Abstract
During growth on ethanolamine, Salmonella enterica synthesizes a multimolecular structure that mimics the carboxysome used by some photosynthetic bacteria to fix CO2. In S. enterica, this carboxysome-like structure (hereafter referred to as the ethanolamine metabolosome) is thought to contain the enzymatic machinery needed to metabolize ethanolamine into acetyl coenzyme A (acetyl-CoA). Analysis of the growth behavior of mutant strains of S. enterica lacking specific functions encoded by the 17-gene ethanolamine utilization (eut) operon established the minimal biochemical functions needed by this bacterium to use ethanolamine as a source of carbon and energy. The data obtained support the conclusion that the ethanolamine ammnonia-lyase (EAL) enzyme (encoded by the eutBC genes) and coenzyme B12 are necessary and sufficient to grow on ethanolamine. We propose that the EutD phosphotransacetylase and EutG alcohol dehydrogenase are important to maintain metabolic balance. Glutathione (GSH) had a strong positive effect that compensated for the lack of the EAL reactivase EutA protein under aerobic growth on ethanolamine. Neither GSH nor EutA was needed during growth on ethanolamine under reduced-oxygen conditions. GSH also stimulated growth of a strain lacking the acetaldehyde dehydrogenase (EutE) enzyme. The role of GSH in ethanolamine catabolism is complex and requires further investigation. Our data show that the ethanolamine metabolosome is not involved in the biochemistry of ethanolamine catabolism. We propose the metabolosome is needed to concentrate low levels of ethanolamine catabolic enzymes, to keep the level of toxic acetaldehyde low, to generate enough acetyl-CoA to support cell growth, and to maintain a pool of free CoA.
Salmonella enterica serovar Typhimurium LT2 (hereafter referred to as S. enterica) can use ethanolamine as a sole source of carbon, nitrogen, and energy (9, 27, 28). In this bacterium, the genetic information needed to use ethanolamine as a carbon, energy, and nitrogen source is encoded in a 17-gene operon known as the ethanolamine utilization operon (eut) (Fig. 1) (17, 35). The eut operon encodes proteins homologous to the shell proteins of the carboxysome structure found in cyanobacteria (2, 24). The carboxysome is proposed to concentrate carbon dioxide, the substrate for the ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) enzyme. Carboxysome-like structures have been visualized in S. enterica cells growing on 1,2-propanediol (5). We refer to these structures as metabolosomes to reflect their broad involvement in metabolism.
FIG. 1.
The ethanolamine utilization (eut) operon of S. enterica. The eutR gene encodes an AraC-type protein that activates eut operon transcription in response to AdoCbl and ethanolamine. eutR expression is controlled independently from the operon.
An understanding of the physiological role of metabolosomes in S. enterica is lacking. Penrod and coworkers recently demonstrated that the EutH protein facilitates the translocation of charged ethanolamine across the inner membrane at a low pH (23). It is not clear, however, whether ethanolamine is transported into the cytosol or directly into the metabolosome where the enzymatic machinery for the degradation of ethanolamine is presumably located.
In principle, the biochemistry underpinning ethanolamine catabolism is simple. Two reactions convert ethanolamine into acetyl coenzyme A (Ac-CoA). First, ethanolamine is converted to acetaldehyde and free ammonia by the adenosylcobalamin (AdoCbl; also known as coenzyme B12)-dependent ethanolamine ammonia-lyase (EC 4.3.1.7) (EAL) encoded by the eutBC genes of the operon (13). Second, acetaldehyde is oxidized to acetate and activated to Ac-CoA in a single step presumably catalyzed by the acetaldehyde dehydrogenase (EutE) enzyme (35). Ammonia generated by the EAL enzyme is used by S. enterica as a source of nitrogen, and Ac-CoA is used to make energy via the tricarboxylic acid cycle and glyoxylate bypass and is a building block for primary and secondary metabolism (34). Eut enzymes catalyze two additional reactions. First, acetaldehyde is reduced to ethanol in a reaction catalyzed by the EutG enzyme with an unclear involvement of the EutJ protein (23). Second, the Ac-CoA is converted to Ac-phosphate (Ac-P) by the EutD phosphotransacetylase (6). Eventually, Ac-P is used to conserve energy via substrate level phosphorylation catalyzed by acetate kinase, yielding ATP and acetate, which is excreted into the medium (34). Excreted acetate is later recaptured by Ac-CoA synthetase (Acs) (33).
The EAL enzyme is prone to suicidal inactivation by ethanol and other compounds (1, 36), and its reactivation requires EutA activity to exchange AdoCbl for hydroxycobalamin from the active site of EAL in the presence of Mg-ATP (21). The EutT enzyme synthesizes AdoCbl (7), which is sensed by the activator EutR protein, triggering transcription of the operon (26, 31).
