deviation. The fluorescence intensity of CENP-50-GFP in the nucleoplasm (small white circles) decreased more rapidly than that in the centromere region (arrows).
Generation of loss-of-function mutants of CENP-50 in DT40 cells.
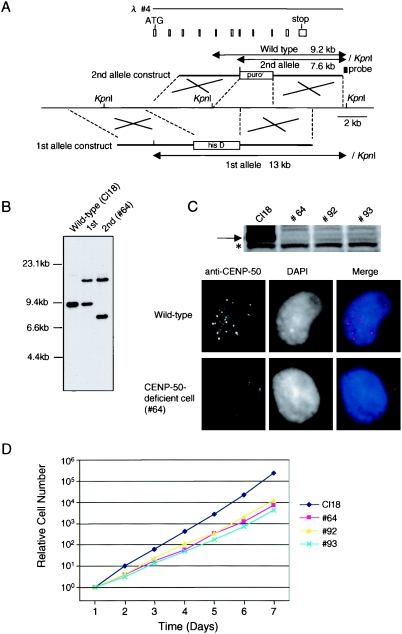
To investigate the function of CENP-50 in higher vertebrate cells, we generated loss-of-function mutants of CENP-50. CENP-50 disruption constructs were generated such that genomic fragments encoding several exons were replaced with a histidinol resistance cassette (first allele construct) or a puromycin resistance cassette (second allele construct) (Fig. 3A). We then sequentially transfected CENP-50 disruption constructs into DT40 cells (Fig. 3A) and isolated CENP-50−/− clones (Fig. 3B). We also generated conditional mutants of CENP-50 in which cells with disruptions of the CENP-50 gene were sustained by expression of the CENP-50 cDNA under the control of a Tet-repressible promoter (CENP-50−/−/CENP-50 transgene genotype). We obtained three clones with the CENP-50−/− genotype (#64, #92, and #93) and two clones with the CENP-50−/−/CENP-50 transgene genotype (#25-4 and #25-6). A 50-kDa band for CENP-50 was not detected on Western blots of lysates from clones #64, #92, and #93 with anti-CENP-50 antibody (Fig. 3C). The 50-kDa form of CENP-50 also was not expressed in clone #25-6 at 60 h after addition of Tet (data not shown); however, we did detect a 13-kDa truncated form of CENP-50. Therefore, we performed immunofluorescence analysis with knockout cells. We did not detect significant CENP-50 signals in the nuclei of these cells (Fig. 3C), and we concluded that these clones were depleted functional CENP-50. We then investigated the growth curves of CENP-50-deficient cells. The growth rates of the various CENP-50-deficient cell lines were slower than those of wild-type DT40 cells (clone Cl18). The doubling time of #93 cells was approximately 12 h, whereas that of wild-type DT40 cells (Cl18) was 8 h (Fig. 3D). Although CENP-50-deficient cells are viable, they appeared to undergo a somewhat abnormal cell cycle progression.
FIG. 3.
Generation of CENP-50-deficient cell lines. (A) Restriction maps of the chicken CENP-50 locus, gene disruption constructs, and targeted loci. White boxes indicate the positions of exons. KpnI restriction sites are shown. The position of the probe used for Southern hybridization is indicated. Novel 7.6- and 13-kb KpnI fragments hybridize to the probe if targeted integration of the construct occurs. his D, histidinol resistance cassette; puror, puromycin resistance cassette. (B) Restriction analysis of genomic DNAs with targeted integration of a CENP-50 disruption construct. Genomic DNAs from wild-type DT40 cells (Cl18), a clone after first-round targeting (+/−), and a clone after second-round targeting (−/−; clone #64) were analyzed by Southern hybridization with the probe indicated in panel A. (C) Western blot analysis of CENP-50-deficient cell extracts with anti-CENP-50 antibody. An asterisk indicates nonspecific bands. Immunofluorescence data with anti-CENP-50 antibody (red) are also shown. The CENP-50 protein was not detected by either technique. (D) Representative growth curves for wild-type cells (Cl18) and CENP-50-deficient cells (clones #64, #92, and #93). The number of cells not stained with Trypan blue was determined. Each experiment was performed twice.
Deletion of CENP-50 causes mitotic delay and leads to chromosome missegregation.
