Abstract
Bacillus subtilis responds to phosphate starvation stress by inducing the PhoP and SigB regulons. While the PhoP regulon provides a specific response to phosphate starvation stress, maximizing the acquisition of phosphate (Pi) from the environment and reducing the cellular requirement for this essential nutrient, the SigB regulon provides nonspecific resistance to stress by protecting essential cellular components, such as DNA and membranes. We have characterized the phosphate starvation stress response of B. subtilis at a genome-wide level using DNA macroarrays. A combination of outlier and cluster analyses identified putative new members of the PhoP regulon, namely, yfkN (2′,3′ cyclic nucleotide 2′-phosphodiesterase), yurI (RNase), yjdB (unknown), and vpr (extracellular serine protease). YurI is thought to be responsible for the nonspecific degradation of RNA, while the activity of YfkN on various nucleotide phosphates suggests that it could act on substrates liberated by YurI, which produces 3′ or 5′ phosphoribonucleotides. The putative new PhoP regulon members are either known or predicted to be secreted and are likely to be important for the recovery of inorganic phosphate from a variety of organic sources of phosphate in the environment.
When Bacillus subtilis encounters phosphate starvation stress, it responds by inducing groups of genes that function to restrict the metabolic consequences of the limited supply of this essential nutrient. These groups of genes are collectively referred to as the phosphate (Pho) stimulon. The phosphate stimulon includes at least two well-described regulons, namely, the sigma B (σB) general stress regulon and the phosphate starvation-specific PhoP regulon. When B. subtilis encounters phosphate starvation, genes of the SigB regulon are induced by the alternative sigma factor, σB, and genes of the PhoP regulon are either induced or repressed by activated PhoP (namely, PhoP∼P).
The σB general stress regulon contains >100 genes (58, 64). These genes provide a nonspecific response to stress by encoding proteins that protect the DNA, membranes, and proteins from the damaging effects of stress. Proteins induced by σB help the cell to survive potentially harmful environmental conditions, such as heat, osmotic, acid, or alkaline shock (6, 21, 23, 26). This protective function is thought to be particularly important in maintaining the viability of nongrowing cells.
The PhoP regulon currently consists of 34 members. Six operons (phoPR [56, 60], phoB-ydhF [7, 14], pstSAC-pstBA-pstBB [3, 67], phoD-tatAD [7, 19], resABCDE [10], and tuaABCDEFGH [40, 72]) and five monocistronic genes (glpQ [7], phoA [30, 31], tatCD [34], ykoL [60], and yttP [62]) are induced and two operons (tagAB and tagDEF (39) are repressed in response to phosphate starvation. phoA and phoB encode alkaline phosphatases (APases) which facilitate the recovery of inorganic phosphate (Pi) from organic sources (11, 30); phoD encodes a phosphodiesterase/APase, putatively involved in cell wall teichoic acid turnover, and is secreted exclusively by the twin arginine transporter (tatCD) pathway (34); the pstSAC-pstBA-pstBB operon encodes a high-affinity phosphate transporter for the uptake of Pi at low Pi concentrations (3, 67); glpQ encodes a glycerophosphoryl diester phosphodiesterase involved in the hydrolysis of deacylated phospholipids (7); the tuaABCDEFGHoperon encodes teichuronic acid biosynthesis; the tagAB and tagDEF operons encode polyglycerolteichoic acid biosynthesis (7, 39, 49); and the phoPR and resABCDE operons encode two-component signal transduction systems PhoP-PhoR and ResD-ResE (29, 30, 37, 40, 49, 51). The functions of three putative Pho regulon genes (ydhF, ykoL, and yttP) are currently unknown (7, 62, 69).
The induction or repression of PhoP regulon genes is mediated by the binding of PhoP∼P to Pho box sequences: direct repeats of TT(A/T/C)ACA with a 5-bp ± 2-bp spacer (18). For efficient binding at promoters where PhoP∼P is essential and sufficient for promoter function, four TT(A/T/C)ACA-like sequences with an 11-bp periodicity on the coding strand between nucleotides −60 and −20 relative to the transcription start site are required. Deletion of a single repeat from the core binding region severely reduces PhoP binding and transcriptional activation in vivo and in vitro (40). In the case of genes induced by PhoP∼P, the PhoP-binding sites are located on the coding strand of the promoter region. Repressed genes usually have consensus sequences on the noncoding strand (39), although the resD promoter is an exception (75).
Recently, we and others have proposed the inclusion of additional genes in the PhoP regulon. Ogura and coworkers (54) analyzed the composition of the PhoP regulon by DNA microarray analysis, after overproduction of PhoP. They identified yycP and yjdB as potential members of the Pho regulon, although they were unable to confirm this observation by lacZ reporter gene studies. Prágai and Harwood (62) putatively identified two additional members of the PhoP regulon, namely, yhbH and yhaX. These genes had the same expression characteristics, since their induction in response to phosphate starvation was dependent on PhoPR and the sporulation-specific sigma factor, SigE. However, more recent studies (60) failed to demonstrate binding of PhoP to the control region of yhaX. This indicates either that yhaX is activated by PhoP indirectly via another regulatory pathway or that binding of PhoP∼P to the yhaX promoter region requires an additional factor(s) (60). Consequently, we have not included these genes as members of the PhoP regulon. PhoP∼P is known to function with EσE holoenzyme, since it enhances transcription at the SigE-dependent PE2 promoter of phoPR. Paul and colleagues (56) have shown that autoregulation of phoPR involves the up-regulation of this lowly expressed promoter as well as the more highly expressed SigA promoters.