The physiological role of the metabolosome during ethanolamine catabolism remains unclear, and the physiological reasons for its evolution remain an open question. The eutSMNLK genes are inferred by homology to encode the shell proteins of the metabolosome (2, 22, 32, 35). To further investigate the role of the metabolosome shell proteins, in-frame deletion mutants of the genes encoding these proteins were constructed and the abilities of the resulting strains to catabolize ethanolamine were assessed under various growth conditions. We have identified conditions that bypass the need for the metabolosome and for the EAL reactivase enzyme.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Salmonella strains used in this study were derivatives of S. enterica serovar Typhimurium LT2. The relevant genotypes of bacterial strains and plasmids used are listed in Table 1. S. enterica strains were grown in a minimal medium (4) supplemented with ethanolamine (30 mM), MgSO4 (1 mM), and l-methionine (0.5 mM), trace minerals (3), and vitamin B12 (cyanocobalamin) (150 nM). Lysogenic broth (LB) was used to cultivate both S. enterica and Escherichia coli strains. When added, oxidized l-glutathione (GSSG) was at 2.5 mM. Working concentrations of antibiotics in LB medium were 100 μg/ml for ampicillin and 12 μg/ml for chloramphenicol (Cm). When necessary, genes under the control of the ParaBAD promoter were induced for expression by addition of l-(+)-arabinose to a final concentration of 100 μM. Expression of genes cloned into vector pTAC-85 was induced by addition of IPTG (isopropyl-β-d-thiogalactoside) to a final concentration of 100 μM. Growth behavior was analyzed in liquid in 16- by 150-mm Kimax borosilicate tubes. Each tube contained 5 ml of fresh medium, which was inoculated with 75 μl (1.5% [vol/vol]) of a culture of an S. enterica strain grown for 24 h in LB medium containing the appropriate antibiotic. Tubes were shaken at 37°C. Growth was monitored as the increase in the absorbance at 650 nm on a Spectronic 20D spectrophotometer (Milton Roy Company, Rochester, NY). Growth behavior was also analyzed by using a microtiter dish (Becton-Dickinson) format using a computer-controlled Ultra Microplate Reader (Bio-Tek Instruments) equipped with the KC4 software package. The temperature of the incubation chamber was set at 37°C. Each well of the plate contained 190 μl of fresh medium supplemented with ethanolamine (30 mM), glycerol (0.5 mM), and ammonium chloride (30 mM), which was inoculated with 10 μl of a 24-h-old inoculum. Growth was monitored as the increase in the absorbance at 630 nm due to fixed-wavelength filters in the microplate reader. Data points were collected every 15 min; cultures were shaken for 870 s between readings.
TABLE 1.
Strains used in this study
| Strain or plasmid | Genotype | Reference/sourcea |
|---|---|---|
| Strains | ||
| TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| Derivatives of TR6583 | ||
| JE2261 | zfa-3648* Tn10* zfa-3649 (Δeut)b | Laboratory collection |
| JE4175 | pBAD30 bla+ | Laboratory collection |
| JE6692 | pKD46 bla+ | Laboratory collection |
| JE7963 | eutE1151::cat+ | |
| JE7973 | ΔeutE1164 | |
| JE7979 | ΔeutE1164/pBAD30 | |
| JE8032 | eutLK1152::cat+ | |
| JE8033 | eutMN1153::cat+ | |
| JE8064 | eutS1154::cat+ | |
| JE8070 | ΔeutLK1158 | |
| JE8071 | ΔeutMN1159 | |
| JE8072 | ΔeutS1160 | |
| JE8087 | eutA1155::cat+ | |
| JE8088 | eutBC1156::cat+ | |
| JE8089 | eutABC1157::cat+ | |
| JE8093 | ΔeutA1161 | |
| JE8094 | ΔeutBC1162 | |
| JE8095 | ΔeutABC1163 | |
| JE8102 | eutLK1152::cat+ ΔeutMN1159 | |
| JE8103 | ΔeutLK1158/pBAD30 | |
| JE8105 | ΔeutMN1159/pBAD30 | |
| JE8133 | ΔeutLK1158 ΔeutMN1159 | |
| JE8135 | ΔeutLK1158 ΔeutMN1159/pBAD30 | |
| JE8138 | ΔeutABC1163/pEUT34 | |
| JE8139 | ΔeutABC1163/pBAD33 | |
| JE8142 | zfa-3648* Tn10* zfa-3649 (Δeut)/pBAD33, pTAC-85 | |
| JE8144 | zfa-3648* Tn10* zfa-3649 (Δeut)/pEUT34, pTAC-85 | |
| JE8191 | ΔeutE1164/pEUT40 | |
| JE8217 | zfa-3648* Tn10* zfa-3649 (Δeut)/pEUT34, pEUT40 | |
| JE8298 | pBAD33 pTAC-85 | |
| JE8327 | zfa-3648* Tn10* zfa-3649 (Δeut)/pEUT42, pEUT40 | |
| JE8392 | pBAD33 cat+ | |
| JE8393 | ΔeutA1161/pEUT33 | |
| JE8395 | ΔeutA1161/pBAD33 | |
| JE8411 | ΔeutBC1162/pEUT42 | |
| JE8412 | ΔeutBC1162/pBAD33 | |
| JE8424 | pTAC-85 bla+ | |
| JE8425 | ΔeutE1164/pTAC-85 | |
| JE8426 | ΔeutS1160/pBAD30 | |
| E. coli strain DH5α | F−f80ΔlacZDM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | 15 |
| Plasmids | ||
| pBAD30 | bla+ | 14 |
| pBAD33 | cat+ | 14 |
| pET22-b | bla+ | Novagen |
| pTAC-85 | bla+ | 20 |
| pEUT31 | eutBC+ in pET22-b | G. Reed |
| pEUT33 | eutA+ in pBAD33 | |
| pEUT34 | eutABC+ in pBAD33 | |
| pEUT40 | eutE+ in pTAC-85 | |
| pEUT42 | eutBC+ in pBAD33 |
Unless otherwise indicated, all strains were constructed during the course of this work.
