Abstract
Sequence analysis of peptide deformylase of Mycobacterium tuberculosis revealed the presence of insertions (residues 74 to 85) and an unusually long carboxy-terminal end (residues 182 to 197). Our results with deletion mutants indicated the contribution of these regions in maintaining enzymatic stability. Furthermore, we showed that the region spanning the insertions was responsible for maintaining resistance to oxidizing agents, like H2O2.
The amino-terminal ends of all nascent polypeptides in eubacteria are formylated. As N-terminal peptidases are unable to utilize formylated peptides as substrates, their removal is a mandatory step during protein synthesis. Since the enzyme peptide deformylase (PDF) is known to deformylate the N-formyl group of the nascent polypeptide chains in the cytoplasm of bacteria (3), its importance has long been appreciated. Available genome sequencing data revealed the presence of a putative gene encoding the peptide deformylase (def) throughout the eubacterial lineage (12), including pathogens (1, 5, 9-11, 14). In this context, we concentrated on the peptide deformylase enzyme of the bacterial pathogen Mycobacterium tuberculosis (mPDF), which causes the dreadful disease tuberculosis.
Recently, we PCR amplified the def gene from M. tuberculosis and, following cloning in pET28c vector, expressed it as a histidine-tagged fusion protein (mPDF) in Escherichia coli strain BL21(DE3). The overexpressed protein was obtained as inclusion bodies and subsequently solubilized with 3 M urea. This was followed by dialysis at 4°C against 20 mM phosphate buffer, pH 7.4, and finally the mPDF protein was purified by using a Ni-nitrilotriacetic acid (NTA) column (17). Although mPDF was a Fe+2-containing enzyme (atomic absorption spectroscopy revealed the presence of 0.94 ± 0.21 mol of iron/mol of mPDF polypeptide, n = 4), its activity was very stable at 30°C, with a half-life of ∼4 h. Furthermore, it maintained its distinction by exhibiting resistance to oxidizing agents, like H2O2 (17). Since conversion of Fe+2 to Fe+3 by environmental oxygen resulted in inactivation of this metalloprotease in E. coli (15), it would really be interesting to know about mPDF, which is insensitive to such a condition. This is in fact a very important aspect to elucidate, because as a successful intracellular pathogen M. tuberculosis has to withstand host defense mechanisms, such as oxidative stress, for its survival.
Irrespective of the difference in behavior between homologues, the sequence of any given protein is derived from the same 20 essential amino acids. Therefore, in the postgenomic era, sequence analysis formed the theoretical basis to gain insight into the functionality of a protein. Analyses of amino acid sequences of all eubacterial PDFs revealed the presence of three (1, GXGXAAXQ; 2, EGCLS; and 3, QHEXXH [where X is any hydrophobic residue]) highly conserved motifs (Fig. 1), despite their broad categorization into type I (gram-negative) and type II (gram-positive) classes (2, 4, 7). Furthermore, motif 1 is the typical signature sequence for any metalloprotease (13). With the CLUSTAL X 1.81 program (19), we compared the nucleotide-derived amino acid sequence of mPDF with well-characterized representatives belonging to type I (E. coli) and type II (Staphylococcus aureus) classes. Although the available literature indicated that an extended C-terminal end is characteristic of type I PDFs (2, 4, 7), interestingly we found that mPDF possessed a similar feature. The unusually long C-terminal end (amino acid residues 182 to 197) of mPDF was very striking compared to the sequences of type II members (Fig. 1) (17), and it is likely to form a helix by the sequence-based secondary-structure prediction program (6). Besides this, like other gram-positive bacteria (type II class), mPDF possessed insertions (amino acid residues 74 to 85) (Fig. 1) between conserved motifs 1 and 2, which are predicted to form a loop (6). We created two deletion mutants of mPDF, one at the insertion sequences (designated ID, where 6 amino acids, MTARRR, were deleted) and the other at the C-terminal end (named TD, where 16 amino acids, PGLSWLPGEDPDPFGH, were removed), employing a PCR-based mutagenesis approach (17, 18) (primer sequences are given in Table 1). Like mPDF, they (both ID and TD) were obtained as inclusion bodies when overexpressed as histidine-tagged fusion proteins. The mutant proteins were solubilized with 3 M urea, refolded, and purified by using Ni-NTA columns, as described for mPDF (17). This was followed by assessment of enzyme activity to evaluate the contribution of these regions to the deformylation ability of mPDF. The enzyme activity of mPDF was determined in the presence of catalase and bovine serum albumin (BSA) by using N-formyl-Met-Ala as the substrate in the trinitrobenzenesulfonic acid (TNBSA) assay (17).