To gain a global perspective on the transcriptional responses of B. subtilis to phosphate starvation, we monitored genome-wide changes in gene expression during phosphate starvation using DNA macroarrays. By comparing the response of the wild-type strain to those of the sigB and phoR mutants, potential new members of the PhoP regulon were identified and subsequently analyzed using a combination of Northern hybridization and reporter gene analyses. The data represent the most comprehensive analysis of the response of B. subtilis to Pi starvation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are listed in Table 1. Strains were grown in Luria-Bertani (LB) medium, low-phosphate medium (LPM) or high-phosphate medium (HPM) (63). The concentration of phosphate was 0.42 mM in LPM and 5.0 mM in HPM. When required, the concentrations of antibiotics were, per milliliter, 0.3 μg of erythromycin, 25 μg of lincomycin, 12.5 μg of tetracycline, and 5 μg of chloramphenicol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | 4 |
| ML6 | trpC2 sigBΔHindIII-EcoRV::Cmr | 32 |
| 168-PR | trpC2 phoRΔBalI::Tcr | 61 |
| 168-MLPR | trpC2 sigBΔHindIII-EcoRV::CmrphoRΔBalI::Tcr | 61 |
| BFS1243 | trpC2 yurI::Emr | MICADOa |
| YFKNd | trpC2 yfkN::Emr | MICADO |
| YBCOdd | trpC2 ybcO::Emr | MICADO |
| BFS436 | trpC2 yjdB::Emr | MICADO |
| BFA1234-PR | trpC2 phoRΔBalI::TcryurI::Emr | This study |
| YFKNd-PR | trpC2 phoRΔBalI::TcryfkN::Emr | This study |
| YBCOdd-PR | trpC2 phoRΔBalI::TcrybcO::Emr | This study |
| BFA1243-ML | trpC2 sigBΔHindIII-EcoRV::CmryurI::Emr | This study |
| YFKNd-ML | trpC2 sigBΔHindIII-EcoRV::CmryfkN::Emr | This study |
| YBCOdd-ML | trpC2 sigBΔHindIII-EcoRV::CmrybcO::Emr | This study |
| YBCOdd-MLPR | trpC2 sigBΔHindIII-EcoRV::CmrphoRΔBalI::TcrybcO::Emr | This study |
| E. coli strain XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 lac [F′ proAB+lacIqlacZΔM15 Tn10 (Tcr)] | Stratagene Europe |
| Plasmids | ||
| pMUTIN4 | Apr EmrspoVG-lacZ Pspac (8.61 kb) | 79 |
| pBluescript II KS(+) | Cloning vector, Apr | Stratagene Europe |
| pphoA | pBluescript II KS(+) containing a 978-bp insert of phoA Apr | This study |
| pyheK | pBluescript II KS(+) containing a 485-bp insert of yheK Apr | This study |
MICADO, Microbial Advanced Database Organization.
DNA manipulations and general methods.
Extraction of plasmid and chromosomal DNA, restriction endonuclease digestion, agarose gel electrophoresis, transformation of Escherichia coli cells, and PCR and bioinformatical analyses were carried out as described previously (61, 63). Enzymes, molecular size markers, and deoxynucleotides were purchased from Roche Diagnostics, Ltd. (Lewes, United Kingdom) and Amersham Pharmacia Biotech, Ltd. (Little Chalfont, United Kingdom).
Transcriptome analysis by DNA macroarray hybridization.
Total RNA was extracted from the wild-type strain and phoR and sigB mutants before, during (T0), and after entry into the stationary growth phase, which was provoked by phosphate starvation. Cell harvesting, preparation of RNA, synthesis of radioactively labeled cDNA, and hybridization of B. subtilis macroarrays (Sigma-Genosys, The Woodlands, Tex.) were performed as described by Eymann and coworkers (22). Each analysis was carried out twice, using two independently isolated RNA preparations and two different array batches. The arrays were exposed to a phosphorescent screen which was subsequently scanned with a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) at a resolution of 50 μm and a 16-bit color depth. For quantification of the hybridization signals and background subtraction, ArrayVision software (version 5.1; Imaging Research, Ontario, Canada) was used. Calculation of normalized intensity values of the individual spots was performed using the overall-spot-normalization function of ArrayVision. To avoid extreme expression ratios for genes close to or below the detection limit, genes with signal intensity values corresponding to less than twice the background were not counted in the analysis. Subsequently, the average of the normalized intensity values of the duplicate spots of each gene was used to calculate the expression level ratios. Data analysis (statistical analysis, visualization, and generation of lists) was performed using the GeneSpring software (version 4.2.12; Silicon Genetics, Redwood City, CA).
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE2667.
Determination of outliers.
Genes whose expression differed significantly in response to phosphate starvation were determined by two independent methods. The data were analyzed by a method similar to the previously described iterative outlier analysis (12, 41). In cases where multiple hybridization reactions were carried out using the same RNA sample, the data were averaged and treated as a single value. The ratios of the values obtained before (control) and after phosphate starvation were transformed to log2. The geometric mean and standard deviation of the entire population were then calculated: any gene which had a ratio of more than 2.5 standard deviations away from the mean was considered an outlier, i.e., significantly induced or repressed. These outlier genes were subsequently removed from the population, and the means and standard deviations were recalculated. Again, any gene more than 2.5 standard deviations away from the mean was considered an outlier. This process was repeated until few or no outliers were detected. In these experiments generally three to four iterations were needed to identify all outliers in the population. The lists of outliers from the two independently grown and prepared samples were compared, and only those genes that were considered to have changed significantly in both data sets were considered further.