Asterisks indicate a Tn10-borne deletion.
Phage P22 transductions.
All transduction crosses were performed as previously described (12) with phage P22 HT105/1 int-210 (29, 30). Transductants were freed of phage as previously described (8).
Construction of nonpolar eut deletion mutants.
In-frame deletion mutants of eut genes constructed during this investigation were constructed by using a modification of the method described previously by Datsenko and Wanner (11). Primers were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Table 2. Briefly, the cat+ cassette was amplified with these primers by using template plasmid pKD3. Manipulations were performed in the S. entericarecipient strain JE6692. The insertion of the cat+ gene was confirmed by P22 transduction and PCR amplification. Removal of the cat+ gene was performed as described previously and the resulting deletion was confirmed by sequencing using BigDye (ABI PRISM) protocols (University of Wisconsin—Madison Biotechnology Center).
TABLE 2.
Primers used for the construction of in-frame, nonpolar eut deletion mutants
| Primer | Sequence |
|---|---|
| eutA5′KO | 5′-GTGAACACTCGCCAGCTACTGAGCGTCGGTATCGATATCGGCACCACCACCGTGTAGGCTGGAGCTGCTTC-3′ |
| eutA3′KO | 5′-TCAGGAAGGAAATGCGAGTGATTTCACCGTCACCGGCACAACCGATCCGCCCATATGAATATCCTCCTTAG-3′ |
| eutB5′KO | 5′-ATGAAACTAAAGACCACATTGTTCGGCAATGTTTATCAGTTTAAGGATGTAGTGTAGGCTGGAGCTGCTTC-3′ |
| eutC3′KO | 5′-TTAACGGGTCATGTTGATGCCGGACGCTTTCTGCTCCAGCATCCGTTTGGCCATATGAATATCCTCCTTAG-3′ |
| eutE5′KO | 5′-ATGAATCAACAGGATATTGAACAGGTGGTGAAAGCGGTACTGCTGAAAATGGTGTAGGCTGGAGCTGCTTC-3′ |
| eutE3′KO | 5′-TTATACAATGCGAAACGCATCCACCAGCACGCATCGACGCAGCCGCACAAACATATGAATATCCTCCTTAG-3′ |
| eutLK5′KO | 5′-ATGCCTGCATTAGATTTAATTCGACCTTCCGTGACTGCCATGCGCGTGATTGTGTAGGCTGGAGCTGCTTC-3′ |
| eutLK3′KO | 5′-TTAATTTTTGATGCGATAGCGACTACTGCGTTTACCGAACGCTCCGTCAGACATATGAATATCCTCCTTAG-3′ |
| eutMN5′KO | 5′-ATGGAAGCATTAGGAATGATTGAAACCCGGGGCCTGGTTGCGCTGATTGAGGTGTAGGCTGGAGCTGCTTC-3′ |
| eutMN3′KO | 5′-CTATTTATGGAAAACCACTTTCCCGCCAGCCACCACTTCATCGACGATGCCCATATGAATATCCTCCTTAG-3′ |
| eutS5′KO | 5′-ATGAATAAAGAACGCATTATTCAGGAATTTGTGCCGGGCAAACAGGTCACGGTGTAGGCTGGAGCTGCTTC-3′ |
| eutS3′KO | 5′-TTAACTTTTTGTTAACTCACACAGGGTAAAGTTTAATAATCGCCCCAGCCCCATATGAATATCCTCCTTAG-3′ |
Construction of a eutMNLK deletion mutant.
Strain JE8071 (ΔeutMN) was transduced to Cm resistance by using phage P22 grown on JE8032 as donor. Cmr transductants were freed of phage and screened by PCR to confirm the presence of the strain with the eutMN allele deleted. Construction of the quadruple deletion mutant from this point was as described above.
Plasmid constructions. (i) Plasmid pEUT31.
S. enterica eutBC+ cloned into the NdeI and BamHI site of pET22-b. A generous gift from G. Reed.
(ii) Plasmid pEUT33.
Allele eutA+ from strain TR6583 was amplified from the chromosome using the forward primer 5′-GAA AGA TGA GCT CGC CCA GGT GAA AA-3′ and the reverse primer 5′-CAA TGT GGT CTC TAG ATT CAT AAG TCG-3′. Bases underlined indicate the SacI and XbaI restriction sites engineered into the primers. The resulting 1.4-kb fragment was A-tailed and gel purified by using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). This product was ligated into plasmid pGEM-T-Easy (Promega, Madison, WI) according to manufacturer's instructions. The resulting plasmid contained the eutA+ gene under the control of the T7 promoter. This eutA+ plasmid was cut with restriction enzymes XbaI and SacI to release a 1.4-kb fragment that was ligated into plasmid pBAD33 (14) cut with the same enzymes. The resulting 6.7-kb plasmid was named pEUT33.