FIG. 1.
Analysis of mPDF sequence. Nucleotide-derived amino acid sequences of iron-containing peptide deformylase from E. coli (representative of type I), S. aureus (representative of type II), and mPDF were aligned using the CLUSTAL X 1.81 program. Gaps in the sequences were introduced for optimum alignment. Asterisks and dots are used to denote identical and similar amino acids, respectively. The insertion of sequences (IR) and extended C-terminal end (TR) of mPDF are underlined. Amino acids deleted for the creation of ID and TD mutants are indicated in bold.
TABLE 1.
List of PCR primers used in this studya
| Primer name | Primer sequence (5′ to 3′) | Amplified gene or region |
|---|---|---|
| CR26 (forward) | GGAATTCCATATGGCAGTCGTACCC | def |
| CR27 (reverse) | CCCAAGCTTTTAGTGACCGAACGG | def |
| CR23 (reverse) | TTAAACGCCCCAGCCATG | TD mutant of def |
| CR25 (internal) | ACCGCGCAGGTGTGGTCATCAAT | ID mutant of def |
| CR24 (internal) | ACCACACCTGCGCGGTCCG | ID mutant of def |
CR26 and CR27 were used as external primers for the amplification of the def open reading frame along with mutation. GGAATTCCAT in CR26 and CCCAAGCTT in CR27 do not correspond to the genome sequences but have been introduced to incorporate NdeI and HindIII sites at respective 5′ and 3′ ends of the PCR-amplified products. Details of PCR amplification, construction of recombinant plasmid, and generation of PDF mutants were described elsewhere (17).
For overexpression of different proteins, BL21(DE3) cells harboring wild-type (WT) or mutant constructs in pET28c vector were grown and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) by following the method described earlier (17). Cells were harvested, suspended in lysis buffer (20 mM phosphate buffer, pH 7.4, containing 5 mM dithiothreitol, 10 μg/ml of catalase, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml of pepstatin, and 1 μg/ml of leupeptin), and sonicated. The pellet fraction (∼12,000 × g for 30 min at 4°C) was resuspended in lysis buffer containing 3 M urea and 2% Triton X-100. Following centrifugation, the supernatant fraction was dialyzed (14 h at 4°C against 20 mM phosphate buffer, pH 7.4) to remove urea and purified on an Ni-NTA column (17). Finally, mPDF or different mutants were eluted in elution buffer (20 mM phosphate buffer, pH 7.4, containing 300 mM NaCl, 250 mM imidazole, and 10 μg/ml of catalase).