Northern blotting.
Total RNA was extracted from B. subtilis strains (168, 168-PR, and ML6) with phenol (42). Northern blot analysis was performed according to the manufacturer's instructions, using 5 μg of total RNA per lane (DIG Northern Starter kit; Roche Diagnostics GmbH, Mannheim, Germany). Digoxigenin (DIG)-labeled probes for yfkN, yjdB, yurI, and vpr were obtained by in vitro transcription from T7 RNA promoter-containing PCR products of the respective genes by T7 RNA polymerase. Synthesis of the templates by PCR was performed using the following pairs of oligonucleotides: for vpr forward (FOR), 5′-CAGCTATTCTCAGGCTTC-3′; for vpr reverse (REV), 5′-CTAATACGACTCACTATAGGG AGAGCTTAATCGTTGGGAC-3′; for yfkN FOR, 5′-AGGTGCAGGATATCGTAG-3′; for yfkN REV, 5′-CTAATACGACTCACTATAGGGAGCCTGATATGTGACACCG-3′); for yjdB FOR, 5′-CTTTATCGATTTCTGCGT-3′; for yjdB REV, 5′-CTAATACGACTCACTATAGGGAGAACAAAGTAATCGTGGCT-3′; for yurI FOR, 5′-CGTATTATCAGCGGACAC-3′; and for yurI REV, 5′-CTAATACGACTCACTATAGGGAGCATTCGAGCAGGACAGA-3′.
Hybridization probes specific for phoA and yheK were DIG labeled by in vitro transcription from HindIII-linearized plasmid pPhoA and pYheK, respectively, with T7 RNA polymerase. The plasmids contained sequences internal to the phoA and yheK genes amplified by PCR with primers phoA FOR (5′-CGCGAAGCTTGCCTTACATAAGAGCCT-3′) and phoA REV (5′-CGCGGATCCTTGATTTCTTCAGATGTG-3′) for pPhoA and primers yheK FOR (5′-CGCGAAGCTTTTCGGGTAGTAGTGTTG-3′) and yheK REV (5′-CGCGGATCCTCCGA ATATTAGCTTTTT-3′) for pYheK. The primers included either BamHI or HindIII restriction sites at their 3′ ends (underlined). The PCR products were cleaved with BamHI and HindIII and inserted into pBluescript KSII linearized with the same enzymes.
Enzyme assays.
Cultures grown in HPM overnight were diluted 500-fold in fresh LPM or HPM. The cultures were grown at 37°C with agitation at 220 rpm. Samples were collected at hourly intervals for the determination of the optical density at 600 nm (OD600), the APase activity (49), and the β-galactosidase activity (45), as described previously (62). The concentration of Pi in the medium was assayed (61) after removal of the cells by filtration through a filter of 0.45-mm pore size.
Genetic analysis.
The whole genome was searched with the sequence pattern TTHACA3-7TTHACA (H = A/C/T) using the Pattern Match function of SubtiList (http://genolist.pasteur.fr/SubtiList/) (47). Targets were reported only if they were located upstream of gene boundaries within 150 bp of the start codon. A deviation of 1 bp was allowed in each repeat of the above sequence.
Final evaluation of the macroarray data included the consideration of putative operon structures derived from the genome sequence as well as previously known operons. Genes induced significantly were analyzed for their transcriptional organization using the SubtiList database (http://genolist.pasteur.fr/SubtiList/) and Artemis software (www.sanger.ac.uk/).
RESULTS
A combination of enzymatic, proteomic, and reporter gene studies have shown that when B. subtilis encounters phosphate starvation, genes of the phosphate stimulon are either induced or repressed. To characterize the changes in gene expression with respect to the onset of phosphate starvation, global transcriptome analyses were performed using DNA macroarrays containing 4,107 protein-encoding genes of the B. subtilis genome (36). In addition to the wild-type B. subtilis strain 168, sigB- and phoR-null mutants were also analyzed to facilitate the tentative assignment of genes to the SigB and PhoR regulons.
Genome-wide analysis of the response of B. subtilis to phosphate starvation.
