Abstract
The Yersinia pestis proteome was studied as a function of temperature and calcium by two-dimensional differential gel electrophoresis. Over 4,100 individual protein spots were detected, of which hundreds were differentially expressed. A total of 43 differentially expressed protein spots, representing 24 unique proteins, were identified by mass spectrometry. Differences in expression were observed for several virulence-associated factors, including catalase-peroxidase (KatY), murine toxin (Ymt), plasminogen activator (Pla), and F1 capsule antigen (Caf1), as well as several putative virulence factors and membrane-bound and metabolic proteins. Differentially expressed proteins not previously reported to contribute to virulence are candidates for more detailed mechanistic studies, representing potential new virulence determinants.
Yersinia pestis, the etiological agent of plague, is a gram-negative bacterium that is both a natural environmental pathogen and a biothreat agent (4, 8, 32). Early studies of Yersinia physiology uncovered the low calcium response (LCR), whereby bacterial cultures grown in rich medium at an elevated temperature (37°C) exhibit a growth defect upon chelation of calcium ions. The growth arrest was shown to be a result of one of the two type III secretion systems (TTSSs) in Y. pestis, the Ysc TTSS, and is responsible for the secretion of virulence factors known as Yersinia outer proteins, or Yops (21, 29; for a review, see reference 61). This TTSS can be activated in vitro and virulence factors can be released into the medium when Y. pestis is grown at 37°C with submillimolar calcium (for a review, see reference 16). Upon interaction with the host, the TTSS enables virulence factors to enter the host cell through a specialized apparatus, the injectisome (15). Once inside the host cell, Yops affect a variety of host pathways, with detectable expression changes in the pathogen as well as the host (14, 52, 82).
The Y. pestis proteome was previously examined using two-dimensional electrophoresis (57, 60, 71, 72). These studies demonstrated that virulence factors were not induced at 26°C or 37°C in the presence of calcium concentrations similar to that found in mammalian plasma (2.5 mM) (71). More recently, the introduction of two-dimensional differential gel electrophoresis (2-D DIGE) has significantly improved the quality of gel-based proteomics through fluorescence-based multiplex analyses providing relative quantitation of expression differences and improved gel-to-gel comparisons (75). Several examples of 2-D DIGE bacterial proteomics have been reported (23), including characterizations of the gram-negative bacterium Escherichia coli (1, 76, 81). Here we report the characterization of the soluble cell-associated proteome of Y. pestis as a function of temperature and calcium, which were used to effect virulence induction. Differentially expressed proteins include virulence-associated factors, membrane-bound proteins, metabolic and housekeeping proteins, and potential new virulence determinants.
Bacterial growth, cell lysis, protein extraction, 2-D DIGE, DeCyder analysis, and mass spectrometry.
Yersinia pestis (KIM5 D27) bacterial cells were grown similarly to a method described previously (52). After two passes on tryptose blood agar, cells were inoculated in 15 ml of best-case-scenario (BCS) medium in a 125-ml flask, except that potassium gluconate was added to 40 mM to achieve full-scale growth in calcium-deficient BCS medium (22). Cells were grown at 26°C with shaking at 200 rpm for two passages of 24 h each until an optical density at 620 nm between 2.3 and 2.5 was reached. Next, 52 ml of this culture was used to inoculate 2.175 liters of BCS medium, which was divided into eight 125-ml cultures grown in 1-liter flasks at 26°C. After 8 h of growth, four of the flasks were shifted to 37°C, and two of the flasks were supplemented with a 0.4 M CaCl2 solution to reach a final concentration of 4 mM. An equal volume of water was added to the other two cultures. Similar additions of CaCl2 or water were repeated for the four flasks growing at 26°C. Cells were grown for an additional 4 h after altering the temperature and calcium concentration, providing four different growth conditions for proteomic analysis. The 125-ml cultures were harvested after 4 h by centrifugation and resuspended in 50 mM ammonium bicarbonate, pH 7.8. Cells were washed two times in the same buffer and pelleted by centrifugation at 4,000 rpm for 10 min. Cell lysis was achieved by bead beating using three 180-s cycles at 4,500 rpm in a Mini-BeadBeater 1 (BioSpec Products, Inc., Bartlesville, OK), with a 5-min incubation on ice between cycles. Lysates were immediately placed on ice, and protease inhibitor (Roche) was added. Protein samples were quantified using the ADV01 reagent (Cytoskeleton) and purified using a 2-D protein cleanup kit (GE Healthcare). Samples for each of the four growth conditions were divided into aliquots (50 μg) and labeled with the fluorescent amine-reactive cyanine dye Cy3 or Cy5 (200 pmol) (GE Healthcare). For an internal pooled standard, equal amounts of all four samples were pooled, and 50-μg aliquots were labeled with 200 pmol of Cy2. This pooled standard was used in each gel to normalize protein abundance measurements across each gel, facilitating intergel spot matching and protein quantitation. After protein labeling for 30 min at 4°C, the reaction was quenched with 10 nmol lysine. In addition to analytical gels containing the fluorescently labeled samples, a “pick” gel containing an additional unlabeled protein sample (50 μg) for each growth condition was prepared to ensure that enough protein was present for subsequent identification by mass spectrometry. Multiplexed samples were separated by charge on 24-cm 3-10 NL Immobiline IPG DryStrips and then separated by size in 12.5% polyacrylamide gels. Following electrophoresis, gels were scanned using a Typhoon 9410 imager (GE Healthcare), and protein spots were analyzed using DeCyder software (27). CyDye images from individual gels were analyzed by differential in-gel analysis, and protein spots were detected using a codetection algorithm to create a single overlay spot map. Differential protein expression was determined between two samples in each gel based on statistically significant differences between the standardized log of abundance of the Cy3-labeled spot and that of the Cy5-labeled spot. The standardized log of abundance is the log of the Cy3- or Cy5-labeled spot volume divided by the log of the Cy2-labeled pooled standard spot volume, creating Cy3/Cy2 and Cy5/Cy2 volume ratios. CyDye images from all the gels within the experiment were then analyzed by biological variation analysis (BVA) (27). Statistical tests (analysis of variance and the t test) were used to determine differential protein expression. The differential spots reported had P values of ≤0.05. DeCyder analysis provided a pick list file containing the pixel locations of the differentially expressed protein spots. Protein spots were picked using a ProPic robotic workstation (Genomic Solutions), using custom software and hardware to integrate DeCyder analysis with the ProPic workstation (R. C. Mahnke and B. A. Chromy, in preparation). Each experiment contained three gel replicates for each of the four growth conditions, and the entire experiment was performed four times. Protein spot digestion and mass spectrometry characterization were performed by Proteomic Research Services (Ann Arbor, MI). Detailed protocols for mass spectrometry techniques are available at http://www.proteomicresearchservices.com/tech_01.html.
Yersinia pestis proteome analysis.