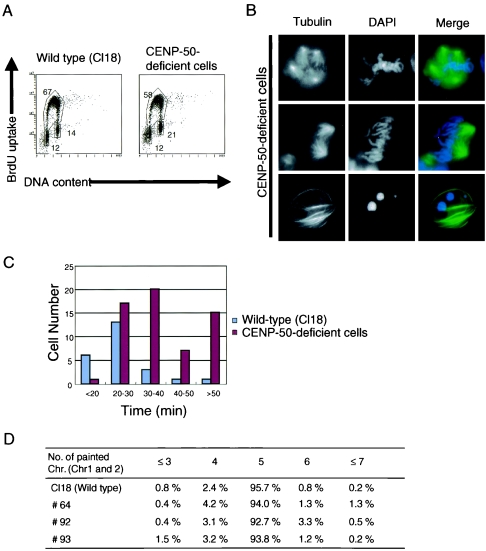
To clarify the reason for the slow growth of CENP-50-deficient cells, we performed cell cycle analysis of these cells. We measured both cellular DNA content and DNA synthesis by fluorescence-activated cell sorting after pulse labeling with BrdU in CENP-50-deficient clone #93 (Fig. 4A). Twenty-one percent of CENP-50-deficient cells accumulated in G2/M phase, whereas 14% of wild-type cells were in G2/M phase, suggesting that it takes a longer time for CENP-50-deficient cells than wild-type cells to progress through G2/M phase. We also obtained similar results for two other knockout clones, #64 and #92 (data not shown).
FIG. 4.
CENP-50-deficient cells show mitotic defects. (A) Cell-cycle distribution of wild-type cells and CENP-50-deficient cells (clone #93). Cells were stained with fluorescein isothiocyanate-anti-BrdU (y axis, log scale) to detect BrdU incorporation (DNA replication) and with propidium iodide to detect total DNA (x axis, linear scale). The lower-left box represents G1-phase cells, the upper box represents S-phase cells, and the lower-right box represents G2/M-phase cells. The numbers given in the boxes indicate the percentages of gated events. (B) Abnormal mitotic morphologies of CENP-50-deficient cells. Cells were stained with α-tubulin (green). DNA was counterstained with DAPI (blue). Multipolar spindles were visible (upper panel). Several chromosomes were not aligned at the metaphase plate (middle panel). Apoptotic cells were also present (lower panel). (C) Quantitation of time to progress from prophase to anaphase for wild-type (n = 24) and CENP-50-deficient (n = 65) cells by living cell observation. Average time to progress from prophase to anaphase was approximately 25 min for wild-type cells. A subset of CENP-50-deficient cells required more than 50 min to progress and eventually died after long mitotic delay. Movies of cell cycles typical for wild-type and CENP-50-deficient cells are presented in Movies S3 and S4 in the supplemental material. (D) Distribution of the number of painted chromosomes per cell. To examine chromosome loss, we used FISH analysis with painting probes specific for chicken chromosomes 1 and 2. Because DT40 cells have three copies of chromosome 2, five painted chromosomes were visible in normal cells. We observed an increase in the number of cells with abnormal numbers of chromosomes in CENP-50-deficient cells.
To determine the exact nature of the G2/M delay, we used DNA staining and immunocytochemical staining of microtubules in CENP-50-deficient cells (Fig. 4B). The mitotic index of CENP-50-deficient cells (8 to 10%) was two times higher than that of wild-type cells (3 to 4%). To analyze the mitotic progression of CENP-50-deficient cells in greater detail, we observed the dynamic behavior of individual living cells (Fig. 4C) (see Movies S3 and S4 in the supplemental material). To visualize microtubules in living cells, the human α-tubulin gene fused with GFP was integrated into the genome of CENP-50-deficient cells. As a control, we integrated the α-tubulin-GFP construct into the genome of wild-type DT40 cells. We then stained the chromosomes of these living cells expressing α-tubulin-GFP with Hoechst 33342 and observed the cells microscopically at 37°C. An example of the time-lapse data for CENP-50-deficient cells is shown in the supplemental material (Movies S3 and S4). We measured the times taken to complete mitosis in living wild-type and CENP-50-deficient cells. Although phototoxicity due to UV light irradiation is known to cause mitotic arrest, we used low-illumination light with a neutral-density filter to avoid such an effect, and cells did not arrest at mitosis in control experiments. Control cells (n = 24) required approximately 25 min to progress from prophase to anaphase (Fig. 4C).We analyzed 65 CENP-50-deficient cells and found that it takes longer for CENP-50-deficient cells than for wild-type cells to complete mitosis (Fig. 4C). This process took more than 50 min in a significant fraction (15 of 65) of CENP-50-deficient cells, and these CENP-50-deficient cells underwent apoptosis after prolonged mitotic arrest. These observations suggest that CENP-50 deficiency causes some mitotic defects.