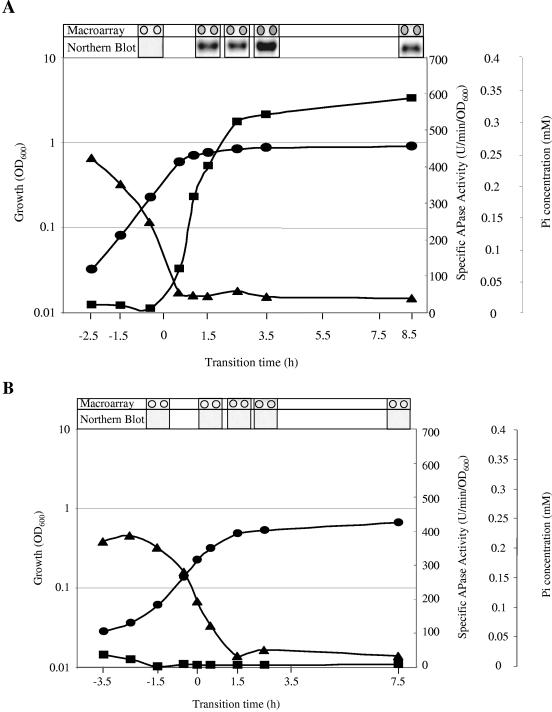
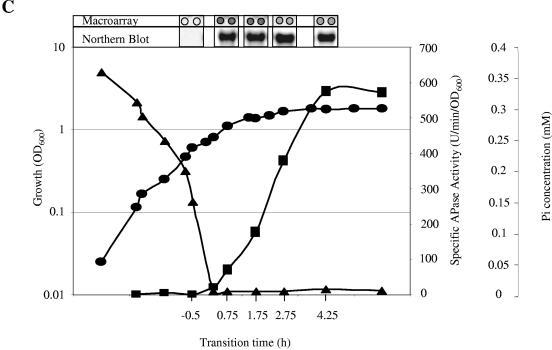
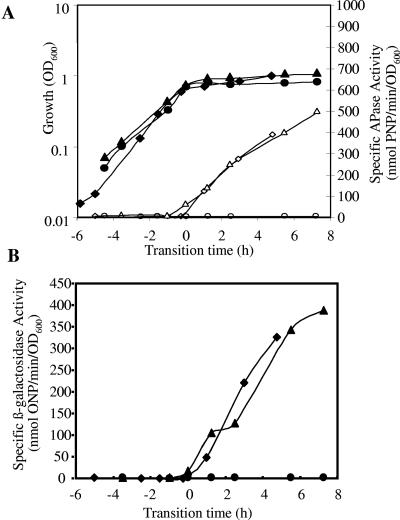
B. subtilis strain 168 (wild type) was grown in LPM, which facilitates phosphate starvation-induced entry into stationary phase. The concentration of inorganic phosphate in the culture medium and APase activity of the culture were monitored throughout growth, confirming the exhaustion of Pi from the medium and the concomitant induction of the PhoP regulon (Fig. 1). Samples of culture were removed before, during, and after the onset of phosphate starvation and used for the extraction of total cell RNA.
FIG. 1.
Representative growth curves and sampling points of various B. subtilis strains grown in LPM. OD600 values (•), APase production (▪), concentration of Pi in the medium (▴), and RNA isolation time points are shown for the wild-type strain 168 (A) and phoR-null (B) and sigB-null (C) mutant strains. Isolated RNA was used for the Northern blots and macroarray experiments. The hybridization for the Northern blot was carried out with DIG-labeled RNA probes specific for the phoA gene. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to phoA-specific riboprobes. For the DNA array analysis, total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex.). Normalization and quantification were performed as described in Materials and Methods.
The quality of the extracted RNA was confirmed by Northern blot analyses using gene-specific riboprobes specific for phoA, which belongs to the PhoP regulon (Fig. 1), and yheK, which belongs to the sigB regulon (data not shown). The RNA was 33P labeled in a copy DNA reaction using reverse transcriptase and gene-specific primers (Sigma Genosys). The resulting labeled target molecules were hybridized to probes spotted in duplicate on DNA macroarrays (Panorama B. subtilis gene array; Sigma Genosys). Data from replicate arrays showed good reproducibility with a typical Pearson correlation coefficient of ∼0.94. Each experiment was performed twice to generate two biologically independent sets of time course data; one set of representative growth curves are shown in Fig. 1. Genes listed in Table 2 were considered significant if they met the following criteria: (i) included by the outlier analysis (see Material and Methods) between pre- and postphosphate starvation in a single time series; (ii) included from a separate outlier analysis in an independent time series; and (iii) at least twofold above the background: if both time points have values less than twice the background, the gene was excluded from the analysis. This analysis was designed to eliminate false-positive results, although this rigorous procedure may lead to false-negative results.
TABLE 2.
List of B. subtilis genes exhibiting significantly increased mRNA levels during phosphate starvation conditions as determined by DNA macroarray analysis
| Gene | Potential Pho box sequences upstream of the start codon | Functiona | Fold induction ratiob
|
Transcriptional organizationa [reference(s)] | Secreted/nonsecreted proteinc | ||
|---|---|---|---|---|---|---|---|
| WT | phoR | sigB | |||||
| Previously identified PhoP regulon members | |||||||
| glpQ | Glycerophosphoryl diester phosphodiesterase | 10.0 | 3.9 | 38.2 | gplQ (7) | Secreted | |
| phoA | ATGATCAACAGCCGCATTTAACAAAG-T37 ATG | Alkaline phosphatase A | 36.9 | 1.2 | 132.1 | phoA (65) | Secreted |
| phoB | ATCTTTAAAATCGATTAATACTAG-N65 TTG | Alkaline phosphatase III | 183.3 | 1.7 | 166.2 | phoB-ydhF (7) | Secreted |
| phoD | TCAGTTCACACTTCTTCACAGTCG-N55 ATG | Phosphodiesterase/alkaline phosphatase | 59.0 | 2.8 | 51.9 | phoD-tatAD-tatCD (34) | Secreted |
| phoP | Two-component response regulator | 1.7 | 1.7 | 4.8 | phoP-phoR (56, 60) | Nonsecreted | |
| phoR | Two-component sensor histidine kinase | 1.9 | 0.5 | 3.6 | phoP-phoR (56, 60) | Nonsecreted | |
| pstA | Phosphate ABC transporter (permease) | 8.8 | 1.5 | 5.4 | pstS-pstC-pstA-pstBA-pstBB (3, 67) | Nonsecreted | |