Following exponential growth of Y. pestis at both 26°C and 37°C, with and without 4 mM calcium, for 4 hours, cells were lysed, and soluble protein fractions were isolated and examined using the Ettan 2-D DIGE system. A representative gel image of the Y. pestis proteome is shown in Fig. 1, with arrows and numbers indicating differentially expressed proteins that were identified by mass spectrometry. 2-D DIGE gel images were analyzed using DeCyder software to determine the relative expression levels of Y. pestis proteins in samples from the same gel (differential in-gel analysis) and among all gels in the experiment (BVA). Proteomic analysis revealed hundreds of differentially expressed protein spots among the four growth conditions. A total of 439 protein spots (P ≤ 0.05) exhibiting differential protein expression for at least one of the four growth conditions were found among the 4,151 unique protein spots. Protein expression changes that were detected between bacterial growth at 26°C with added 4 mM calcium and that at 37°C with no calcium added were further characterized by mass spectrometry (Table 1).
FIG. 1.
2-D DIGE overlay image of the Y. pestis soluble proteome showing differential protein expression between two in vitro growth conditions containing different temperatures and calcium levels. Colored spots represent proteins that are more abundant at 26°C with 4 mM calcium (red), at 37°C with 0 mM calcium (blue), or in the pooled standard (green). White spots represent proteins with unaltered expression across the gel. Arrows with numbers show the locations of differentially expressed proteins identified by mass spectrometry (listed in Table 1).
TABLE 1.
Differentially expressed Y. pestis proteins related to virulence
| Spot no. | Protein | Accession no. | Mola | pIa | Methodb | Fold change
|
|||
|---|---|---|---|---|---|---|---|---|---|
| 26°C, 4 mM Ca2+ vs 37°C, 0 mM Ca2+ | 26°C vs 37°C at 4 mM Ca2+ | 4 vs 0 mM Ca2+ | 26°C vs 37°C | ||||||
| 1 | Catalase-peroxidase (α-KatY) | NP_668204 | 81.5 | 6.9 | MALDI | 2.5 | 5.1 | −1.7 | 3.0 |
| 2 | Catalase-peroxidase (α-KatY) | NP_668204 | 81.5 | 6.9 | MALDI | 3.5 | 4.5 | −1.3 | 3.3 |
| 3 | Catalase-peroxidase (α-KatY) | NP_668204 | 81.5 | 6.9 | MALDI | 4.1 | 6.1 | −1.4 | 4.0 |
| 4 | Clp ATPase (ClpB) | NP_668245 | 95.8 | 5.6 | MALDI | 2.9 | 2.6 | 1.2 | 3.1 |
| 5 | Clp ATPase (CIpB) | NP_668245 | 95.8 | 5.6 | MALDI | 2.3 | 1.7 | 1.0 | 1.6 |
| 6 | 60-Da chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | MALDI | 1.7 | 1.3 | 1.3 | 1.3 |
| 7 | Murine toxin (Ymt) | NP_857852 | 67.5 | 5.6 | LC/MS/MS | 2.3 | 1.3 | 1.5 | 1.9 |
| 8 | Catalase-peroxidase (β-KatY) | NP_668204 | 53.6 | 5.8 | LC/MS/MS | 3.3 | 8.0 | −1.1 | 7.0 |
| 9 | Catalase-peroxidase (β-KatY) | NP_668204 | 53.6 | 5.9 | MALDI | 7.4 | 3.5 | −1.3 | 3.2 |
| 10 | Aspartate aminotransferase (AspC) | NP_670061 | 43.7 | 5.1 | MALDI | 3.3 | 2.5 | 1.3 | 2.5 |
| 11a | High-temperature protein G (HtpG) | NP_668394 | 70.8 | 5.0 | MALDI | 3.1 | 2.4 | 1.2 | 2.7 |
| 11b | Phosphoglycerate kinase (Pgk) | NP_670607 | 41.2 | 5.3 | MALDI | 3.1 | 2.4 | 1.2 | 2.7 |
| 12 | Maltose binding protein periplasmic protein precursor (MalE) | NP_667372 | 43.8 | 6.2 | De novo sequencing | −1.3 | −1.4 | 1.3 | −1.6 |
| 13 | Fructose-bisphosphate aldolase class II (Fba) | NP_670606 | 39.3 | 5.7 | LC/MS/MS | −1.5 | −1.3 | 1.2 | −1.6 |
| 14 | 6-Phosphogluconate dehydrogenase (Gnd) | NP_669932 | 53.4 | 5.3 | De novo sequencing | 3.5 | 1.9 | 1.4 | 2.3 |
| 15 | α-Enolase (Eno) | NP_668150 | 45.5 | 5.2 | LC/MS/MS | 4.9 | 4.4 | 1.6 | 4.2 |
| 16 | d-3-Phosphoglycerate (SerA) | NP_670600 | 44.5 | 5.8 | LC/MS/MS | 5.9 | 4.5 | 1.1 | 4.7 |
| 17 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | 4.1 | 3.7 | −1.1 | 3.7 |
| 18 | Plasminogen activator (Pla) | NP_857784 | 34.6 | 6.1 | MALDI | 2.8 | 2.4 | 1.1 | 2.4 |
| 19 | F1 capsule antigen (Caf1) | NP_857692 | 17.7 | 4.8 | De novo sequencing | 2.4 | 2.2 | 1.0 | 2.1 |
| 20 | Succinyl-CoA synthetase, alpha chain (SucD) | NP_992413 | 30.0 | 6.1 | De novo sequencing | 4.2 | 5.1 | −1.0 | 4.0 |
| 21 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | 4.2 | 4.3 | 1.0 | 6.6 |
| 22 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | 7.2 | 3.8 | 1.2 | 3.1 |
| 23 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | MALDI | 8.2 | 5.3 | 1.2 | 4.6 |
| 24 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | 5.3 | 3.8 | 1.1 | 6.6 |
| 25 | Outer membrane porin A (OmpA) | NP_992692 | 38.0 | 6.8 | LC/MS/MS | 12.3 | 8.0 | 1.3 | 6.2 |
| 26 | Catalase-peroxidase (γ-KatY) | NP_668204 | 36.0 | 5.6 | MALDI | 8.9 | 6.0 | 1.3 | 6.0 |
| 27 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | MALDI | 3.2 | 2.0 | 1.8 | 1.5 |
| 28 | Probable N-acetylmuramoyl-l-alanine amidase (AmpD3) | NP_993160 | 28.7 | 5.5 | MALDI | 2.7 | 2.3 | 1.2 | 1.8 |
| 29 | Murine toxin (Ymt) | NP_857852 | 67.7 | 5.6 | LC/MS/MS | 3.3 | 1.8 | 1.7 | 1.4 |
| 30 | 50S ribosomal protein L3 (RplC) | NP_991606 | 22.3 | 9.9 | LC/MS/MS | 8.3 | 4.8 | 1.7 | 4.6 |
| 31 | 50S ribosomal protein L3 (RplC) | NP_991606 | 22.3 | 9.9 | LC/MS/MS | 10.6 | 4.2 | 2.0 | 7.5 |
| 32 | 50S ribosomal protein L3 (RplC) | NP_991606 | 22.3 | 9.9 | LC/MS/MS | 2.5 | 1.7 | 1.0 | 1.6 |
| 33 | Elongation factor Ts (EF1B) | NP_670435 | 30.8 | 5.2 | LC/MS/MS | 3.3 | 2.6 | 1.0 | 2.5 |
| 34a | 50S ribosomal protein L3 (RplC) | NP_991606 | 22.3 | 9.9 | LC/MS/MS | 1.8 | 1.4 | 1.0 | 1.8 |
| 34b | Elongation factor Ts (EF1B) | NP_670435 | 30.8 | 5.2 | LC/MS/MS | 1.8 | 1.4 | 1.0 | 1.8 |
| 35 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | −1.2 | −1.1 | −1.1 | −1.1 |
| 36 | 30S ribosomal protein SI (RpsA) | NP_670081 | 61.4 | 5.0 | LC/MS/MS | 2.0 | 1.6 | 1.1 | 1.9 |
| 37 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | LC/MS/MS | 1.5 | 1.1 | 1.2 | 1.5 |
| 38 | Catalase-peroxidase (δ-KatY) | NP_668204 | 34.0 | 5.6 | MALDI | 5.5 | 3.5 | 1.4 | 4.6 |
| 39 | Trp repressor binding protein (WrbA) | NP_669755 | 20.8 | 8.0 | LC/MS/MS | 1.4 | 1.4 | −1.3 | 1.7 |