During the course of cytological analysis of CENP-50-deficient cells, we observed abnormal mitotic cells with chromosomes that failed to congress normally at the metaphase plate, although we detected ∼5% of cell with abnormal mitosis (Fig. 4B, middle panel). We also observed cells with multiple spindles (Fig. 4B, upper panel). As mentioned above, live cell imaging of knockout cells revealed that approximately 1 h is required for knockout cells to complete mitosis, whereas the process takes only 25 min in control cells. During the delay of mitosis, we observed abnormal mitotic cells. However, after the delay of mitosis, the abnormalities were fixed, and the cells entered the next cell cycle (see Movie S4 in the supplemental material). We detected abnormalities during the 30- to ∼40-mindelay, which was consistent with the number (∼5%) observed with fixed images. We also observed an increased number of apoptotic cells in CENP-50-deficient cells in fixed cells and stained with antitubulin antibody (Fig. 4B, lower panel), supporting the view that abnormal mitosis leads to cell death. We next examined whether mitotic abnormalities caused by CENP-50 depletion were associated with the induction of aneuploidy. We performed FISH analysis of metaphase spreads with chromosome painting probes specific for chicken chromosomes 1 and 2. Because DT40 cells contain three copies of chromosome 2, five fluorescent signals are expected for each wild-type cell. The percentage of cells with five signals was lower for CENP-50-deficient cells than for wild-type cells (Fig. 4D), suggesting that the degree of aneuploidy increased in knockout cells and that CENP-50 deficiency leads to chromosome missegregation.
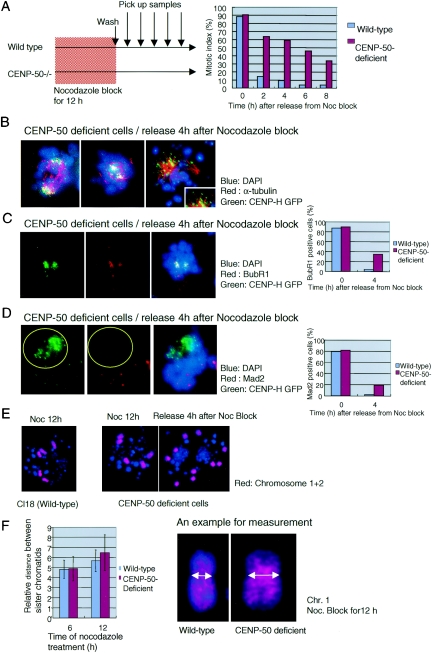
CENP-50-deficient cells do not exit mitosis after release from nocodazole block due to severe mitotic defects.
During the course of living cell analysis, we found that some CENP-50-deficient cells died after long mitotic delay. Therefore, we hypothesized that CENP-50 deficiency adversely affects mitotic exit after prolonged mitotic delay. We treated CENP-50-deficient cells with nocodazole for 12 h, then washed cells, and examined entry of the cells into the next cell cycle with a cytogenetic method (Fig. 5A). Control cells progressed to G1 phase immediately, and 8% of cells were mitotic 4 h after release from nocodazole block (Fig. 5A). In contrast, 60% of CENP-50-deficient cells showed mitotic figures at 4 h after the release, and most cells did not exit mitosis and eventually died (Fig. 5A). To understand why CENP-50-deficient cells do not exit mitosis after long mitotic delay, we analyzed individual cells cytologically (Fig. 5B to F). We found that most kinetochore signals were concentrated near the spindle pole in mitotically arrested cells, which did not exit mitosis after release of nocodazole block (Fig. 5B). These cells appeared to be in anaphase (Fig. 5B, left and center panels). However, 63% of mitotic cells showed BubR1 signals at kinetochores, indicating that these cells are arrested at (pro)metaphase (Fig. 5C), although many cells had an anaphase-like morphology (see below). We also stained arrested cells with anti-Mad2 antibody (Fig. 5D). Thirty-three percent of mitotic cells showed Mad2 signals at the kinetochore (Fig. 5D). However, even in Mad2-positive cells, we could not detect Mad2 signals in the kinetochores, which were concentrated near the spindle pole (Fig. 5D, yellow circle), whereas Mad2 signals were visible on unaligned chromosomes. We found that many cells with BubR1 signals had an anaphase-like morphology, and most of these cells were negative for Mad2. We believe that the sister chromatids of these chromosomes were separated even when BubR1 was still associated with the kinetochores. We also detected microtubule attachment to kinetochores in these mitotically arrested cells (Fig. 5B, right panel). The data suggest that many of the sister chromatids had separated prematurely. Consistent with these observations, we found several separated chromosomes at this time point in chromosome spreads of CENP-50-deficient cells by FISH analysis (Fig. 5E).
FIG.5.