| pstBA | Phosphate ABC transporter (ATP-binding protein) | 85.1 | 1.8 | 48.5 | pstS-pstC-pstA-pstBA-pstBB (3, 67) | Nonsecreted | |
| pstBB | Phosphate ABC transporter (ATP-binding protein) | 8.6 | 1.8 | 6.9 | pstS-pstC-pstA-pstBA-pstBB (3, 67) | Nonsecreted | |
| pstC | Phosphate ABC transporter (permease) | 31.0 | N/A | 18.8 | pstS-pstC-pstA-pstBA-pstBB (3, 67) | Nonsecreted | |
| pstS | CCTTTTTACATAGAACCTTTACTCTAT-N45 ATG | Phosphate ABC transporter (binding protein) | 32.1 | 1.5 | 18.1 | pstS-pstC-pstA-pstBA-pstBB (3, 67) | Secreted |
| resA | AAATTTCACATAACCTTCAAAAAGT-N64 GTG | Essential protein similar to cytochrome c biogenesis protein | 2.2 | 0.9 | 1.0 | resA-resB-resC-resD-resE (10) | Nonsecreted |
| resB | Required for cytochrome c synthesis | 0.8 | 0.6 | 0.7 | resA-resB-resC-resD-resE (10) | Nonsecreted | |
| resC | Required for cytochrome c synthesis | 1.1 | 0.8 | N/A | resA-resB-resC-resD-resE (10) | Nonsecreted | |
| resD | ACAGTTCTCAAACTTTCTCACGAT-N41 ATG | Two-component response regulator | 1.1 | 0.9 | 0.9 | resA-resB-resC-resD-resEresD-resE (10) | Nonsecreted |
| resE | Two-component sensor histidine kinase | 1.3 | 0.9 | 0.6 | resA-resB-resC-resD-resE resD-resE (10) | Nonsecreted | |
| tagA | Teichoic acid biosynthesis | 0.5 | 0.64 | N/A | tagA-tagB (44, 66) | Nonsecreted | |
| tagB | Involved in polyglycerol phosphate teichoic acid biosynthesis | 1.6 | 0.71 | N/A | tagA-tagB (44, 66) | Nonsecreted | |
| tagD | Glycerol-3-phosphate cytidylyltransferase | 0.4 | 0.47 | 1.7 | tagD-tagE-tagF (44, 66) | Nonsecreted | |
| tagE | UDP-glucose:polyglycerol phosphate glucosyltransferase | 0.4 | N/A | N/A | tagD-tagE-tagF (44, 66) | Nonsecreted | |
| tagF | N/A | N/A | N/A | tagD-tagE-tagF (44, 66) | Nonsecreted | ||
| tuaA | AGTATTAACAACATTTATCAGAAA-N11 GTG | Biosynthesis of teichuronic acid | 19.8 | 0.6 | 5.5 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted |
| tuaB | Biosynthesis of teichuronic acid | 3.1 | 0.6 | 5.1 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaC | Biosynthesis of teichuronic acid | 23.1 | 1.8 | 7.6 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaD | Biosynthesis of teichuronic acid | 14.4 | 1.3 | 10.0 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaE | Biosynthesis of teichuronic acid | 10.4 | 0.8 | N/A | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaF | Biosynthesis of teichuronic acid | 24.9 | 1.2 | 16.4 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaG | Biosynthesis of teichuronic acid | 15.3 | 2.6 | 18.1 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tuaH | Biosynthesis of teichuronic acid | 3.8 | 0.9 | 13.7 | tuaA-tuaB-tuaC-tuaD-tuaE-tuaF-tuaG-tuaH (37) | Nonsecreted | |
| tatAd | Similar to hypothetical proteins | 2.7 | 1.5 | 1.5 | phoD-tatAD-tatCD (34) | Nonsecreted | |
| ydhF | Similar to unknown proteins from B. subtilis | 76.9 | 0.72 | 43.9 | phoB-ydhF (7) | Secreted | |
| ykoL | ATTCTTTACATTAGATTCATACCAC-N56 ATG | Unknown | 3.6 | N/A | 3.2 | ykzB-ykoL ykoL (60) | Nonsecreted |
| yttP | Similar to unknown proteins | 1.2 | 0.8 | 1.3 | yttP | Nonsecreted | |
| Remaining wild-type genes | |||||||
| bpr | GTAATTCAGATTGTCTACAGTTA-N88 ATG | Bacillopeptidase F | 9.7 | 2.4 | 14.2 | bpr | Secreted |
| lytB | Modifier protein of major autolysin LytC | 52.8 | N/A | N/A | lytA-lytB-lytC (38) | Secreted | |
| rapA | Response regulator aspartate phosphatase | 8.7 | 6.0 | 7.6 | rapA-phrA (57) | Nonsecreted | |
| glcU | Glucose uptake | 5.3 | 1.3 | N/A | glcU-gdh (52) | Nonsecreted | |
| ydbD | Similar to manganese-containing catalase | 9.2 | 3.8 | 1.1 | ydbE-ydbD (8) | Nonsecreted | |
| cotP | Spore coat protein | 5.4 | 0.6 | N/A | cotP (68) | Nonsecreted | |
| yfkN | AATAGTTACAAAATATTCTTACAATAG-N3 GTG | Similar to 2′,3′-cyclic-nucleotide 2′-phosphodiesterase | 6.3 | 1.1 | 17.3 | yfkN | Secreted |
| yukJ | Unknown | 5.5 | 2.7 | 4.7 | yuxI-yukJ | Nonsecreted | |
| yxnB | Unknown | 9.8 | 3.5 | 18.5 | yxbB-yxbA-yxnB-asnH-yxaM (83) | Nonsecreted | |
| Genome-wide analysis of a phoR-null mutant | |||||||
| csbD | σB-transcribed gene | 1.7 | 29.7 | N/A | csbD (2) | Nonsecretoryprotein | |