| 40 | Glyceraldehyde 3-phosphate dehydrogenase A (GapA) | NP_669476 | 36.0 | 6.2 | LC/MS/MS | 2.0 | 1.5 | 1.2 | 1.8 |
| 41 | d-Ribose-5-phosphate 3-epimerase (Rpe) | NP_671231 | 25.0 | 5.4 | LC/MS/MS | 1.2 | 1.3 | 1.1 | 1.1 |
| 42 | 50S ribosomal protein L6 (RplF) | NP_991621 | 19.0 | 9.7 | LC/MS/MS | 4.1 | 3.1 | 1.2 | 4.7 |
| 43 | 60-kDa chaperonin (Cpn60) | NP_667946 | 57.4 | 4.9 | De novo sequencing | 3.5 | 1.5 | 1.6 | 2.5 |
pI values and molecular weights in italics represent reported values for the full-length protein.
MALDI, matrix-assisted laser desorption ionization; LC/MS/MS, liquid chromatography-tandem mass spectrometry.
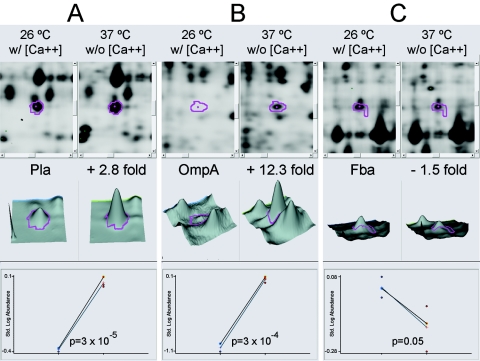
To illustrate the differential expression of representative proteins, Fig. 2 shows DeCyder BVA images. Protein expression changes for two proteins that were upregulated (Pla [Fig. 2A] and OmpA [Fig. 2B]) and one that was downregulated (fructose-bisphosphate aldolase class II [Fig. 2C]) at 37°C without calcium compared to growth at 26°C with calcium are shown. The majority of differentially expressed proteins detected in this study were upregulated following an increase in temperature and a decrease in calcium, consistent with an earlier report that overall protein production is greatly increased during Y. pestis invasion of host immune cells, which correlates with the experimental increases in temperature and decreases in calcium concentration used here (61, 71).
FIG. 2.
DeCyder BVA output images showing differential expression of Pla (A), OmpA (B), and fructose-bisphosphate aldolase class II (C). Each panel shows three types of data output from the DeCyder software. The top sections show magnified regions of the gel image containing the protein spots. The left side shows the results at 26°C with added calcium, and the right side shows the results at 37°C without added calcium. Borders for the selected spots are shown in magenta. The middle sections show three-dimensional fluorescence intensity profiles for the individual spot volumes. The bottom sections show graphs of the normalized spot volumes from replicate gels. The circles displayed for each condition on the graphs represent single normalized spot volumes from one gel, and lines were plotted corresponding to the averaged values for the particular growth condition from multiple experiments. Data are representative of the proteins identified in Table 1.
Differential protein expression following change in calcium concentration.
To address protein expression changes due to the LCR, we examined the soluble Y. pestis proteome with and without supplementation of 4 mM calcium. BVA revealed that 94 protein spots were differentially expressed for growth at 0 mM and 4 mM calcium at both temperatures. This result suggests a larger role for calcium regulation than has previously been reported. Earlier gene expression studies using similar growth parameters showed that no chromosomally carried pPCP1 or pMT1 genes were affected by calcium; only genes found on the pCD1 plasmid that mediate the TTSS were modulated by calcium (52). The present proteomic study did not focus on calcium-regulated proteins; however, several proteins that were identified as being differentially expressed between conditions of 26°C with calcium and 37°C without calcium were also differentially expressed due to calcium only, as noted in Table 1, including an α-KatY isoform and several chaperone and translation-related proteins. A closer evaluation of these proteins in future studies may yield new mechanistic information regarding the LCR and the TTSS of Y. pestis.
Differential protein expression following change in temperature.
A comparison of the Y. pestis proteome following growth at 26°C and 37°C revealed 267 differentially expressed protein spots by BVA. When differential expression due to temperature was separated from the effect of calcium, 139 proteins were differentially expressed at 0 mM calcium and 68 proteins were differentially expressed at 4 mM calcium. This supports the classical observation that the increase in protein expression due to the LCR is also dependent on the temperature change, as would occur during interaction with a mammalian host.
Differential protein expression following change from 26°C with calcium to 37°C with and without added calcium.
The differential protein expression due to the temperature change from 26°C to 37°C with calcium resulted in a relatively small number of differential proteins (68 proteins). However, 375 differential protein spots were detected between growth at 26°C with calcium and that at 37°C without added calcium. Table 1 lists Y. pestis proteins with differential expression in terms of the fold change in level for the four BVA comparisons in this study. The accession number, putative pI, and molecular weight of each full-length protein are shown. The molecular weights and pI values for protein spots with altered migration from that of the full-length proteins are shown in italics. The method of mass spectrometry used for protein identification is also listed. The differentially expressed Y. pestis proteins identified include proteins known to be associated with virulence, including Pla, the F1 antigen, Ymt, and KatY. These proteins were determined to be upregulated at 37°C, with and without calcium, compared to their expression at 26°C with calcium. Also, metabolic, structural, and enzymatic proteins were differentially expressed, including some potentially involved in virulence, such as OmpA, AmpD3, and α-enolase. Other differentially expressed proteins are associated with the Y. pestis inner and outer membranes, including Pla, the maltose binding protein periplasmic protein precursor, Ymt, KatY, and OmpA (24, 37, 47). The detection of multiple membrane-associated proteins suggests that membrane proteins can be studied by the current methodology, without the need for more complex fractionation and sample processing.
Plasminogen activator (Pla).