CENP-50-deficient cells do not exit mitosis after release from nocodazole block. (A) Schematic representation of the experiments and the mitotic index. Wild-type and CENP-50-deficient cells were treated with nocodazole for 12 h. After nocodazole was removed from cells by washing, the percentage of mitotic cells was measured. In control cells, the mitotic index decreased, indicating rapid exit from mitosis. The mitotic index of CENP-50-deficient cells remained high after the release from nocodazole block. (B) Images of abnormal mitotic CENP-50-deficient cells, which did not exit mitosis. CENP-50-deficient cells expressing CENP-H-GFP were fixed and stained with anti-α-tubulin, and DNA was counterstained with DAPI. Many minichromosomes were separated to two spindle poles. This morphology is similar to that of anaphase cells. (C) Image of abnormal mitotic CENP-50-deficient cells stained with anti-BubR1 antibody (red) 4 h after release from nocodazole block. BubR1-positive cells comprise 38% of the total cells. Mitotic cells are 60% of the total cells, and therefore, 63% (38/60) of mitotic cells are positive for BubR1. (D) Image of abnormal mitotic CENP-50-deficient cells stained with anti-Mad2 antibody (red) 4 h after release from nocodazole block. Even in Mad2-positive cells, Mad2 signals were not detected in the kinetochores, which were concentrated near the spindle pole (yellow circle). Mad2 signals were visible on unaligned chromosomes. Mad2-positive cells constitute approximately 20% of the total cells. Because approximately 60% of the total cells were mitotic, 33% (20/60) of mitotic cells were positive for Mad2. (E) Separated sister chromatids as revealed by FISH with painting probes (chromosomes 1 and 2). (F) The distance between sister chromatids in CENP-50-deficient and wild-type cells after nocodazole treatment for 6 h and 12 h. A typical example for measurement was shown. The mean distance in CENP-50-deficient cells is significantly larger than that in wild-type cells.
We found that premature sister chromatid separation occurred during the lengthy delay in mitosis of CENP-50-deficient cells. Therefore, we measured the distance between the sister chromatids after prolonged treatment of CENP-50-deficient cells with nocodazole (Fig. 5F). We measured the distance between sister chromatids in 194 (control, 6 h), 277 (CENP-50-deficient cells, 6 h), 254 (control, 12 h), and 257 (CENP-50-deficient cells, 12 h) cells and statistically analyzed these data by using the t test. We observed a significantly larger (P value = 1.28 × 10−9) mean distance between sister chromatids of CENP-50-deficient cells than that for wild-type cells. These results suggest that sister chromatid adhesion weakened during mitotic delay in CENP-50-deficient cells. Paired chromosomes with loose adhesions separate prematurely after release from mitotic block, and this prevents the cells from entering the next cell cycle. Our findings suggest that CENP-50 is required for maintenance of sister chromatid adhesion during mitotic checkpoint activation.
Centromere localization of CENP-50 is dependent on CENP-H and CENP-I but not the Nuf2 complex.
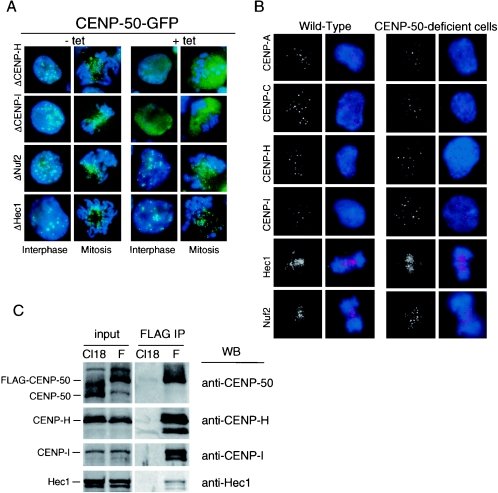
CENP-50 is a constitutive centromere protein, and we are interested in how CENP-50 is involved in centromere assembly. We previously generated mutants for several centromere proteins (9, 10, 18, 28, 35), and these mutants have been useful in analyzing the involvement of CENP-50 in centromere assembly. We made a CENP-50-GFP expression construct and introduced it into CENP-H-, CENP-I-, Nuf2-, and Hec1-conditional knockout cells. In these conditional knockout cells, expression of target proteins is inactivated when Tet is added to the culture medium. We analyzed CENP-50-GFP signals in these cells in both the presence and absence of Tet. Typical results are shown in Fig. 5A. In the absence of Tet, CENP-50-GFP gave discrete signals in interphase nuclei and at centromeres similar to the pattern in wild-type cells. In the presence of Tet, CENP-50-GFP signals were diffusely distributed in CENP-H and CENP-I conditional knockout cells (Fig. 6A). In contrast, centromeric signals for CENP-50-GFP were observed in Nuf2- and Hec1-deficient cells, and the signal intensities were similar to those in control cells (Fig. 6A). These results indicate that localization of CENP-50 to the centromere is dependent on CENP-H and CENP-I but not on Nuf2 and Hec1.