| csbX | σB-transcribed gene | 1.1 | 6.0 | N/A | csbX-bofC(24) | Nonsecreted | |
| dps | Stress- and starvation-induced gene controlled by σB | 1.6 | 6.0 | N/A | dps (6) | Nonsecreted | |
| gsiB | General stress protein | 6.0 | 63.5 | 1.4 | gsiB (35) | Nonsecreted | |
| gspA | General stress protein | 2.6 | 19.8 | N/A | gspA (5) | Nonsecreted | |
| rsbW | Negative regulation of σB-dependent gene expression | 2.3 | 10.7 | 0.7 | rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbX rsbV-rsbW-sigB-rsbX (82) | Nonsecreted | |
| rsbX | Indirect negative regulation of σB-dependent gene expression | 1.8 | 5.8 | N/A | rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbXrsbV-rsbW-sigB-rsbX (82) | Nonsecreted | |
| skfA | GGGTTAAAATATTCTATTTATACTAA-N124 ATG | Unknown | 9.9 | 5.1 | 32.8 | skfA-skfB-skfC-skfD-skfE-skfF-skfG-skfH (25) | Nonsecreted |
| ybyB | Unknown | 8.5 | 29.0 | N/A | ybyB | Nonsecreted | |
| ycsD | Similar to hydroxymyristoyl-(acyl carrier protein) dehydratase | 6.0 | 8.6 | N/A | ycsD-ycsE | Nonsecreted | |
| ydaD | Similar to alcohol dehydrogenase | 2.0 | 10.2 | N/A | ydaD-ydaE (59) | Nonsecreted | |
| ydaE | Unknown | 3.3 | 97.6 | N/A | ydaD-ydaE (59) | Nonsecreted | |
| ydaP | Unknown | 2.8 | 11.2 | N/A | mutT-ydaP | Nonsecreted | |
| yfkJ | Similar to protein-tyrosine phosphatase | 1.9 | 6.9 | 1.5 | yfkJ-yfkI-yfkH | Nonsecreted | |
| yfkM | Similar to hypothetical proteins | 1.2 | 29.6 | N/A | yfkK-yfkL-yfkM (59) | Nonsecreted | |
| yflT | Unknown | 8.1 | 22.7 | 0.5 | yfmA-yflT | Nonsecreted | |
| ygxB | Similar to hypothetical proteins from B. subtilis | 2.2 | 16.7 | N/A | ygxB | Nonsecreted | |
| yheK | Similar to hypothetical proteins | 4.1 | 27.3 | N/A | yheK (61) | Nonsecreted | |
| yjbC | Induced by phosphate starvation in a σB-dependent and PhoPR-independent manner | 1.3 | 4.8 | 2.1 | yjbC-spx (yjbD) (7) | Nonsecreted | |
| ykzA | TTATTTTCCATTTTGTTCACCAACT-N124 ATG | Similar to general stress protein | 10.7 | 13.7 | N/A | ykzA (81) | Nonsecreted |
| yocB | Unknown | 2.3 | 7.4 | N/A | yocB | Nonsecreted | |
| yqgZ | Similar to hypothetical proteins | 5.1 | 9.1 | N/A | yqgZ | Nonsecreted | |
| yrzI | Unknown | 1.6 | 4.6 | N/A | yrzI | Nonsecreted | |
| ysnF | TATATTCACACATTTTTCACCTT-N68 ATG | Unknown | 1.6 | 27.0 | N/A | ysnF | Nonsecreted |
| ytxG | TATGTATACAGCCCAGTACACATGTT-N55 ATG | Similar to general stress protein | 1.1 | 21.7 | 1.7 | ytxG-ytxH-ytxJ | Nonsecreted |
| ytzE | Similar to transcriptional regulator | 4.7 | 12.9 | 7.7 | ytzE | Nonsecreted | |
| yvgO | Unknown | 1.5 | 5.0 | 0.6 | yvgO | Secreted | |
| yvyD | AAAGTTCACTGAATTTTCACAAA-N79 ATG | Similar to ribosomal protein S30AE family | 1.4 | 22.2 | 1.1 | yvyD (17) | Nonsecreted |
| ywzA | Similar to hypothetical proteins from B. subtilis | 4.9 | 36.6 | N/A | ywzA | Nonsecreted | |
| Genome-wide analysis of a sigB-null mutant | |||||||
| asnH | Asparagine synthetase | N/A | N/A | 259.4 | yxbB-yxbA-yxnB-asnH-yxaM (83) | Nonsecreted | |
| bpr | GTAATTCAGATTGTCTACAGTTA-N88 ATG | Bacillopeptidase F | 9.7 | 2.4 | 14.4 | Bpr | Secreted |
| metC | Methionine biosynthesis | 1.8 | 0.6 | 13.1 | metI-metC (9) | Nonsecreted | |
| pel | Pectate lyase | 4.0 | 2.2 | 4.0 | pel (53) | Secreted | |
| pksE | Involved in polyketide synthesis | 2.3 | 0.6 | 3.4 | pksB-pksC-pksD-pksE-acpK-pksF | Nonsecreted | |
| ppsA | Plipastatin synthetase | N/A | 1.6 | 4.7 | ppsA-ppsB-ppsC-ppsD-ppsE (77) | Nonsecreted | |
| rapA | Response regulator aspartate phosphatase | 8.7 | 6.2 | 7.6 | rapA-phrA (57) | Nonsecreted | |
| skfA | GGGTTAAAATATTCTATTTATACTAA-N124ATG | Unknown | 9.9 | 5.1 | 32.8 | skfA-skfB-skfC-skfD-skfE-skfF-skfG-skfH | Nonsecreted |
| spoIIAA | Stage II sporulation | N/A | N/A | 5.7 | spoIIAA-spoIIAB-sigF (70) | Nonsecreted | |
| spoIIB | Stage II sporulation | 4.3 | N/A | 9.9 | spoIIB | Nonsecreted | |
| srfAA | TTAGTTCATAAGAATTAAAAGCTG-N70 ATG | Surfactin production and competence | 2.2 | 1.0 | 3.3 | srfAA-srfAB-srfAC-srfAD | Nonsecreted |