Pla, a multifunctional outer membrane protease, is believed to contribute to the highly invasive nature of Y. pestis (45, 68). Notably, Pla shares homology with other proteolytic plasminogen activators, and in addition to plasminogen, the Pla protease cleaves complement component C3 (63, 70), suggesting a role for Pla in the host bloodstream. However, previous experiments with pla deletion mutants have shown that the virulence role of Pla may be strain dependent and related to the route of infection (79). Strains possessing a pla mutant phenotype failed to cause disease in mice infected by the subcutaneous route but were fully virulent by the intravenous (9) and intraperitoneal (79) routes of infection.
The fibrinolytic activity of Pla is believed to contribute to virulence by facilitating bacterial metastasis through enhanced bacterial attachment to the mammalian extracellular matrix (38, 70). Pla fibrinolytic activity is temperature dependent, increasing from 26°C to 37°C (45), and Pla synthesis continues at 37°C during growth restriction in calcium-deficient medium (46). In this study, Pla was upregulated at 37°C with calcium (2.4-fold) and at 37°C without calcium (2.8-fold) compared to the case at 26°C with calcium (Table 1). Previous studies have shown that neither temperature-dependent transcription nor translation has been observed for the pla gene, suggesting that the temperature dependence of the fibrinolytic activity may instead be due to temperature-induced modifications or conformational changes in the Pla protein (11, 45). However, earlier gene expression studies using similar growth conditions in BCS medium showed approximately twofold increases in pla gene expression at 37°C, with and without calcium, compared to that at 26°C without calcium (52), correlating well with our proteomic results. Since Pla is able to bind several noncollagenous matrix proteins and is involved in the degradation of these proteins, a possible virulence role for Pla is aiding Y. pestis entry into host cells, resulting in altered signal transduction and cytoskeletal rearrangement (6).
F1 antigen (Caf1).
Caf1 is a major constituent of the F1 capsule (12, 13) that, in concert with secreted Yops, functions to prevent the uptake of Y. pestis by macrophages. Caf1 was previously reported to be a virulence factor due to its role in inhibiting the invasiveness of HeLa cells (17), and the expression of Caf1 prevented the association of Y. pestis with phagocytes, presumably by preventing adhesion receptor interactions (19). The protein spot identified as Caf1 in this study (Fig. 1, spot 19) agrees with the theoretical molecular weight and charge of the Caf1 dimer. Although native Caf1 is a homo-oligomer of megadalton size, the mature Caf1 protein monomer contains 149 amino acids, after cleavage of a 21-amino-acid leader sequence, and has a molecular mass of 15.5 kDa with a calculated pI of 4.3.
In this study, Caf1 expression was upregulated 2.2-fold at 37°C with calcium compared to that at 26°C with calcium, consistent with the formation of the F1 capsule under host physiological conditions (77). The 2.4-fold increase in Caf1 protein expression at 37°C without calcium compared to that at 26°C with calcium is also consistent with existing studies. For example, Caf1 was previously shown to be stable over several hours following pulse-chase experiments at 37°C in calcium-deficient medium (46). Furthermore, the temperature regulation of Caf1 was demonstrated at the transcriptional level, using a luciferase construct where caf-1 was overexpressed 20- to 40-fold at 37°C compared to its expression at 26°C (18). Increased gene expression is consistent with the protein expression changes found at 37°C (66). In this study, a decrease in calcium concentration had no effect on Caf1 protein expression. Our studies showed that a temperature increase resulted in a 2.1-fold increase in expression at 37°C compared to that at 26°C, consistent with reports of temperature-dependent Caf1 expression (46). The earlier gene expression results showed a temperature-dependent increase in caf-1 of 84-fold (52). Although the trends in expression in these studies are similar, the dramatic difference in fold changes between the genomic and proteomic studies may be due to a large increase in gene expression resulting in smaller increases in each of the F1 protein isoforms, of which only one was identified in this study.
Murine toxin (Ymt).
Murine toxin (Ymt) is a phospholipase D superfamily member, containing two phospholipase D motifs (62), which hydrolyzes host phospholipids upon invasion. Although Ymt has not been shown to be required for virulence, Ymt is a transmission factor necessary for host colonization of the flea midgut (30, 31). In this study, two protein spots identified as Ymt were upregulated at 37°C, with and without added calcium, compared to the case at 26°C with calcium. The full-length, soluble form of Ymt, with a molecular mass of 67 kDa and a theoretical pI of 5.6, identified as spot 7 in Fig. 1, showed a 1.8-fold increase with calcium and a 2.3-fold increase without calcium at 37°C (Table 1). Ymt is known to have three start methionines, at amino acid 1, producing a soluble species, and at amino acids 42 and 56, producing insoluble species. The Ymt species found at spot 7 appears to start at the first methionine and is not a C-terminal truncation, based on the peptides identified by mass spectrometry, the protein solubility, the molecular weight, and the pI.
Previously, full-length ymt gene expression was shown to be greater at 26°C than at 37°C by use of a lux promoter (18) and in microarray studies (52). However, there are conflicting data on the temperature regulation of the Ymt protein (18). The results from the present study showing a 1.9-fold increase in expression due solely to a temperature increase further suggest a temperature-dependent regulation of Ymt at the protein level. In addition to the three Ymt monomer species, smaller breakdown products of Ymt exist. At least four lower-molecular-mass isoforms of Ymt have been reported, including 12-kDa and 24-kDa isoforms (18, 51) as well as two truncated forms that are stable during LCR (46). The peptides identified in the second Ymt protein spot (Fig. 1, spot 29) are consistent with a Ymt C-terminal fragment containing only one phospholipase D motif. Although these smaller forms of Ymt have not been specifically tied to function, the importance of discussing each of the Ymt isoforms is evident when comparing the proteome data to gene expression data for similar growth conditions. In a microarray study, the ymt transcript level was 1.7-fold higher at 26°C than at 37°C (52). While an increase in temperature downregulated ymt, here the protein expression level was found to be increased for two isoforms. Stability of the RNA transcript or a posttranslational proteolysis event may account for the discrepancy between the gene and expression differences. Since there is no consensus on the functional role or stability of the smaller Ymt species, the presence of these isoforms in the present proteomic study suggests a need to further address the role of Ymt proteolysis in virulence.
Catalase-peroxidase (KatY).
KatY, one of two catalases in Y. pestis, binds LcrF, a transcription activator responsible for the upregulation of Yops, and is one of the major stable proteins synthesized during the LCR (46). In this study, KatY was identified in seven distinct protein spots, consistent with reports that KatY proteolysis is necessary for its activity (24). Previously, KatY was detected in both the cytoplasmic and the periplasmic fractions of Y. pestis (24, 58). The presence of multiple KatY proteins may be due to posttranslational modifications and/or a requirement for a cellular location.