(iii) Plasmid pEUT34.
Plasmid pEUT31 was cut with SalI and XbaI to release a 2.3-kb fragment containing allele eutBC+ from Salmonella enterica. The eutBC+ fragment was ligated into plasmid pEUT33 cut with the same enzymes. The resulting 9-kb plasmid containing eutABC+ was named pEUT34.
(iv) Plasmid pEUT40.
The eutE+ allele from S. enterica was amplified from the chromosome using the forward primer 5′-CAT AAA TAG GAT CCA ACA TCA TGA ATC AAC AG-3′ and the reverse primer 5′-GTT CGT CGT GCG TCT AGA GTC ATC-3′. Underlined bases indicate the BamHI and XbaI restriction sites engineered into the primers. The resulting 1.4-kb fragment was A-tailed and gel purified by using the QIAquick gel extraction kit (QIAGEN). The product was ligated into plasmid pGEM-T-Easy (Promega) according to manufacturer's instructions. The resulting plasmid contained the eutE+ allele under the control of the T7 promoter. The eutE+ allele was cut out of this plasmid by using BamHI and SalI and ligated into the same sites of plasmid pTAC-85. The resulting 6.5-kb plasmid was named pEUT40.
(v) Plasmid pEUT42.
Plasmid pEUT31 was cut with SalI and XbaI to release a 2.3-kb fragment containing allele eutBC+ from Salmonella enterica. The eutBC+ fragment was ligated into pBAD33 cut with the same enzymes. This resulting 7.6-kb plasmid was named pEUT42.
TEM.
Strains used for transmission electron microscopy (TEM) were grown in minimal medium supplemented with the appropriate carbon source. When cultures attained an optical density (650 nm) of 1 or greater, 3 ml of culture was pelleted at 18,000 × g in a Microfuge 18 microcentrifuge (Beckman-Coulter) for 3 min. Samples were fixed in 2.5% glutaraldehyde-2.0% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h at room temperature. They were then postfixed in 1% osmium tetroxide for 1 h at room temperature in the same buffer. Next, the samples were dehydrated in a graded ethanol series, rinsed in propylene oxide and embedded in Spurr's epoxy resin. Following resin polymerization, the samples were cut into 60- to 90-nm sections for transmission electron microscopy using a Reichert-Jung Ultra-Cut E Ultramicrotome. Prior to transmission microscopic observation, the sectioned samples were stained with routing concentrations of ethanoic uranyl acetate and Reynolds' lead citrate. The specimens were observed by using a Philips CM 120 transmission electron microscope, and images were documented with a SIS Mega-View III digital camera.
RESULTS
Strains lacking EutS, EutMN, or EutLK proteins can grow on ethanolamine as the sole source of carbon and energy; a strain lacking EutMNLK cannot.
Although the conversion of ethanolamine to Ac-CoA is straightforward, the genetic information encoded within the eut operon suggests that physiological problems arise during ethanolamine catabolism. To investigate the role of the proteins exhibiting similarity to the carboxysome shell proteins of CO2-fixing prokaryotes, in-frame deletion mutants of the genes encoding these proteins were constructed, and the ability of the resulting strains to grow on ethanolamine was assessed. In-frame deletion mutants of the genes eutS, eutMN, and eutLK had little effect on ethanolamine-dependent growth. A 14-h doubling time was measured for cultures of ΔeutS, ΔeutL, ΔeutMN, and wild-type strains. In contrast, a strain with eutLKMN deleted failed to grow on ethanolamine (data not shown). Although the ΔeutS, ΔeutL, and ΔeutMN strains lacked one or two of the five proteins that comprise the ethanolamine metabolosome, they grew on ethanolamine suggesting that some type of structure was still formed by the remaining proteins. Recent crystallographic studies performed with carboxysome shell proteins CcmK2 and CcmK4 showed that these proteins form hexameric units of the polyhedral shell (16). It appears that the loss of one or two of the metabolosome proteins does not prevent the assembly of a variant of the metabolosome capable of functioning.
The simplest explanation for the lack of growth of the eutLKMN strain on ethanolamine was that in the absence of a functional metabolosome, ethanolamine catabolism did not occur because the concentration of the ethanolamine catabolic enzymes was diluted to the point of inefficiency.
TEM.