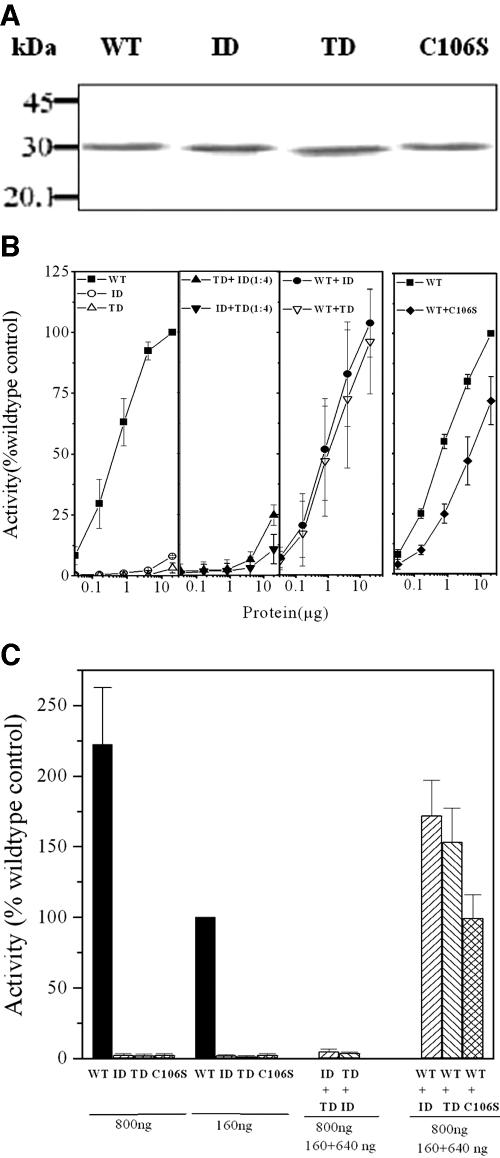
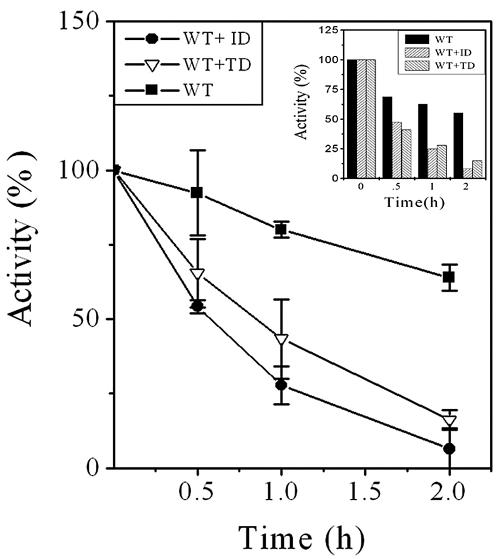
The expressed mutant proteins (ID and TD) were recognized by the anti-His-tag antibody, as evidenced by Western blotting (Fig. 2A). As reported earlier, deletion of TR (Fig. 1), resulting in the mutant TD, affected deformylase activity of mPDF (Fig. 2B, left panels) (17). Like TD, even with the use of an excess amount of protein (20 μg incubated with 5 mM of N-formyl-Met-Ala) in the assays, the ID mutant hardly showed any deformylase activity (Fig. 2B, left, panel 1). Loss in enzyme activity has often been correlated with its stability. To address this question, we mixed both of the mutant proteins (keeping the total protein concentration of stock the same for each mutant) with each other (ID:TD and TD:ID at 1:4, named ID+TD and TD+ID, respectively) after denaturation with urea. Following refolding (removing urea by slow dialysis [17]) and subsequent purification through Ni-NTA resins, their deformylation abilities were assessed as the function of increasing concentration of total proteins. Among them, the use of 20 μg of “cofolded” protein displayed marginal deformylation ability (for ID+TD, 11 ± 6%, and for TD+ID, 25 ± 4%) compared to the wild type (100%), suggesting the abilities of both mutants in partially complementing enzyme activity (Fig. 2B, left, panel 2). To further confirm this aspect, mutants were mixed with the wild type (WT:ID and WT:TD at 1:4, named WT+ID and WT+TD, respectively) and used to assess enzymatic activity. Interestingly, both WT+ID and WT+TD displayed the ability to deformylate N-formyl-Met-Ala more efficiently than the corresponding amounts of the wild-type protein in the mixtures (Fig. 2B, left, panel 3). To highlight the authenticity of such an observation, we carried out a similar experiment by using a C106S mutant of mPDF (mutation at the metal ion coordinating Cys at motif 2 of mPDF), which did not exhibit any deformylase activity (17). Unlike WT+ID and WT+TD, the C106S mutant (recognized by the anti-His-tag antibody [Fig. 2A]), when mixed with the wild type (WT:C106S at 1:4, named WT+C106S), showed enzyme activity at the level corresponding to the amount of wild-type protein present in the mixture (Fig. 2B, right). These results argued that mPDF was catalytically active as a multimer, and it is therefore logical to presume that heteromonomeric units of the wild type and ID/TD exhibited cooperativeness among themselves for rendering enzymatic activity of cofolded proteins. As shown in Fig. 2C, the extent of deformylation with 800 ng of total protein (amounts of wild-type and TD/ID proteins were 160 ng and 640 ng, respectively) was increased by 58 to 75% compared to that of the wild-type