| thrC | Threonine biosynthesis | 0.7 | 0.6 | 7.7 | hom-thrC-thrB (55) | Nonsecreted | |
| yfkN | ATAGTTACAAAATATTCTTACAATAG-N38GTG | Similar to 2′,3′-cyclic-nucleotide 2′-phosphodiesterase | 6.3 | 1.1 | 17.4 | yfkN | Secreted |
| yhcR | Similar to 5′-nucleotidase | 1.7 | 1.5 | 4.3 | yhcR-yhcS | Nonsecreted | |
| yhzC | CCGGTTTACGGCATTTTGCAGGAT-N105 ATG | Unknown | 2.0 | 1.2 | 6.5 | yhzC | Nonsecreted |
| yjdB | ATTATTAACATTTATTTACAAGGA-N63 ATG | Unknown | 2.6 | 0.8 | 5.1 | yjdB | Secreted |
| ykzF | Unknown | 1.5 | 2.9 | 3.9 | ykuJ-ykuK-ykzF-ykuL-ykuM | Nonsecreted | |
| yoeB | TATTGTAACATTTGTAACATAAG-N29 ATG | Unknown | 0.7 | N/A | 4.0 | yoeB | Nonsecreted |
| ytzE | Similar to transcriptional regulator (DeoR family) | 4.7 | 12.9 | 7.6 | ytzE | Nonsecreted | |
| yuiB | Similar to hypothetical proteins from B. subtilis | 2.7 | 2.3 | 11.6 | yuiA-yuiB | Nonsecreted | |
| yukJ | Unknown | 5.5 | 2.7 | 4.7 | yuxI-yukJ | Nonsecreted | |
| yuxI | Unknown | N/A | 1.8 | 8.4 | yuxI-yukJ | Nonsecreted | |
| ywkC | Unknown | 3.0 | 1.7 | 4.4 | ywkD-ywkC | Nonsecreted | |
| yxbB | Similar to hypothetical proteins | 4.9 | 6.2 | 26.9 | yxbB-yxbA-yxnB-asnH-yxaM (83) | Nonsecreted | |
| yxnB | Unknown | 9.8 | 3.5 | 18.2 | yxbB-yxbA-yxnB-asnH-yxaM (83) | Nonsecreted | |
| yycO | Unknown | 3.6 | N/A | 15.0 | yycO-yycP (20) | Secreted | |
Function and putative transcriptional organization are according to the associated reference. When no reference has been detected in the literature, the organization is according to the SubtiList database (http://genolist.pasteur.fr/SubtiList) (48). Gene names in parentheses are alternative names.
Calculated fold induction ratios (between pre- and postphosphate starvation) for genes averaged from two biologically independent experiments. Genes that fulfill the criteria for the outlier analysis are given in bold type and underlined. Genes that did not fulfill the outlier analysis criteria are shown as N/A. WT, wild type.
Whether the protein is predicted to be secreted or nonsecreted according to Tjalsma and coworkers (76).
Analysis of the DNA macroarray data identified 24 genes in the wild type that were induced in response to phosphate starvation, several of which were members of the PhoP and σB regulons (Table 2, previously identified PhoP regulon members and remaining wild-type genes). phoB, a member of the PhoP regulon that encodes a secreted APase, had the highest induction factor (183-fold). Other well-established members of the PhoP regulon, such as phoA (APase) and phoD (phosphodiesterase/APase) were also significantly induced (Table 2, previously identified PhoP regulon members). However, some members of the PhoP regulon did not show a significant level of induction. These included the resABCDE operon, which has been shown previously (5) to be induced in response to phosphate starvation in a PhoP∼P-dependent manner. Another omission is phoPR, which we have recently confirmed to be a member of the PhoP regulon but which is only weakly induced (approximately twofold) in response to phosphate starvation (60). It is likely that this level of induction is not sufficient to be included in the stringent outlier analysis.
The PhoP regulon also includes the divergent operons tagAB and tagDEF, encoding proteins responsible for the synthesis of the anionic polymer, teichoic acid. These genes have been shown previously to be repressed about twofold in response to phosphate starvation (39). Together with the concomitant induction of the tua operon, these genes bring about a change in the composition of the cell wall from one predominately consisting of the phosphate-containing teichoic acid to one composed mainly of the non-phosphate-containing teichuronic acid (37). The array analysis was not sensitive enough to detect repression of the tag genes, since their expression was not significantly down-regulated in response to phosphate starvation (44). However, several, but not all of the genes in the tua operon were included in the outlier analysis as being induced in response to phosphate starvation.