The growth phenotypes of strains lacking eutS, eutMN, eutLK, or eutMNLK suggested that a variant of the ethanolamine metabolosome capable of supporting growth on ethanolamine occurred in all of them except the EutMNLK-deficient strain. To address this possibility, we performed TEM studies with strains grown in 30 mM ethanolamine to induce synthesis of the metabolosome or 22 mM glycerol as the negative control. In all cases, cells were harvested from stationary-phase cultures (optical density of ≥1 at 650 nm). When grown in a minimal medium containing ethanolamine as the sole source of carbon and energy, a strain with a fully intact eut operon (JE8392 eut+) showed electron-dense structures likely to be the metabolosome shell (Fig. 2A). These structures were reminiscent of those observed by Bobik et al. during growth of S. enterica on 1,2-propanediol (5). These structures were absent when the same strain was grown on glycerol (Fig. 2B), and in a strain lacking the entire eut operon but harboring genes encoding ethanolamine ammonia-lyase and acetaldehyde dehydrogenase (JE8217 Δeut/peutABC+ peutE+). Strain JE8217 did exhibit, however, a large electron-dense region present in almost every cell viewed in the microscope field (Fig. 2C). Strains lacking one or more of the ethanolamine metabolosome shell proteins also exhibited these electron-dense zones. The nature of the latter is unknown.
FIG. 2.
Transmission electron micrographs of S. enterica strains grown on ethanolamine as carbon and energy source. A. JE8392 (eut+) grown in 30 mM ethanolamine viewed at 40,000-fold magnification. B. JE8392 grown in 22 mM glycerol viewed at 40,000-fold magnfication. C. Strain JE8217 (Δeut peutABC+ peutE+) grown in 30 mM ethanolamine, viewed at 53,000-fold magnification.
Complementation studies with strains lacking functions critical for ethanolamine catabolism.
If the idea of the metabolosome serving as a concentrator of catabolic enzymes were correct, one would predict that raising the level of the ethanolamine catabolic enzymes would circumvent the need for the metabolosome. To test this possibility, we constructed in-frame deletion mutants of genes encoding the ethanolamine ammonia-lyase, its reactivase and acetaldehyde dehydrogenase, and performed complementation analyses with plasmids carrying the appropriate genes to verify that the deletion mutants generated did not have any negative effect on the expression of downstream genes.
A culture of a strain lacking EAL (EutBC) and the EAL reactivase (EutA) protein (ΔeutABC, JE8139) failed to grow on ethanolamine as carbon and energy source (Fig. 3A). Introduction of plasmid pEUT34 (ParaBAD-eutABC+) into strain JE8095 and inclusion of arabinose (100 μM) in the medium restored growth on ethanolamine with a doubling time of 14 h and the culture reached full density (Fig. 3A). Similarly, a culture of a strain carrying an in-frame deletion mutant of eutBC (JE8412) behaved like strain JE8139 (ΔeutABC) (Fig. 3B). Introduction of plasmid pEUT42 (ParaBAD-eutBC+) into strain JE8094 restored growth with a doubling time of 22 h, and the culture reached full density (Fig. 3B).
FIG. 3.
Complementation studies. In all panels VOC means vector-only control. A. A strain lacking the eutABC genes encoding the EAL reactivase EutA and the large and small subunits of EAL, respectively, was tested for the ability to use ethanolamine as sole carbon and energy source. Expression of genes encoded by plasmid pEUT34 (ParaBAD-eutABC+) was induced with l-(+)-arabinose (JE8138) (100 μM) (solid diamonds); empty cloning vector pBAD33 was introduced into JE8095 (ΔeutABC1163) and the growth behavior of the resulting strain (JE8139) was also analyzed in the presence of l-(+)-arabinose (100 μM) (solid squares). Strain JE8392 (eut+/pBAD33) (solid circles) was used as a positive control. B. Strain JE8094 (ΔeutBC) was tested for its ability to grow on ethanolamine. Strain JE8411 (JE8094/pEUT42 (ParaBAD-eutBC+) was grown on ethanolamine in the absence of l-(+)-arabinose (solid diamonds). Strain JE8412 (JE8094/pBAD33) (solid squares) was used negative control. Strain JE8392 (eut+/pBAD33) (solid circles) was used as positive control. C. Strain JE7973 (ΔeutE) was tested for the ability to grow on ethanolamine. Strain JE8191 (JE7973/pEUT40 Ptac-eutE+) was grown in ethanolamine medium containing 100 μM IPTG (solid diamonds). Strain JE8425 (JE7973/pTAC-85) (solid squares) was used as the negative control, and strain JE8424 (TR6583/pTAC-85) (solid circles) was used as the positive control.
An in-frame deletion mutant of the gene encoding the putative aldehyde dehydrogenase eutE was also constructed. Strain JE8425 (ΔeutE) failed to grow on ethanolamine (Fig. 3C). Introduction of plasmid pEUT40 (Ptac-eutE+) into strain JE7973 and inclusion of IPTG (100 μM) in the medium restored growth of strain JE7973 on ethanolamine with a doubling time of 10 h, and the culture reached full density (Fig. 3C).
Collectively, the results described above make two points. First, they confirmed that the deletions of eutABC, eutBC, and eutE were in-frame (as suggested by DNA sequencing), since they could be complemented by the missing function(s) on a plasmid. Second, they showed that the eutABCE functions were critical to ethanolamine catabolism.
The metabolosome is dispensable when ethanolamine ammonia-lyase (EutBC) and acetaldehyde dehydrogenase (EutE) enzyme levels are elevated.