control (160 ng protein). On the other hand, such an increase in enzyme activity was not evident with the WT+C106S mixture (Fig. 2C). Therefore, the obvious explanation of such an observation was the contribution of either ID or TD towards the enzymatic activity of wild-type mPDF. Furthermore, this increase in the enzyme activity could not be seen if the purified mutant (ID or TD) or any other proteins (catalase/BSA) were mixed with the wild type at the same ratio during the assay (data not shown). Interestingly, following mixing with the wild type (WT+ID or WT+TD; amount of total protein used per assay was 70 ng), both of the mutants lost deformylation ability within 2 h (Fig. 3). Even the use of an increased amount of total protein (350 ng, where the amount of WT protein was 70 ng) in the assay yielded similar results (Fig. 3, inset, showing a representative experiment). Thus, all of this evidence argues that the loss in enzyme activity for the ID or TD mutant was the result of the enzymatic instability of the protein.
FIG. 2.
Assessment of deformylase activities of mPDF mutants. (A) Western blotting of WT and mutant (ID, TD, and C106S) proteins using anti-His-tag antibody. (B) Deformylase activities of mPDF mutants. WT and mutant proteins were mixed with each other (ID+TD, TD+ID, WT+ID, WT+TD, or WT+C106S) at the ratio of 1:4 after denaturation with urea. They were refolded and subsequently purified as mentioned above. Enzyme activities were monitored with increasing concentrations of total proteins in the presence of catalase and BSA by using 5 mM of N-formyl-Met-Ala as the substrate in the TNBSA assay (17). (Left, panels 1 to 3) Assays with WT, ID, TD, ID+TD, TD+ID, WT+ID, and WT+TD proteins. (Right) Assays with WT and WT+C106S proteins. Results are expressed as percentages of enzyme activity obtained with 20 μg WT protein. (C) Enzyme activities with cofolded proteins. Assays were carried out with indicated amount of proteins (WT, ID, TD, ID+TD, TD+ID, WT+ID, WT+TD, or WT+C106S) in the presence of catalase and BSA by using 5 mM of N-formyl-Met-Ala as the substrate (17). Results are expressed as the percentages of enzyme activity obtained with 160 ng WT protein (100% = 3.24 ± 0.7 μmol free amino group produced/min/mg protein).
FIG. 3.
Assessment of enzymatic stability of mPDF mutants. Overexpressed and purified proteins were obtained as described in the text. Enzyme activities were monitored with 17.5 ng of WT or 70 ng of WT+ID or WT+TD proteins in the presence of catalase and BSA by using 5 mM of N-formyl-Met-Ala as the substrate in the TNBSA assay (17). (Inset) Representative experiment showing the enzyme activity with 5 mM of N-formyl-Met-Ala using 70 ng of WT or 350 ng of WT+ID or WT+TD protein.