The average upregulation of the seven KatY spots at 37°C with calcium compared to that at 26°C with calcium was 5.2-fold, while the average upregulation at 37°C without calcium compared to that at 26°C with calcium was 5.0-fold. A previous gene expression analysis demonstrated a 17-fold temperature induction for katY (52). In this study, three of the KatY protein spots (Fig. 1, spots 1, 2, and 3) represent differently charged isomers or small proteolytic cleavage products of full-length α-KatY, a 78.8-kDa protein with a calculated pI of 6.43. A known false translational start site produces the β isoform, which is 488 amino acids long and has a molecular mass of 53.6 kDa and a pI of 5.87 (24). β-KatY begins at amino acid 250, starting downstream of the second peroxidase motif of the full-length protein. Spots 8 and 9 of Fig. 1 are consistent with the molecular size for β-KatY; however, these two spots appear to be charge isomers based on their gel migration, potentially due to posttranslational modifications. The γ and δ forms of KatY are Pla-mediated degradation products of the larger α and β isoforms (24). Spots 26 and 38 in Fig. 1 likely represent the γ-KatY (36 kDa) and δ-KatY (34 kDa) isoforms, respectively, based on their size, migration, and mass spectrometric peptide coverage.
KatY expression has not previously been shown to be regulated by calcium, as it is expressed in both calcium-containing and calcium-deficient medium. However, KatY has been shown to be temperature dependent, as increased KatY expression occurs almost immediately after a temperature shift to 37°C (24). The increased expression of full-length KatY is also consistent with an earlier study that showed increased KatY expression by chip-based mass spectrometry analysis of Y. pestis cultured under the same growth conditions used for this study (74). Published data on KatY proteolysis and KatY function in general are limited, and further study of KatY, including the use of katY deletion mutants, will help to elucidate the role of this protein in Y. pestis virulence.
The absence of other Yops in the present study is likely due to the particular conditions used, which can result in Yop secretion. It is known that in vitro stimulation of the TTSS results in the secretion of Yops into the culture medium (49); however, this study did not address the extracellular proteome. Alternatively, it is known that Pla proteolyzes Yops (69), which could result in degradation and therefore a lack of detection of Yops under the experimental growth conditions used. Finally, many of the TTSS components, in particular Syc proteins, have relatively small molecular masses (13.5 to 19 kDa) and may not be detected under the electrophoresis conditions used (12.5% acrylamide gels), which preferentially resolve proteins in the 20- to 120-kDa range.
It should also be noted that the choice of BCS medium may have affected the extent of Yop detection. BCS medium was chosen to counteract the known growth restriction at 37°C without calcium. The medium was experimentally derived to elicit the best growth parameters for vegetative Y. pestis cells and was previously shown to permit full-scale growth under all the conditions used here (22). Therefore, BCS medium ensures that differences in protein expression are not due to growth arrest, and the growth parameters of temperature and calcium used here have previously been employed to study changes in gene (52) and protein (54, 74) expression. Taken together, the observed changes in protein expression detected in this study reflect adaptations to growth conditions rather than changes due to a cessation of growth. Future experiments will address whether the choice of growth medium and the lack of growth restriction play a role in the amount of Yop expression at the protein level and the ability to detect other virulence factors by 2-D DIGE.
Putative virulence factors.
The Y. pestis outer membrane porin protein OmpA was found to be upregulated at 37°C with calcium (8.0-fold) and at 37°C without calcium (12.3-fold) compared to its expression at 26°C with calcium. Several lines of evidence suggest that OmpA may be implicated in virulence. First, in host-pathogen interaction studies, OmpA was detected in monocyte cell lysates following exposure to Y. pestis (B. A. Chromy and S. L. McCutchen-Maloney, unpublished data). Secondly, the OmpA protein from Klebsiella pneumoniae (KpOmpA) was found to induce an immunogenic response, triggering dendritic cell migration to infected lymph nodes (35). Macrophages and dendritic cells are known to bind KpOmpA (36) through scavenger receptors (33), and these cells are activated in a Toll-like receptor 2-dependent manner (34). Finally, OmpA from the human pathogen E. coli K1 appears to reduce the complement-mediated attack (59) and binds to host cells (65). Although an essential function of OmpA is outer membrane stabilization (78), the finding of OmpA in host cell lysates and reports of other bacterial OmpA proteins as virulence factors suggest that the Y. pestis OmpA protein may also play a role in virulence.
AmpD3, a putative N-acetylmuramoyl-l-alanine amidase, was found to be upregulated at 37°C with calcium (2.3-fold) and at 37°C without calcium (2.7-fold) compared to its expression at 26°C with calcium. The amidase activity cleaves the amide bond between the lactyl group of muramic acid residues and the α-amino group of l-alanine residues of bacterial cell wall glycopeptides (26). Bacterial proteins possessing this activity are often autolysins, which are able to cleave muramyl-containing glycopeptides on their own surfaces. Autolysins are believed to contribute to virulence via pathogen adhesion and amplification of the host inflammatory response (28, 50). For example, an aut deletion mutant of Listeria monocytogenes showed reduced invasiveness and virulence following intravenous inoculation of mice and oral infection of guinea pigs (10). Other autolysins from pathogens include the atl gene product of Staphylococcus aureus (53), AmiC from Bacillus anthracis (48), murein hydrolase from Pseudomonas aeruginosa (40), and 14 other proteins from gram-negative bacteria, including five autolysins from E. coli that produce external membrane vesicles (39). Future studies of Y. pestis ampD3 mutant strains should elucidate the potential invasive and virulence roles of AmpD3.
α-Enolase, a cytoplasmic enzyme active in the glycolytic pathway, was found to be upregulated at 37°C with calcium (4.4-fold) and at 37°C without calcium (4.9-fold) compared to its expression at 26°C with calcium. α-Enolase has been shown to be a multifunctional protein, possessing heat shock, cytoskeletal binding, and transcriptional modulation activities (for a review, see reference 55). Plasminogen activity has been attributed to α-enolase both in vitro and in vivo (20), and the expression of α-enolase on the surfaces of the outer membranes of gram-negative bacteria is thought to provide access to host plasminogen (64). The bacterial α-enolase and host plasminogen complex aids in the dissemination of invasive pathogens by the degradation of fibrin blots, laminin, and fibronectin (25, 42). In fact, the human pathogen Staphylococcus pneumoniae binds both human plasminogen and plasmin via α-enolase (7), suggesting a possible virulence role for the Y. pestis α-enolase. Thus, the expression increases in both α-enolase and Pla found here are consistent with earlier literature, and these two Y. pestis proteins may work in concert to permit increased growth and dissemination of Y. pestis in the host.