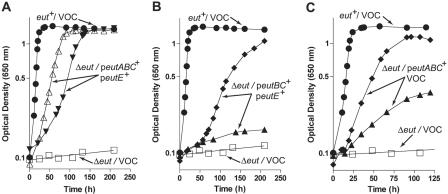
Once reliable mutant strains and complementing plasmids were available, we tested the idea that elevated levels of ethanolamine catabolic enzymes would bypass the need for the metabolosome. For this purpose, two compatible plasmids, pEUT34 (ParaBAD-eutABC+) and pEUT40 (Ptac-eutE+), were moved into strain JE2261 carrying a chromosomal deletion of the entire eut operon (Δeut), and growth of the resulting strain on ethanolamine was monitored over time.
In the absence of the metabolosome, plasmids pEUT34 (ParaBAD-eutABC+) and pEUT40 (Ptac-eutE+) restored growth of strain JE2261 (Δeut) on ethanolamine, reaching full density with a doubling time of 67 h (Fig. 4A). The addition of glutathione to the medium shortened the lag and reduced the doubling time to 56 h (Fig. 4A). When plasmid pEUT42 (ParaBAD-eutBC+) was substituted for plasmid pEUT34 (ParaBAD-eutABC+), the resulting strain (JE8327) grew only when the medium was supplemented with glutathione with a doubling time of 100 h (Fig. 4B). In all instances, control experiments showed that the stimulatory effect of glutathione was not due to its use as a carbon and energy source (data not shown).
FIG. 4.
Minimal functions for the ethanolamine catabolism. For all panels, VOC means vector-only control, strain JE8142 (Δeut/pBAD33 pTAC-85) was used as the negative control (open squares), and strain JE8298 (eut+/pBAD33 pTAC-85) (solid circles) was used as the positive control. When added, arabinose or IPTG was at 100 μM. A. Strain JE2261 (Δeut) was transformed with plasmids pEUT34 (ParaBAD-eutABC+) and pEUT40 (Ptac-eutE+) and phenotypes assessed. For growth on ethanolamine as the sole source of carbon and energy, 2.5 mM oxidized GSSG (open triangles) or no GSSG (inverted solid triangles) was added to the medium. B. Strain JE2261 (Δeut) was transformed with plasmids pEUT42 (ParaBAD-eutBC+) and plasmid pEUT40 (Ptac-eutE+), and phenotypes were assessed. For growth on ethanolamine as the sole source of carbon and energy: plus GSSG (solid diamonds) or without GSSG (solid triangles) addition to the medium. C. Strain JE2261 (Δeut) was transformed with plasmids pEUT34 (ParaBAD-eutABC+) and pTAC-85 and phenotypes were assessed: plus GSSG (solid diamonds) or without GSSG (solid triangles) addition to the medium.
EutBC reactivase (EutA) is conditionally required, and nonspecific acetaldehyde dehydrogenases partially compensate for the lack of EutE.
Under some growth conditions, the absence of EutE protein was partially compensated by nonspecific acetaldehyde dehydrogenase activities. For example, when plasmid pEUT34 (ParaBAD-eutABC+) was moved into strain JE2261 (Δeut) but plasmid pEUT40 (Ptac-eutE+) was not, the resulting strain (JE8144) struggled to grow (doubling time, 167 h) and the cell density of the culture was poor (∼0.35) (Fig. 4C). The absence of EutE protein may lead to the accumulation of acetaldehyde to toxic levels, hence arresting growth. This idea was consistent with the effect obtained by the effect of the addition of glutathione to the medium, which drastically stimulated growth, allowing strain JE8144 to reach near full density after 96 h of incubation (doubling time, 58 h) (Fig. 4C). These results revealed the existence of alternative nonspecific acetaldehyde dehydrogenases that can compensate for the absence of EutE. Possible explanations for the stimulatory effect of glutathione are discussed below.
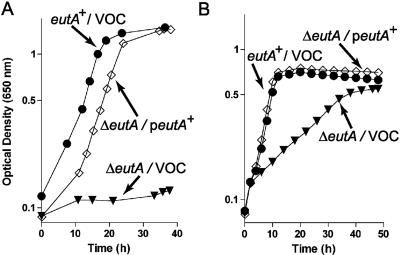
We also found conditions under which EutA activity was not required for growth on ethanolamine. Strain JE8395 (ΔeutA) displayed a severe growth defect on ethanolamine (Fig. 5A). These results suggested that under the aerobic conditions used, EAL was deactivated and EutA activity was needed to keep EAL functional. Introduction of plasmid pEUT33 (ParaBAD-eutA+) and inclusion of arabinose (100 μM) in the medium restored wild-type growth of strain JE8093 (Fig. 5A), indicating that the plasmid-encoded EutA was functional, and that the eutA mutation in the chromosome did not affect the expression genes downstream of it. It is noteworthy that when strain JE8093 (ΔeutA) was inoculated into ethanolamine medium in a microtiter plate, the culture grew to full density with a doubling time of 53 h, compared to 10 h for the strain carrying plasmid pEUT33 (ParaBAD-eutA+) (Fig. 5B).
FIG. 5.