The activity as well as the stability of an enzyme often depends on its intermolecular association status. PDF from E. coli has been reported to be active as a monomer and that of Leptospira interrogans as a dimer (9, 16). To gain insight on this aspect, we determined the molecular masses of the purified recombinant mPDF and its deletion mutants (ID and TD) through dynamic light-scattering studies performed at 25°C on a Dyna Pro instrument (Protein Solutions) equipped with DYNAMICS V6 software by using a microcuvette. Samples (10 to 15 μl) used for this study were always filtered through a 0.1-μM filtration device (Millipore). The majority of the wild-type mPDF in solution showed a molecular mass of 100 kDa, with a radius of the molecules of 4.3 nm (Table 2). Since the calculated molecular mass of the protein was 22.6 kDa (although by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, the expressed protein exhibited an anomalous migration of ∼31 kDa [Fig. 2A] [17]), our results argued the multimeric nature of the catalytically active mPDF. Interestingly, in the ID mutant these parameters remained unaltered. On the other hand, considering the deletion of 16 amino acids from mPDF, in the TD mutant both radius and molecular mass of the protein were less than those of the wild type, but the multimeric nature was evident (Table 2). By gel filtration chromatography (AKTAprime Plus chromatography system with a Sephacryl 200 column; GE Biosciences), mPDF exhibited two peaks, one at the void volume and the other at ∼44 kDa (data not shown). We found that only the ∼44-kDa protein had the deformylation ability (kcat/Km of∼1,084 M−1 s−1, as opposed to ∼1,220 M−1 s−1 reported in reference 17 with the supernatant fraction obtained following low-speed centrifugation) and that further mixing of these fractions (high and low molecular masses) did not have any effect on the enzyme activity of mPDF. As expected, by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the ∼44-kDa protein showed a molecular mass of ∼31 kDa and glutaraldehyde cross-linking (0.0125% at 25°C for 30 to 240 min) indicated the existence of high-molecular-mass (∼100-kDa) forms (data not shown). Notably, the dynamic light-scattering data suggested that the mPDF was predominantly tetramer, while gel filtration chromatography indicated its dimeric status. Since both methods rely on shape and hydrodynamic volume and do not always coincide, we prefer to leave this issue untouched for the time being. However, our results unequivocally established the multimeric nature of the active protein, and mutations at IR or TR did not affect the intermolecular association status of mPDF.
TABLE 2.
Molecular masses of wild-type and mutant mPDFs
| Sample | Species | Radius (nm) | Polydispersity (%) | Molecular massa
|
|
|---|---|---|---|---|---|
| kDa | % | ||||
| Wild type | Multimer | 4.3 | 0.0 | 100 | 85.4 |
| Soluble aggregate | 20.4 | 11.4 | 3,907 | 13.4 | |
| ID | Multimer | 4.3 | 0.0 | 100 | 85.3 |
| Soluble aggregate | 17.7 | 19.9 | 2,808 | 12.3 | |
| TD | Multimer | 3.9 | 15.4 | 82 | 95.3 |
| Soluble aggregate | 23.1 | 17.3 | 5,239 | 4.7 | |
Molecular masses were determined by dynamic light-scattering studies, as mentioned in the text. All entries in the table are the means of values derived from a minimum of 20 independent measurements.
Analysis of the crystal structure of PDF from L. interrogans, which is a Zn+2-containing metalloprotease, indicated that part of the C-terminal end was involved in anchoring of the substrate to the active site of the protein (20). A similar role of this region in mPDF is rather difficult to predict from our study, as insertion of the C-terminal end to the active site of the neighboring molecules might occur due to crystal packing. However, it is apparent from our results that the extended C-terminal end of mPDF definitely influenced enzyme stability. IR, on the other hand, has been predicted to form a loop in mPDF (6). For other gram-positive bacteria, this loop was found to be flexible but had no role in substrate binding (7). The enzymatic stability, especially in Fe+2-containing PDFs, is a perplexing issue. The iron was coordinated with histidine and cysteine (H148, H152, and C106S in the M. tuberculosis enzyme) residues in all PDFs (2, 4, 7, 15). The metal-coordinating cysteine Sγ has been shown to undergo oxidation to cysteine-sulfonic acid and/or cysteine-sulfinic acid in E. coli, S. aureus, Streptococcus pneumoniae, and Thermotoga maritima, resulting in severe consequences or even complete loss of enzyme activity (7, 15). Thus, despite structural identity near the active site metal, stabilities of PDFs in different bacteria were very different (2). These findings led to the postulation that such variation might be due to protein dynamics and/or alteration in sequences beyond the conserved regions, which could affect the rate of oxidation of iron or amino acid side chains that are metal ion ligands (2).