FIG. 6.
CENP-50 interacts with the CENP-H/CENP-I complex. (A) Localization of CENP-50-GFP (green) in CENP-H, CENP-I, Nuf2, or Hec1 conditional knockout cells. Conditional knockout cells were cultured in the absence (-tet) or presence (+tet) of tetracycline. Expression of each target protein was blocked completely after addition of Tet, as described previously. DNA was counterstained with DAPI (blue). (B) Immunofluorescence analysis of wild-type and CENP-50-deficient cells stained with anti-CENP-A, anti-CENP-C, anti-CENP-H, anti-CENP-I, anti-Hec1, or anti-Nuf2 antibody. Antibody signals were detected with Cy3-conjugated antibodies (red). DNA was counterstained with DAPI (blue). (C) Coimmunoprecipitation of centromere proteins with CENP-50-FLAG. A cell line (F3) in which expression of CENP-50 was replaced with that of CENP-50-FLAG was created. The chromatin fraction of wild-type (Cl18) and F3 (F) cells was immunoprecipitated with anti-FLAG antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-CENP-50, anti-CENP-H, anti-CENP-I, and anti-Hec1 antibodies.
We then investigated the localization of other centromere proteins in the CENP-50-deficient cells (Fig. 6B). Immunocytochemical analyses were performed with wild-type DT40 and CENP-50-deficient cells. We stained cells with antibodies against CENP-A, -C, -H, -I, Hec1, and Nuf2. As shown in Fig. 6B, all antibodies tested gave typical centromere signals in wild-type cells. In CENP-50-deficient cells these antibodies gave centromere signals that were of intensities similar to those observed in control cells. The data indicate that the centromeric localizations of the proteins examined were not affected by CENP-50 depletion. Although CENP-50 depletion causes mitotic defects and slow growth of cells, CENP-50-deficient cells progressed through cell division under normal conditions. This is consistent with our findings that the localization and intensities of signals for the centromere proteins examined in this experiment are not changed in CENP-50-deficient cells.
Because centromere localization of CENP-50 is dependent on CENP-H and CENP-I, we next examined whether CENP-50 forms complexes with other centromere proteins. We created the F3 cell line, which was created by introduction of chicken CENP-50-FLAG under control of a cytomegalovirus promoter into cells of the #25-6 clone, which is a conditional knockout cell line of CENP-50 under the control of a Tet-responsive promoter. In F3 cells, expression of CENP-50 is replaced with that of CENP-50-FLAG after Tet addition. After addition of Tet, we isolated the chromatin fraction from F3 cells and then performed immunoprecipitation of both fractions of F3 cells with anti-FLAG antibody. As a control, we carried out immunoprecipitation with wild-type cells (Cl18), which do not express CENP-50-FLAG. Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with antibodies against several centromere proteins (Fig. 6C). We detected CENP-H and CENP-I in anti-FLAG immunoprecipitates. These data in combination with the immunocytological data suggest that CENP-50 interacts with the CENP-H/CENP-I complex at the kinetochore.
Hec1 localization was not changed in CENP-50-deficient cells, and CENP-50 localized to centromeres in Hec1-deficient cells (Fig. 6A and B). However, Hec1 was present in immunoprecipitates with FLAG antibody. We confirmed the interaction between CENP-50 and Hec1 by yeast two-hybrid analysis (data not shown). Hec1 may interact with several proteins in the inner kinetochore, including CENP-50 and CENP-H (24). We did not detect Mad2 or Mis12 in immunoprecipitates of the chromatin fraction of F3 cells (data not shown).
DISCUSSION
CENP-50 as a constitutive centromere component.
We isolated CENP-50 as a protein that interacts with MgcRacGAP, which is an evolutionarily conserved Rho GTPase-activating protein involved in the completion of cytokinesis (17, 19, 25). Recently, Oceguera-Yanez et al. (31) reported that MgcRacGAPalso functions during mitosis through activation of Cdc42. Furthermore, Obuse et al. (29) found MgcRacGAP in the centromere complex by proteomics analysis. MgcRacGAP is distributed throughout the nucleus during interphase (17), and a portion of this protein must be targeted to the centromere. Although it is presently unclear how MgcRacGAP is involved in centromere function via its interaction with CENP-50, CENP-50 may participate in the signaling pathway regulated by MgcRacGAP.