Conditional requirement of eutA function. Strain JE8093 (ΔeutA) was transformed with either plasmid pEUT33 [gene expression induced with 100 μM l-(+)-arabinose] (open diamonds) or pBAD33 (solid inverted triangles). Strain JE8392 (solid circles) was used as a positive control. In both panels, VOC means vector-only control. A. The experiment was performed in 5-ml cultures in 16- by 150-mm borosilicate tubes. B. The experiment was performed in a microtiter plate with medium supplemented with ethanolamine (30 mM) and glycerol (0.5 mM). Glycerol was needed to prime cell growth; in its absence the onset of exponential growth was drastically delayed. Strain JE8142 (Δeut/pBAD33 pTAC-85) was used as the negative control.
DISCUSSION
The availability of a large collection of in-frame deletion mutant strains and plasmids carrying functional eut alleles allowed us to query the role of the metabolosome multimolecular complex during growth of Salmonella enterica on ethanolamine.
Results from experiments reported here clearly show that a strain of S. enterica lacking the genes encoding the ethanolamine metabolosome can grow on ethanolamine as the sole source of carbon and energy as long as ethanolamine ammonia-lyase (EutBC) and acetaldehyde dehydrogenase (but not necessarily EutE) enzymes are present in the cell at elevated levels. The metabolosome is not involved in the biochemistry of ethanolamine catabolism and may represent a strategy for reducing the level of enzymes needed to support growth on a poor carbon and energy source while serving as a containment structure for controlling the level of toxic acetaldehyde. By using the ethanolamine metabolosome, S. enterica may effectively reduce the level of acetaldehyde by concentrating within the metabolosome two enzymes that would rapidly consume acetaldehyde, i.e., acetaldehyde dehydrogenase (EutE) and alcohol dehydrogenase (presumably EutG).
The need for EutA reactivase function can be bypassed by glutathione and reduced-oxygen conditions.
Given the known role of EutA in maintaining EAL activity (21), it was not unexpected to see the lack of growth of an EutA-deficient strain on ethanolamine (Fig. 5A). It was very surprising, however, to see that the effect of the lack of EAL reactivase could be largely circumvented by glutathione (Fig. 4B). It was not surprising to learn that the EAL enzyme was more prone to inactivation under aerobic conditions than under reduced-oxygen (microtiter dish format) growth conditions (Fig. 5A versus B), since adenosylcoblamin (the coenzyme of EAL) can be readily inactivated by oxidation to Co2+ cobalamin (18). The meaning of these results may be explained by studies of EAL inactivation under aerobic and microaerophilic conditions as a function of substrate limitation.
The role of glutathione.
The requirement for glutathione during growth of S. enterica on ethanolamine was reported a decade ago, when the activity of EAL in a glutathione-deficient gshA strain was noted to be >90% reduced relative to the activity of the EAL enzyme in the gshA+ strain (25). Although it is tempting to speculate that glutathione may be involved in the EutA reaction, a role for this tripeptide in EutA-dependent EAL reactivation was not evident from the results recently reported by Toraya and coworkers (21).
The role of glutathione in ethanolamine catabolism may be more complex than previously thought. The striking positive effect of glutathione on the growth of an EutE-deficient strain on ethanolamine was different than the effect on the EutA-deficient strain, since the EutE-deficient strain contained a wild-type eutA allele. It is possible that in the case of the EutE-deficient strain, glutathione enhances toxic acetaldehyde quenching (10, 19) or the substrate for the alternative acetaldehyde dehydrogenase is glutathionyl hemithioacetal. Furthermore, GSH may be a required cofactor for the alternative acetaldehyde dehydrogenase. The role of glutathione in ethanolamine catabolism requires further investigation.
A new perspective on ethanolamine catabolism and the role of the metabolosome.
Fig. 6 shows our current model for the catabolism of ethanolamine. Ethanolamine enters the cell either by diffusion or via the EutH permease (23), depending on its protonation state. Within the metabolosome multimolecular complex (EutSMNLK), ethanolamine is converted to Ac-CoA via acetaldehyde by the concerted actions of EAL and EutE enzymes.
FIG. 6.
Model for ethanolamine catabolism in S. enterica. The metabolosome representation is not to scale.
We hypothesize that the rate of Ac-CoA synthesis in the metabolosome may exceed the rate of Ac-CoA consumption, leading to the accumulation of Ac-CoA and probably acetaldehyde. Two safety valves appear to be built into the system to avoid problems created by flux variations. One is the putative EutG alcohol dehydrogenase, which would detoxify acetaldehyde by reducing it to ethanol. The latter, not redox balance, is likely to be the role of EutG, as S. enterica respires ethanolamine aerobically by using the electron transport chain. Although at present the kinetic analysis of EutG is lacking and the protein has not been studied in any detail, we predict that EutG will be a very efficient enzyme, so the level of toxic acetaldehyde can be maintained low.
The second safety valve is EutD, which converts Ac-CoA to Ac-P (6). The latter is used to conserve energy by the substrate level phosphorylation reaction catalyzed by acetate kinase (Ack) with the concomitant excretion of acetate, which is later recaptured by the Ac-CoA synthetase (33). Although the EutD phosphotransacetylase may play additional a role in ATP generation, we see EutD as an efficient strategy to maintain free CoA levels from becoming limiting.