Other differentially expressed proteins identified here are enzymes involved in sugar metabolism. Expression increases in phosphoglycerate kinase, 6-phosphogluconate dehydrogenase, d-3-phosphoglycerate, succinyl-coenzyme A synthetase, ribulose-5-phosphate epimerase, and glyceraldehyde-3-phosphate dehydrogenase alpha, with decreases detected for fructose-bisphosphate aldolase class II and the maltose binding protein periplasmic protein precursor, were found at 37°C without calcium compared to the case at 26°C with calcium. The differential expression of these proteins supports a model whereby different types and/or amounts of sugars are utilized in the transition from 26°C with calcium to 37°C without calcium. Notably, the E. coli ribulose-5-phosphate epimerase (Rpe) is essential for growth on single pentose sugars, and deletion of the rpe gene caused a loss of catalytic activity, rendering the mutant strain unable to utilize single pentose sugars (44). The specific utilization of sugars in different milieus is suggestive of a metabolic adaptive mechanism that may promote virulence. Alternatively, these glycolytic proteins may play additional roles in virulence, similar to the hypothesized role for α-enolase. Multiple functions have now been attributed to the former housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase alpha, separate from its role in glycolysis. These alternate functions include roles in apoptosis, microtubule binding, and membrane transport and fusion (67), any of which could account for the upregulation of the protein noted in this study. Moreover, glyceraldehyde-3-phosphate dehydrogenase alpha has been reported to function as a plasminogen activator (56, 67), and thus may also contribute to the invasive nature of Y. pestis. Although additional studies are required, these results indicate that expression changes in housekeeping and metabolic proteins may have a more significant role in virulence than was previously appreciated.
The TTSS is believed to be the most complex protein translocation process in biological systems (61). In order to induce the TTSS, protein production machinery must be activated to express all of the necessary proteins for the delivery of virulence factors into the host cell. Therefore, the increased expression of elongation factor 1B and several ribosomal proteins following growth at 37°C without calcium is likely due to increased protein production and suggests that these proteins may be involved in the regulation of virulence factor expression.
It is well known that chaperone proteins in Y. pestis aid the formation and injection of virulence factors into the host cell. Three chaperone proteins not previously linked to the TTSS, namely, ClpB, HtpG, and Cpn60, exhibited increased expression at 37°C, with and without calcium, compared to that at 26°C with calcium. Two isoforms of ClpB were identified here (Fig. 1, spots 4 and 5; Table 1), with 2.6- and 1.7-fold increases at 37°C with calcium and 2.9- and 2.3-fold increases at 37°C without calcium compared to their expression at 26°C with calcium. HtpG, coidentified in the same protein spot as phosphoglycerate kinase, demonstrated a 3.1-fold increase at 37°C without calcium compared to its expression at 26°C with calcium. Future studies aimed at resolving this gel region are needed to elucidate the individual expression levels. Cpn60 was found in 10 distinct spots, potentially due to posttranslational regulation affording multiple chaperone functions or protein degradation due to experimental processing (3). Nine spots were upregulated an average of 4.3-fold and one spot was downregulated 1.2-fold at 37°C without calcium compared to the case at 26°C with calcium. Interestingly, ClpB is known to play a role in the regulation of the virulence factor invasin and in bacterial motility in Yersinia enterocolitica (5). Furthermore, ClpB and HtpG participate in cellular protein folding under mild heat shock conditions (73). An increased expression of both HtpG and Cpn60 may reflect their roles as heat shock proteins (43). Alternatively, Cpn60 may be recognized by host cells as antigenic, similar to the Cpn60 chaperonin homolog of Helicobacter pylori, Hsp60, which functions in the pathogenesis of gastric mucosa-associated lymphoma (2, 80). HtpG has also been shown to function as an immunogen and potentially contributes to the virulence of the periodontal pathogen Porphyromonas gingivalis (41). Thus, it is possible that Cpn60 and HtpG may not only serve as chaperones for virulence factors, but also may play a role in modulating the host immune response.
Concluding comments.
This study represents the first 2-D DIGE proteomic analysis of Y. pestis cultured under conditions that are relevant to the induction of virulence. The proteomic map of Y. pestis as a function of temperature and calcium changes provides knowledge of expression levels of virulence-associated factors, putative virulence factors, and metabolic and housekeeping proteins as well as potential novel virulence determinants. Future proteomic studies addressing secreted and membrane-bound Y. pestis proteins, using altered gel formulations to examine the lower-molecular-weight Y. pestis proteome, and experiments to control the proteolytic activity of Pla will more fully address the Y. pestis proteome. In addition, the data and methods established here are guiding proteomic comparisons of diverse and mutant strains of Y. pestis as well as studies using different growth media to better understand growth and virulence in the life cycle of Y. pestis. While inferences can be made about the functions of the differentially expressed proteins identified, future studies of gene-disrupted and mutant strains of Y. pestis are needed to address the biological functions of putative virulence proteins. In summary, the proteomic map of Y. pestis reported here paves the way for future comparisons of diverse clinical isolates, strains with different virulence attributes, and mutant strains, which will provide an improved mechanistic understanding of virulence, lead to pathogen signatures for biodefense-related detection and threat assessment, and identify new therapeutic targets for plague.
Acknowledgments
We thank Anne Clatworthy and Kurt Schesser for critical reviews of the manuscript, Greg Plano and Janet Foley for helpful discussions, and Emilio Garcia and Vladimir Motin for the kind gift of Y. pestis.
This work was funded by the Lawrence Livermore National Laboratory (Laboratory Directed Research and Development) and the Department of Homeland Security (Biological Countermeasures Program). This work was performed under the auspices of the U.S. Department of Energy by University of California Lawrence Livermore National Laboratory under contract no. W-7405-Eng-48 (UCRL-JRNL-209060).
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed] [Google Scholar]
- 2.Amini, H. R., F. Ascencio, A. Cruz-Villacorta, E. Ruiz-Bustos, and T. Wadstrom. 1996. Immunochemical properties of a 60 kDa cell surface-associated heat shock-protein (Hsp60) from Helicobacter pylori. FEMS Immunol. Med. Microbiol. 16:163-172. [DOI] [PubMed] [Google Scholar]
- 3.Amini, H. R., F. Ascencio, E. Ruiz-Bustos, M. J. Romero, and T. Wadstrom. 1996. Cryptic domains of a 60 kDa heat shock protein of Helicobacter pylori bound to bovine lactoferrin. FEMS Immunol. Med. Microbiol. 16:247-255. [DOI] [PubMed] [Google Scholar]
- 4.Anisimov, A. P., L. E. Lindler, and G. B. Pier. 2004. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badger, J. L., B. M. Young, A. J. Darwin, and V. L. Miller. 2000. Yersinia enterocolitica ClpB affects levels of invasin and motility. J. Bacteriol. 182:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedek, O., G. Nagy, and L. Emody. 2004. Intracellular signalling and cytoskeletal rearrangement involved in Yersinia pestis plasminogen activator (Pla) mediated HeLa cell invasion. Microb. Pathog. 37:47-54. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2004. Characterization of plasmin(ogen) binding to Streptococcus pneumoniae. Indian J. Med. Res. 119(Suppl.):29-32. [PubMed] [Google Scholar]
- 8.Brubaker, R. R. 2001. Yersinia pestis, p. 2033-2058. In M. Sussman (ed.), Molecular medical microbiology, vol. 3. Academic Press, London, United Kingdom. [Google Scholar]
- 9.Brubaker, R. R., E. D. Beesley, and M. J. Surgalla. 1965. Pasteurella pestis: role of pesticin I and iron in experimental plague. Science 149:422-424. [DOI] [PubMed] [Google Scholar]
- 10.Cabanes, D., O. Dussurget, P. Dehoux, and P. Cossart. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601-1614. [DOI] [PubMed] [Google Scholar]
- 11.Cavanaugh, D. C. 1971. Specific effect of temperature upon transmission of the plague bacillus by the oriental rat flea, Xenopsylla cheopis. Am. J. Trop. Med. Hyg. 20:264-273. [DOI] [PubMed] [Google Scholar]
- 12.Cavanaugh, D. C., and J. E. Williams. 1980. Plague: some ecological interrelationships, p. 245-256. In Proceedings of the International Congress on Fleas. A. A. Balkema, Rotterdam, The Netherlands.