CENP-50 was also reported as the nuclear protein KLIP1/MLF1IP (14, 33). Although CENP-50 was detected in soluble nuclear fraction in the present study (Fig. 2), we found that CENP-50 colocalizes with CENP-A and CENP-H, which are constitutive kinetochore components, throughout the cell cycle in human and chicken cells (Fig. 1). Because CENP-50 is a constitutive centromere protein and associates tightly with the CENP-H/CENP-I complex (Fig. 6), we believe that CENP-50 is a component of the inner kinetochore. Surprisingly, CENP-50-deficient cells are viable, unlike mutants of other inner kinetochore components, such as CENP-A, CENP-C, CENP-H, and CENP-I (9, 10, 28, 35). This suggests that the function of CENP-50 is distinct from those of other components of the inner kinetochore and is not essential for normal progression of the cell cycle. However, CENP-50-deficient cells show significant mitotic delay (Fig. 4), and the kinetochore structure of CENP-50-deficient cells is not normal. During mitotic delay in CENP-50-deficient cells, the time necessary for chromosome movement after chromosomes are aligned at the metaphase plate is significantly longer than that for wild-type cells (see Movies S3 and S4 in the supplemental material). It is possible that microtubule attachment to the kinetochore is weak in CENP-50-deficient cells.
Role of CENP-50 in inner kinetochore assembly.
CENP-50 associates tightly with the CENP-H/CENP-I complex, which is thought to localize in or near the inner kinetochore plate (28, 40). Our iFRAP experiments indicate that CENP-50 associates stably with the centromere, suggesting that CENP-50 is a structural component for inner kinetochore assembly. However, it is unclear how CENP-50 is involved in the inner kinetochore assembly. We previously reported that formation of CENP-A-containing nucleosomes occurs first in the kinetochore assembly, and CENP-H and CENP-I are then targeted to these structures (11, 28, 35). In the present study, we showed that centromere localization of CENP-50 is abolished in CENP-H- and CENP-I-deficient cells and that localization of CENP-H and CENP-I is not changed in CENP-50-deficient cells (Fig. 6). Our present data, together with our previously reported data, suggest that CENP-50 is targeted to the CENP-A/CENP-H/CENP-I structure. We also found, by coimmunoprecipitation experiments and yeast two-hybrid analysis, that CENP-50 interacts with Hec1, a member of the Nuf2 complex (45). However, centromere localization of CENP-50 is not altered in Nuf2- and Hec1-deficient cells, and localization of Nuf2 and Hec1 is not altered in CENP-50-deficient cells. These findings suggest that the Nuf2 complex interacts with other proteins in the inner kinetochore. Consistent with this hypothesis, we previously reported that the Nuf2 complex interacts with CENP-H (24). The Nuf2 complex should interact with CENP-H in CENP-50-deficient cells. In the present study, we did not detect proteins of the Mis12 complex (2, 30), another component of the inner kinetochore, in CENP-50 immunoprecipitates. This supports the idea that Mis12 is independent of the CENP-A loading pathway (13).
CENP-50 plays a backup role for sister chromatid adhesion during activation of the mitotic checkpoint pathway.
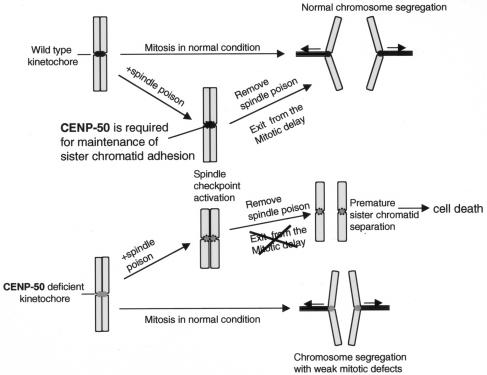
We observed that CENP-50-deficient cells did not exit mitosis after release from nocodazole block due to severe mitotic defects. We examined premature sister chromatid separation in asynchronous and synchronous cultures with other reagents in CENP-50-deficient cells. We observed premature sister chromatid separation in CENP-50-deficient cells only after the release of the block in M phase. This finding is important to understanding the role of CENP-50 (Fig. 7). During mitotic arrest, most kinetochore signals in CENP-50-deficient cells were concentrated near the spindle pole. This anaphase-like morphology indicated that the sister chromatids were prematurely separated (Fig. 5). On the basis of these observations, we believe that the main function of CENP-50 is to promote sister chromatid adhesion and thereby prevent premature sister chromatid separation during activation of the mitotic checkpoint pathway. If mitotic accidents occur in cells, the cell cycle stops due to activation of a mitotic checkpoint pathway. While the checkpoint pathway is activated, mitotic abnormalities should be fixed. Cells must have a system to prevent premature sister chromatid separation during the cell cycle delay. We propose that CENP-50 is involved in maintenance of sister chromatid adhesion during this delay. We obtained similar results in human HeLa cells by small interfering RNA analysis (data not shown). This finding suggests that the backup system involving CENP-50 is conserved in vertebrate cells.
FIG. 7.