We view the ethanolamine metabolosome as a strategy to concentrate low levels of ethanolamine catabolic enzymes. If the ethanolamine metabolosome were not made, the cell would have to synthesize large amounts of Eut enzymes, increasing the level of acetaldehyde generated by EAL in the absence of a containment structure. We propose that the metabolosome is also an effective means to concentrate acetaldehyde for the EutG and EutE enzymes that use it as a substrate. The same argument applies to the buildup of Ac-CoA. In other words, by confining ethanolamine catabolism to the metabolosome, the cell can swiftly respond to increases in the concentrations of these intermediates of the pathway beyond tolerable levels. We posit that the need to respond to even low levels of these intermediates may have been the selective pressure for the evolution of the EutD phosphotransacetylase, which has much higher affinity for its substrates and a higher catalytic efficiency than the housekeeping phosphotransacetytlase (Pta) enzyme (6), hence EutD is poised to maintain the level of Ac-CoA low. It is unclear why EutD activity is necessary, since the Pta enzyme present in the cell could replace EutD in the conversion of Ac-CoA to Ac-P. We speculate that Ac-CoA levels during ethanolamine catabolism are kept low, since in the absence of EutD function, acetate excretion is abolished in spite of the presence of the Pta enzyme (34), suggesting that the levels of Ac-CoA are not high enough to serve as the substrate for Pta.
Acknowledgments
This work was supported by grant GM40313 to J.C.E.-S.
We thank George Reed for plasmids. We thank Ben August and Randall Massey at the University of Wisconsin—Madison medical school electron microscopy facility for assistance with TEM imaging.
REFERENCES
- 1.Abend, A., V. Bandarian, R. Nitsche, E. Stupperich, J. Retey, and G. H. Reed. 1999. Ethanolamine ammonia-lyase has a “base-on” binding mode for coenzyme B12. Arch. Biochem. Biophys. 370:138-141. [DOI] [PubMed] [Google Scholar]
- 2.Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buan, N. R., S. J. Suh, and J. C. Escalante-Semerena. 2004. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J. Bacteriol. 186:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 9.Chang, G. W., and J. T. Chang. 1975. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254:150-151. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, K. O., G. Witz, and C. M. Witmer. 1987. Mutagenicity and toxicity of several α,β-unsaturated aldehydes in the Salmonella typhimurium mutagenicity assay. Environ. Mut. 9:289-295. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A Manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Faust, L. P., J. A. Connor, D. M. Roof, J. A. Hoch, and B. M. Babior. 1990. Cloning, sequencing and expression of the genes encoding the adenosylcobalamin-dependent ethanolamine ammonia-lyase of Salmonella typhimurium. J. Biol. Chem. 265:12462-12466. [PubMed] [Google Scholar]
- 14.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 16.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 17.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lexa, D., J. M. Saveant, and J. Zickler. 1977. Electrochemistry of vitamin B12. 2. Redox and acid-base equilibria in the B12a/B12r system. J. Am. Chem. Soc. 99:2786-2790. [DOI] [PubMed] [Google Scholar]
- 19.Marinari, U. M., M. Ferro, L. Sciaba, and A. M. Bassi, G. Brambilla. 1984. DNA-damaging activity of biotic and xenobiotic aldehydes in chinese hamster ovary cells. Cell Biochem. Funct. 2:243-248. [DOI] [PubMed] [Google Scholar]
- 20.Marsh, P. 1986. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 14:3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori, K., R. Bando, N. Hieda, and T. Toraya. 2004. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J. Bacteriol. 186:6845-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orus, M. I., M. L. Rodriguez, F. Martinez, and E. Marco. 1995. Biogenesis and ultrastructure of carboxysomes from wild type and mutants of Synechococcus sp. strain PCC 7942. Plant Physiol. 107:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penrod, J. T., C. C. Mace, and J. R. Roth. 2004. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J. Bacteriol. 186:6885-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, G. D., S. M. Howitt, K. Harrison, and M. R. Badger. 1993. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol. 175:2871-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rondon, M. R., R. Kazmierczak, and J. C. Escalante-Semerena. 1995. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J. Bacteriol. 177:5434-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roof, D. M., and J. R. Roth. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmieger, H. 1971. A method for detection of phage mutants with altered transduction ability. Mol. Gen. Genet. 100:378-381. [DOI] [PubMed] [Google Scholar]
- 30.Schmieger, H., and H. Bakhaus. 1973. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants). Mol. Gen. Genet. 120:181-190. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard, D. E., and J. R. Roth. 1994. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J. Bacteriol. 176:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 33.Starai, V. J., and J. C. Escalante-Semerena. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell Mol. Life Sci. 61:2020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starai, V. J., J. Garrity, and J. C. Escalante-Semerena. 2005. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD), and acetate recapture depends on acetyl-CoA synthetase activity. Microbiology 151:3793-3801. [DOI] [PubMed] [Google Scholar]
- 35.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutj eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toraya, T. 2000. Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell Mol. Life Sci. 57:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]