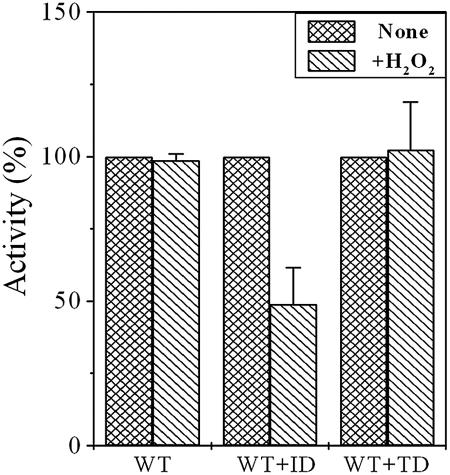
We have recently reported that preincubation of mPDF with an oxidizing agent, like H2O2, had no significant effect on its deformylating ability, despite the presence of Fe+2 at its metal binding core (17). This seems to be an important observation, considering the fact that M. tuberculosis has to cope with oxidative stress for its survival within the host as a successful pathogen. To know the contributions of IR or TR (Fig. 1) of the mPDF towards its resistance to oxidizing agents, we utilized WT+ID and WT+TD proteins (total protein, 70 ng/reaction) and monitored the effect of H2O2 (500 mM) on enzyme activity. As shown in Fig. 4, preincubation with H2O2 for 15 min exhibited a significant decrease in the deformylase activity of WT+ID compared to that of the wild type or WT+TD. Under our assay conditions, even the use of an increased amount of total protein displayed similar results (data not shown). Thus, our findings argued for a contribution of the IR region, specifically MTARRR sequences, in protecting Fe+2 in the metal binding core of mPDF from oxidation to the Fe+3 form. Interestingly, sequence analysis of known iron-containing PDFs revealed that three consecutive arginines in the IR of mPDF are typical of different mycobacterial species (M. avium, M. bovis, M. leprae, M. smegmatis, and M. tuberculosis). Furthermore, interaction of oxygen with the side chain of arginine has already been established (8) and, therefore, it could play a crucial role in preventing oxidation of Fe+2 in mycobacterial PDFs. However, structure-function analysis needs to be carried out to unravel the mystery. Nonetheless, the results we present here unequivocally establish the association of IR and TR of mPDF in maintaining its enzymatic stability.
FIG. 4.
Effect of H2O2 on deformylase activity of mPDF mutants. Overexpressed and purified proteins were obtained as described in the text. WT (17.5 ng/reaction), WT+ID, or WT+TD (each cofolded protein, 70 ng/reaction) proteins were preincubated with or without H2O2 (final concentration, 500 mM) at 30°C for 15 min. This was followed by an enzyme assay in the presence of catalase and BSA with 5 mM of N-formyl-Met-Ala as the substrate, following the method described earlier (17).
Acknowledgments
We acknowledge the kind help of P. Guptasarma for dynamic light-scattering studies. We thank D. Sarkar and Meghna Thakur for critical reading of the manuscript. We are indebted to J. Prasad for providing us with excellent technical assistance.
One of the authors (R.S.) is the recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), New Delhi, India. This project is financially supported by Network Project grant SMM 003 from CSIR.