Model of CENP-50 function in the centromere. When cells progress through the cell cycle without problems, CENP-50 is dispensable for chromosome segregation. However, when problems such as spindle abnormalities or kinetochore defects occur during mitosis, cell cycle progression is arrested due to activation of a mitotic checkpoint pathway. At this point, CENP-50 promotes sister chromatid adhesion and prevents premature sister chromatid separation. We propose that CENP-50 functions as a backup mechanism to prevent chromosome abnormalities during delay due to activation of mitotic checkpoints. If this backup system is disrupted, the cells remain in mitosis for a prolonged period and eventually die.
We also observed mitotic delay in CENP-50-deficient cells without nocodazole block, suggesting that there were partial defects in the inner kinetochore structure (Fig. 4). This finding suggested that CENP-50-deficient cells could not exit mitosis after release of nocodazole block because the microtubules failed to attach to the kinetochores. We stained arrested cells with anti-Mad2 antibody; however, we could not detect Mad2 signals in the kinetochores, which were concentrated near the spindle pole, whereas Mad2 signals were visible on unaligned chromosomes. Mad2 signals disappear from the kinetochore after microtubules attach to the kinetochore (26). We also observed microtubule attachment to the kinetochores directly (Fig. 5B, right panel). Our observations suggest that microtubules attach to kinetochores of chromosomes in mitotically arrested CENP-50-deficient cells.
We are interested in the molecular mechanism by which this backup system is activated. We observed that sister chromatid adhesion was weakened during mitotic arrest (Fig. 5F), and it is possible that the conventional cohesion complex (27) is involved in CENP-50 function. We previously created a conditional knockout cell line for Rad21/Scc1, which is a member of the cohesion complex (39). The phenotype of Rad21-deficient cells is different from that of CENP-50-deficient cells. Complete sister chromatid separation was visible in chromosome spreads of Rad21-deficient cells, whereas sister chromatid adhesion was simply weakened in CENP-50-deficient cells. We believe that Rad21 has an essential role in the establishment and maintenance of sister chromatid cohesion and that CENP-50 is required only for the maintenance of sister chromatid adhesion during mitotic checkpoint activation. Furthermore, we did not observe any change in the localization of Rad21 in CENP-50-deficient cells, and CENP-50 does not appear to interact with Rad21 as determined by an immunoprecipitation analysis (data not shown). The function of CENP-50 in maintenance of sister chromatid adhesion during mitotic arrest may be independent of the conventional cohesion system. CENP-50 may target specific proteins at the kinetochores, and then those proteins may function there to maintain sister chromatid adhesion while the mitotic checkpoint is activated. Recent proteomics approaches have revealed that CENP-50 interacts with many proteins whose functions are not known (our unpublished data). The kinetochore structure may be divided into several subcomplexes in vertebrate cells, similar to the kinetochore structure in Saccharomyces cerevisiae (44). Molecular analyses of each complex will help to clarify the components and functions of this backup system.
Supplementary Material
Acknowledgments
We are very grateful to K. Suzuki and K. Kita for technical assistance. We thank R. Y. Tsien for mRFP plasmids.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas “Cancer Cell Biology,” “Cell Cycle,” and “Nuclear Dynamics” from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman, I. M., S. Niessen, S. Anderson, F. Hyndman, J. R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo, K. H. 2001. Domain organization at centromere and neocentromere. Dev. Cell 1:165-177. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407-421. [DOI] [PubMed] [Google Scholar]
- 5.Craig, J. M., W. C. Earnshaw, and P. Vagnarelli. 1999. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp. Cell Res. 246:249-262. [DOI] [PubMed] [Google Scholar]
- 6.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 7.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meier, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukagawa, T., and W. R. A. Brown. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301-2308. [DOI] [PubMed] [Google Scholar]
- 9.Fukagawa, T., C. Pendon, J. Morris, and W. Brown. 1999. CENP-C is necessary but not sufficient to induce formation of functional centromere. EMBO J. 18:4196-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukagawa, T., Y. Mikami, A. Nishihashi, V. Regnier, T. Haraguchi, Y. Hiraoka, N. Sugata, K. Todokoro, W. Brown, and T. Ikemura. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukagawa, T. 2004. Assembly of kinetochore in vertebrate cells. Exp. Cell Res. 296:21-27. [DOI] [PubMed] [Google Scholar]
- 12.Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784-791. [DOI] [PubMed] [Google Scholar]