- 13.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 83:348-363. [PubMed] [Google Scholar]
- 14.Chromy, B. A., J. Perkins, J. L. Heidbrink, A. D. Gonzales, G. A. Murphy, J. P. Fitch, and S. L. McCutchen-Maloney. 2004. Proteomic characterization of host response to Yersinia pestis and near neighbors. Biochem. Biophys. Res. Commun. 320:474-479. [DOI] [PubMed] [Google Scholar]
- 15.Cornelis, G. R. 1998. The Yersinia deadly kiss. J. Bacteriol. 180:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du, Y., E. Galyov, and A. Forsberg. 1995. Genetic analysis of virulence determinants unique to Yersinia pestis. Contrib. Microbiol. Immunol. 13:321-324. [PubMed] [Google Scholar]
- 19.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehinger, S., W. D. Schubert, S. Bergmann, S. Hammerschmidt, and D. W. Heinz. 2004. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343:997-1005. [DOI] [PubMed] [Google Scholar]
- 21.Forsberg, A., I. Bolin, L. Norlander, and H. Wolf-Watz. 1987. Molecular cloning and expression of calcium-regulated, plasmid-coded proteins of Y. pseudotuberculosis. Microb. Pathog. 2:123-137. [DOI] [PubMed] [Google Scholar]
- 22.Fowler, J. M., and R. R. Brubaker. 1994. Physiological basis of the low calcium response in Yersinia pestis. Infect. Immun. 62:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gade, D., J. Thiermann, D. Markowsky, and R. Rabus. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. Soluble proteins of the marine bacterium Pirellula sp. strain 1. J. Mol. Microbiol. Biotechnol. 5:240-251. [DOI] [PubMed] [Google Scholar]
- 24.Garcia, E., Y. A. Nedialkov, J. Elliott, V. L. Motin, and R. R. Brubaker. 1999. Molecular characterization of KatY (antigen 5), a thermoregulated chromosomally encoded catalase-peroxidase of Yersinia pestis. J. Bacteriol. 181:3114-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge, J., D. M. Catt, and R. L. Gregory. 2004. Streptococcus mutans surface alpha-enolase binds salivary mucin MG2 and human plasminogen. Infect. Immun. 72:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genereux, C., D. Dehareng, B. Devreese, J. Van Beeumen, J. M. Frere, and B. Joris. 2004. Mutational analysis of the catalytic centre of the Citrobacter freundii AmpD N-acetylmuramyl-l-alanine amidase. Biochem. J. 377:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gharbi, S., P. Gaffney, A. Yang, M. J. Zvelebil, R. Cramer, M. D. Waterfield, and J. F. Timms. 2002. Evaluation of two-dimensional differential gel electrophoresis for proteomic expression analysis of a model breast cancer cell system. Mol. Cell. Proteomics 1:91-98. [DOI] [PubMed] [Google Scholar]
- 28.Gupta, D., Y. P. Jin, and R. Dziarski. 1995. Peptidoglycan induces transcription and secretion of TNF-alpha and activation of Lyn, extracellular signal-regulated kinase, and Rsk signal transduction proteins in mouse macrophages. J. Immunol. 155:2620-2630. [PubMed] [Google Scholar]
- 29.Heesemann, J., U. Gross, N. Schmidt, and R. Laufs. 1986. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect. Immun. 54:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 31.Hinnebusch, J., P. Cherepanov, Y. Du, A. Rudolph, J. D. Dixon, T. Schwan, and A. Forsberg. 2000. Murine toxin of Yersinia pestis shows phospholipase D activity but is not required for virulence in mice. Int. J. Med. Microbiol. 290:483-487. [DOI] [PubMed] [Google Scholar]
- 32.Inglesby, T. V., D. T. Dennis, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, J. F. Koerner, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, M. Schoch-Spana, and K. Tonat. 2000. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA 283:2281-2290. [DOI] [PubMed] [Google Scholar]
- 33.Jeannin, P., B. Bottazzi, M. Sironi, A. Doni, M. Rusnati, M. Presta, V. Maina, G. Magistrelli, J. F. Haeuw, G. Hoeffel, N. Thieblemont, N. Corvaia, C. Garlanda, Y. Delneste, and A. Mantovani. 2005. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity 22:551-560. [DOI] [PubMed] [Google Scholar]
- 34.Jeannin, P., G. Magistrelli, L. Goetsch, J. F. Haeuw, N. Thieblemont, J. Y. Bonnefoy, and Y. Delneste. 2002. Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen presenting cells—impact on vaccine strategies. Vaccine 20(Suppl. 4):A23-A27. [DOI] [PubMed] [Google Scholar]
- 35.Jeannin, P., G. Magistrelli, N. Herbault, L. Goetsch, S. Godefroy, P. Charbonnier, A. Gonzalez, and Y. Delneste. 2003. Outer membrane protein A renders dendritic cells and macrophages responsive to CCL21 and triggers dendritic cell migration to secondary lymphoid organs. Eur. J. Immunol. 33:326-333. [DOI] [PubMed] [Google Scholar]
- 36.Jeannin, P., T. Renno, L. Goetsch, I. Miconnet, J. P. Aubry, Y. Delneste, N. Herbault, T. Baussant, G. Magistrelli, C. Soulas, P. Romero, J. C. Cerottini, and J. Y. Bonnefoy. 2000. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat. Immunol. 1:502-509. [DOI] [PubMed] [Google Scholar]
- 37.Kukkonen, M., and T. K. Korhonen. 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 294:7-14. [DOI] [PubMed] [Google Scholar]
- 38.Lahteenmaki, K., R. Virkola, A. Saren, L. Emody, and T. K. Korhonen. 1998. Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect. Immun. 66:5755-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Z., A. J. Clarke, and T. J. Beveridge. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180:5478-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Z., A. J. Clarke, and T. J. Beveridge. 1996. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division, and secretion in surface membrane vesicles. J. Bacteriol. 178:2479-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopatin, D. E., C. E. Shelburne, N. Van Poperin, C. J. Kowalski, and R. A. Bagramian. 1999. Humoral immunity to stress proteins and periodontal disease. J. Periodontol. 70:1185-1193. [DOI] [PubMed] [Google Scholar]
- 42.Lottenberg, R., D. Minning-Wenz, and M. D. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20-24. [DOI] [PubMed] [Google Scholar]
- 43.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 44.Lyngstadaas, A., G. A. Sprenger, and E. Boye. 1998. Impaired growth of an Escherichia coli rpe mutant lacking ribulose-5-phosphate epimerase activity. Biochim. Biophys. Acta 1381:319-330. [DOI] [PubMed] [Google Scholar]
- 45.McDonough, K. A., and S. Falkow. 1989. A Yersinia pestis-specific DNA fragment encodes temperature-dependent coagulase and fibrinolysin-associated phenotypes. Mol. Microbiol. 3:767-775. [DOI] [PubMed] [Google Scholar]