REFERENCES
- 1.Apfel, C. M., H. Locher, S. Evers, B. Takacs, C. Hubschwerlen, W. Pirson, M. G. Page, and W. Keck. 2001. Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob. Agents Chemother. 45:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, E. T., M. S. Harris, A. W. Yem, C. L. Wolfe, A. F. Vosters, K. A. Curry, R. W. Murray, J. H. Bock, V. P. Marshall, J. I. Cialdella, M. H. Merchant, G. Choi, and M. R. Deibel, Jr. 2002. Crystal structure of type II peptide deformylase from Staphylococcus aureus. J. Biol. Chem. 277:31163-31171. [DOI] [PubMed] [Google Scholar]
- 3.Giglione, C., M. Perrie, and T. Meinnel. 2000. Peptide deformylase as a target for new generation, broad-spectrum antibacterial agents. Mol. Microbiol. 36:1197-1205. [DOI] [PubMed] [Google Scholar]
- 4.Guilloteau, J. P., M. Mathieu, C. Giglione, V. Blanc, A. Dupuy, M. Chevrier, P. Gil, A. Famechon, T. Meinnel, and V. Mikol. 2002. The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents. J. Mol. Biol. 320:951-962. [DOI] [PubMed] [Google Scholar]
- 5.Han, C., Q. Wang, L. Dong, H. Sun, S. Peng, J. Chen, Y. Yang, J. Yue, X. Shen, and H. Jiang. 2004. Molecular cloning and characterization of a new peptide deformylase from human pathogenic bacterium Helicobacter pylori. Biochem. Biophys. Res. Commun. 319:1292-1298. [DOI] [PubMed] [Google Scholar]
- 6.Kaur, H., and G. P. S. Raghava. 2003. Prediction of β-turns in proteins from multiple alignment using neural network. Protein Sci. 12:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreusch, A., G. Spraggon, C. C. Lee, H. Klock, D. McMullan, K. Ng, T. Shin, J. Vincent, I. Warner, C. Ericson, and S. A. Lesley. 2003. Structure analysis of peptide deformylases from Streptococcus pneumoniae, Staphylococcus aureus, Thermotoga maritima and Pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase. J. Mol. Biol. 330:309-321. [DOI] [PubMed] [Google Scholar]
- 8.Lass, A., A. Suessenbacher, G. Wolkart, B. Mayer, and F. Brunner. 2002. Functional and analytical evidence for scavenging of oxygen radicals by l-arginine. Mol. Pharmacol. 61:1081-1088. [DOI] [PubMed] [Google Scholar]
- 9.Li, Y., Z. Chen, and W. Gong. 2002. Enzymatic properties of a new peptide deformylase from pathogenic bacterium Leptospira interrogans. Biochem. Biophys. Res. Commun. 295:884-889. [DOI] [PubMed] [Google Scholar]
- 10.Margolis, P., C. Hackbarth, S. Lopez, M. Maniar, W. Wang, Z. Yuan, R. White, and J. Trias. 2001. Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob. Agents Chemother. 45:2432-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margolis, P. S., C. J. Hackbarth, D. C. Young, W. Wang, D. Chen, Z. Yuan, R. White, and J. Trias. 2000. Peptide deformylase in Staphylococus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob. Agents Chemother. 44:1825-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazel, D., E. Coic, S. Blanchard, and W. Saurin. 1997. A survey of polypeptide deformylase function throughout the eubacterial lineage. J. Mol. Biol. 266:939-949. [DOI] [PubMed] [Google Scholar]
- 13.Meinnel, T., C. Lazennec, S. Villoing, and S. Blanquet. 1997. Structure-function relationship within the peptide deformylase family. Evidence for a conserved architecture of the active site involving three conserved motifs and a metal ion. J. Mol. Biol. 267:749-761. [DOI] [PubMed] [Google Scholar]
- 14.Newton, D. T., C. Creuzenet, and D. Mangroo. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 274:22143-22146. [DOI] [PubMed] [Google Scholar]
- 15.Rajagopalan, P. T., and D. Pei. 1998. Oxygen-mediated inactivation of peptide deformylase. J. Biol. Chem. 273:22305-22310. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopalan, P. T., A. Datta, and D. Pei. 1997. Purification, characterization, and inhibition of peptide deformylase from Escherichia coli. Biochemistry 36:13910-13918. [DOI] [PubMed] [Google Scholar]
- 17.Saxena, R., and P. K. Chakraborti. 2005. The carboxy-terminal end of the peptide deformylase from Mycobacterium tuberculosis is indispensable for its enzymatic activity. Biochem. Biophys. Res. Commun. 332:418-425. [DOI] [PubMed] [Google Scholar]
- 18.Shirley, K., Y. C. Sheng, J. S. John, and W. Alice. 1995. Design and use of mismatched and degenerate primers, p. 143-155. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, Z., X. Song, Y. Li, and W. Gong. 2004. Unique structural characteristics of peptide deformylase from pathogenic bacterium Leptospira interrogans. J. Mol. Biol. 339:207-215. [DOI] [PubMed] [Google Scholar]