- 13.Goshima, G., T. Kiyomitsu, K. Koda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanissian, S. H., U. Akbar, B. Teng, Z. Janjetovic, A. Hoffmann, J. K. Hitzler, N. Iscove, K. Hamre, X. Du, Y. Tong, S. Mukatira, J. H. Robertson, and S. W. Morris. 2004. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene 23:3700-3707. [DOI] [PubMed] [Google Scholar]
- 15.Haraguchi, T., T. Kaneda, and Y. Hiraoka. 1997. Dynamics of chromosomes and microtubules visualized by multiple-wavelength fluorescence imaging in living mammalian cells: effects of mitotic inhibitors on cell cycle progression. Genes Cells 2:369-380. [DOI] [PubMed] [Google Scholar]
- 16.Haraguchi, T., D.-Q. Ding, A. Yamamoto, T. Kaneda, T. Koujin, and Y. Hiraoka. 1999. Multiple-color fluorescence imaging of chromosomes and microtubules in living cells. Cell Struct. Funct. 24:291-298. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, K., T. Kawashima, I. Iwamoto, T. Nosaka, and T. Kitamura. 2001. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276:5821-5828. [DOI] [PubMed] [Google Scholar]
- 18.Hori, T., T. Haraguchi, Y. Hiraoka, H. Kimura, and T. Fukagawa. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347-3362. [DOI] [PubMed] [Google Scholar]
- 19.Jantsch-Plunger, V., P. Gonczy, A. Romano, H. Schnabel, D. Hamill, R. Schnabel, A. A. Hyman, and M. Glotzer. 2000. CYK-4: a Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalitsis, P., K. J. Fowler, E. Earle, J. Hill, and K. H. A. Choo. 1998. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. USA 95:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 22.Liu, S. T., J. C. Hittle, S. A. Jablonski, M. S. Campbell, K. Yoda, and T. J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341-345. [DOI] [PubMed] [Google Scholar]
- 23.Mellone, B. G., and R. C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Cell Biol. 13:191-198. [DOI] [PubMed] [Google Scholar]
- 24.Mikami, Y., T. Hori, H. Kimura, and T. Fukagawa. 2005. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25:1958-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minoshima, Y., T. Kawashima, K. Hirose, Y. Tonozuka, A. Kawajiri, Y. C. Bao, X. Deng, M. Tatsuka, S. Narumiya, W. S. May, Jr., T. Nosaka, K. Semba, T. Inoue, T. Satoh, M. Inagaki, and T. Kitamura. 2003. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 4:549-560. [DOI] [PubMed] [Google Scholar]
- 26.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 28.Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W. C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2:463-476. [DOI] [PubMed] [Google Scholar]
- 29.Obuse, C., H. Yang, N. Nozaki, S. Goto, T. Okazaki, and K. Yoda. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9:105-120. [DOI] [PubMed] [Google Scholar]
- 30.Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135-1141. [DOI] [PubMed] [Google Scholar]
- 31.Oceguera-Yanez, F., K. Kimura, S. Yasuda, C. Higashida, T. Kitamura, Y. Hiraoka, T. Haraguchi, and S. Narumiya. 2005. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J. Cell Biol. 168:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, D. K., and R. L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, H.-Y., T.-J. Zhang, X.-P. Wang, J.-H. Deng, F.-C. Zhou, and S.-J. Gao. 2003. Identification of a novel cellular transcriptional repressor interacting with the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:9758-9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pluta, A. F., A. M. Mackay, A. M. Ainsztein, I. G. Goldberg, and W. C. Earnshaw. 1995. The centromere: hub of chromosomal activities. Science 270:1591-1594. [DOI] [PubMed] [Google Scholar]
- 35.Regnier, V., P. Vagnarelli, T. Fukagawa, T. Zerjal, E. Burns, D. Trouche, W. Earnshaw, and W. Brown. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitoh, H., J. Tomkiel, C. A. Cooke, H. Ratrie, M. Maure, N. F. Rothfield, and W. C. Earnshaw. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70:115-125. [DOI] [PubMed] [Google Scholar]
- 37.Shah, J. V., E. Botvinick, Z. Bonday, F. Furnari, M. Berns, and D. W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942-952. [DOI] [PubMed] [Google Scholar]
- 38.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromere chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonoda, E., T. Matsusaka, C. Morrison, P. Vagnarelli, O. Hoshi, T. Ushiki, K. Nojima, T. Fukagawa, I. C. Waizenegger, J. M. Peters, W. C. Earnshaw, and S. Takeda. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1:759-770. [DOI] [PubMed] [Google Scholar]
- 40.Sugata, N., S. Li, W. C. Earnshaw, T. J. Yen, K. Yoda, H. Masumoto, E. Munekata, P. E. Warburton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 9:2919-2926. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan, B. A., and S. Schwartz. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4:2189-2197. [DOI] [PubMed] [Google Scholar]
- 42.Tomkiel, J., C. A. Cooke, H. Saitoh, R. L. Bernat, and W. C. Earnshaw. 1994. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 125:531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a novel nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 44.Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates III, D. G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.