- 46.Mehigh, R. J., and R. R. Brubaker. 1993. Major stable peptides of Yersinia pestis synthesized during the low-calcium response. Infect. Immun. 61:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehigh, R. J., A. K. Sample, and R. R. Brubaker. 1989. Expression of the low calcium response in Yersinia pestis. Microb. Pathog. 6:203-217. [DOI] [PubMed] [Google Scholar]
- 48.Mesnage, S., and A. Fouet. 2002. Plasmid-encoded autolysin in Bacillus anthracis: modular structure and catalytic properties. J. Bacteriol. 184:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michiels, T., P. Wattiau, R. Brasseur, J. M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212-1224. [DOI] [PubMed] [Google Scholar]
- 51.Montie, T. C. 1981. Properties and pharmacological action of plague murine toxin. Pharmacol. Ther. 12:491-499. [DOI] [PubMed] [Google Scholar]
- 52.Motin, V. L., A. M. Georgescu, J. P. Fitch, P. P. Gu, D. O. Nelson, S. L. Mabery, J. B. Garnham, B. A. Sokhansanj, L. L. Ott, M. A. Coleman, J. M. Elliott, L. M. Kegelmeyer, A. J. Wyrobek, T. R. Slezak, R. R. Brubaker, and E. Garcia. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmblad, M., M. Ramstrom, C. G. Bailey, S. L. McCutchen-Maloney, J. Bergquist, and L. C. Zeller. 2004. Protein identification by liquid chromatography-mass spectrometry using retention time prediction. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 803:131-135. [DOI] [PubMed] [Google Scholar]
- 55.Pancholi, V. 2001. Multifunctional alpha-enolase: its role in diseases. Cell Mol. Life Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry, R. D., P. A. Harmon, W. S. Bowmer, and S. C. Straley. 1986. A low-Ca2+ response operon encodes the V antigen of Yersinia pestis. Infect. Immun. 54:428-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perry, R. D., T. S. Lucier, D. J. Sikkema, and R. R. Brubaker. 1993. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect. Immun. 61:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prasadarao, N. V., A. M. Blom, B. O. Villoutreix, and L. C. Linsangan. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352-6360. [DOI] [PubMed] [Google Scholar]
- 60.Price, S. B., and S. C. Straley. 1989. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect. Immun. 57:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed] [Google Scholar]
- 62.Rudolph, A. E., J. A. Stuckey, Y. Zhao, H. R. Matthews, W. A. Patton, J. Moss, and J. E. Dixon. 1999. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 274:11824-11831. [DOI] [PubMed] [Google Scholar]
- 63.Sample, A. K., and R. R. Brubaker. 1987. Post-translational regulation of Lcr plasmid-mediated peptides in pesticinogenic Yersinia pestis. Microb. Pathog. 3:239-248. [DOI] [PubMed] [Google Scholar]
- 64.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34:195-204. [DOI] [PubMed] [Google Scholar]
- 65.Shin, S., G. Lu, M. Cai, and K. S. Kim. 2005. Escherichia coli outer membrane protein A adheres to human brain microvascular endothelial cells. Biochem. Biophys. Res. Commun. 330:1199-1204. [DOI] [PubMed] [Google Scholar]
- 66.Simpson, W. J., R. E. Thomas, and T. G. Schwan. 1990. Recombinant capsular antigen (fraction 1) from Yersinia pestis induces a protective antibody response in BALB/c mice. Am. J. Trop. Med. Hyg. 43:389-396. [DOI] [PubMed] [Google Scholar]
- 67.Sirover, M. A. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432:159-184. [DOI] [PubMed] [Google Scholar]
- 68.Sodeinde, O. A., and J. D. Goguen. 1988. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect. Immun. 56:2743-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sodeinde, O. A., A. K. Sample, R. R. Brubaker, and J. D. Goguen. 1988. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect. Immun. 56:2749-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sodeinde, O. A., Y. V. Subrahmanyam, K. Stark, T. Quan, Y. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 258:1004-1007. [DOI] [PubMed] [Google Scholar]
- 71.Straley, S. C., and R. R. Brubaker. 1981. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc. Natl. Acad. Sci. USA 78:1224-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Straley, S. C., and R. R. Brubaker. 1982. Localization in Yersinia pestis of peptides associated with virulence. Infect. Immun. 36:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas, J. G., and F. Baneyx. 2000. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 36:1360-1370. [DOI] [PubMed] [Google Scholar]
- 74.Thulasiraman, V., S. L. McCutchen-Maloney, V. L. Motin, and E. Garcia. 2001. Detection and identification of virulence factors in Yersinia pestis using SELDI ProteinChip system. BioTechniques 30:428-432. [DOI] [PubMed] [Google Scholar]
- 75.Tonge, R., J. Shaw, B. Middleton, R. Rowlinson, S. Rayner, J. Young, F. Pognan, E. Hawkins, I. Currie, and M. Davison. 2001. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1:377-396. [DOI] [PubMed] [Google Scholar]
- 76.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18:2071-2077. [DOI] [PubMed] [Google Scholar]
- 77.Vorontsov, E. D., A. G. Dubichev, L. N. Serdobintsev, and A. V. Naumov. 1990. Association-dissociation processes and supermolecular organisation of the capsule antigen (protein F1) of Yersinia pestis. Biomed. Sci. 1:391-396. [PubMed] [Google Scholar]
- 78.Wang, Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396-401. [DOI] [PubMed] [Google Scholar]
- 79.Welkos, S. L., A. M. Friedlander, and K. J. Davis. 1997. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain C092. Microb. Pathog. 23:211-223. [DOI] [PubMed] [Google Scholar]
- 80.Yamasaki, R., K. Yokota, H. Okada, S. Hayashi, M. Mizuno, T. Yoshino, Y. Hirai, D. Saitou, T. Akagi, and K. Oguma. 2004. Immune response in Helicobacter pylori-induced low-grade gastric-mucosa-associated lymphoid tissue (MALT) lymphoma. J. Med. Microbiol. 53:21-29. [DOI] [PubMed] [Google Scholar]
- 81.Yan, J. X., A. T. Devenish, R. Wait, T. Stone, S. Lewis, and S. Fowler. 2002. Fluorescence two-dimensional difference gel electrophoresis and mass spectrometry based proteomic analysis of Escherichia coli. Proteomics 2:1682-1698. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, C. G., A. D. Gonzales, M. W. Choi, B. A. Chromy, J. P. Fitch, and S. L. McCutchen-Maloney. 2005. Subcellular proteomic analysis of host-pathogen interactions using human monocytes exposed to Yersinia pestis and Yersinia pseudotuberculosis. Proteomics 5:1877-1888. [DOI] [PubMed] [Google Scholar]