The changes in induction ratios of PhoP-regulated genes range from 2 to >100. The differences are often accounted for by the basal levels of expression. For example, the phoPR operon exhibits a low change in induction value because of a significant level of expression during phosphate-replete growth. This basal expression ensures that phosphate-replete cells synthesize the signal transduction pathway required to detect and respond to declining phosphate concentrations.
The SigB regulon currently contains approximately 120 genes, identified using DNA array analysis and promoter prediction (58, 64). However, only one σB-regulated gene, ydbD, was significantly up-regulated in the wild type in response to phosphate starvation. YdbD is a manganese-containing catalase that is thought to prevent oxidative damage by destroying peroxides and related oxidizing compounds (64). We have shown that several members of the SigB regulon are induced in response to phosphate starvation using reporter genes (62) and Northern blotting (Fig. 1) and confirmed that phosphate starvation activates the SigB regulon exclusively via the RsbP phosphatase (data not shown). The failure of the DNA arrays to detect significant levels of induction of other members of the SigB regulon in the wild type may reflect the relatively low sensitivity and incremental response of the RsbP energy-sensing pathway (discussed below).
Eight additional genes, namely, yfkN, bpr, glcU (previously ycxE), cotP (previously ydfT), lytB, rapA, yukJ, and yxnB, which are not currently members of the PhoP and SigB regulons were significantly induced in the wild type in response to phosphate starvation (Table 2, remaining wild-type genes). These are discussed in detail later. In addition, yhaX and yhbH are included by the outlier analysis if a later time point is used (T5).
Sixty-nine genes are significantly repressed in the wild type in response to phosphate starvation. Many of these genes are involved in nucleic acid and protein synthesis, including 11 rps genes, 11 rpl and 5 rpm genes (encoding small and large ribosome subunit proteins), 4 rpo genes (encoding components of RNA polymerase), and 5 genes involved in purine synthesis. The data confirm previous studies showing that genes associated with the information processing machinery are down-regulated in response to growth arrest (27).
Genome-wide analysis of a phoR-null mutant.
The global response of a phoR-null mutant to phosphate starvation (Fig. 1B) was determined to identify PhoPR-dependent genes and to monitor the cell's response to the additional stress imposed by the lack of a phosphate-specific stress response. Twenty-nine genes were significantly induced in the phoR-null mutant in response to phosphate starvation according to the outlier analysis (Table 2, genome-wide analysis of a phoR-null mutant). As expected, none were current members of the PhoP regulon; however, in contrast to the response of the wild-type strain, 24 of the phosphate starvation-induced genes were members of the SigB regulon (7, 58, 64). These data indicate that, in the absence of a functional PhoP regulon, the extent of the energy stress detected via RsbP is enhanced, leading to the significant induction of about one quarter of the genes in the SigB regulon. This response was reflected in a markedly higher level of expression of the genes in the SigB regulon in the phoR-null strain compared to the wild type (Table 3).
TABLE 3.
Total expression of genes in the PhoP and SigB regulons
| Genes | Total expression of genes in straina:
|
||
|---|---|---|---|
| Wild-type | sigB-null mutant | phoR-null mutant | |
| PhoP regulon members | 3.9 | 4.0 | 1.1 |
| SigB regulon members | 3.4 | 1.9 | 9.4 |
Most of the members of the SigB regulon that are induced in response to phosphate starvation are of unknown or experimentally unconfirmed function. dps (6) encodes a homologue of the Dps/PexB protein of E. coli which protects the chromosome from acid, heat, and oxidative stress (43); a dps-null mutant exhibits severely reduced resistance to oxidative stress (6). yjbC is involved in salt tolerance, and a yjbC-null mutant is almost as sensitive to salt as a sigB-null mutant (58). Other SigB-induced genes appear to have a detoxification role; ykzA (80) encodes a protein that is similar to Ohr, an organic hydrogen peroxide resistance protein of Xanthomonas campestris (46), while yqgZ encodes a putative arsenate reductase (64). gsiB, encoding a general stress protein of unknown function, showed the highest level of induction (>200-fold) in response to phosphate starvation. The exceptional stability (half-life of ∼20 min) of gsiB mRNA has been attributed to the presence of a strong ribosome binding site (35).
Four additional genes were induced significantly in the phoR-null mutant in response to phosphate starvation but are not currently members of the PhoP and SigB regulons: skfA (previously ybcO), yrzI, ytzE, and ycsD.
Genome-wide analysis of a sigB-null mutant.
The global response of a sigB-null mutant to phosphate starvation (Fig. 1C) was determined to facilitate the identification of SigB-dependent genes and to monitor the cell's response to the additional stress imposed by the lack of the general stress response. Forty-two genes were significantly induced in the sigB-null mutant in response to phosphate starvation (Table 2, genome-wide analysis of a sigB-null mutant). As expected, none were members of the SigB regulon but 16 of the genes were members of the PhoP regulon (Table 2, previously identified PhoP regulon members). In confirmation of the findings of Prágai and Harwood (62), members of the PhoP regulon were generally up-regulated in the sigB-null mutant compared to the wild type (Fig. 2A). A number of genes that are induced in the wild type in response to phosphate starvation are also induced in the sigB-null mutant (yfkN, bpr, yjdB, yukJ, ynxB, and rapA). These are good candidates for inclusion in the Pho regulon and are discussed in detail below.
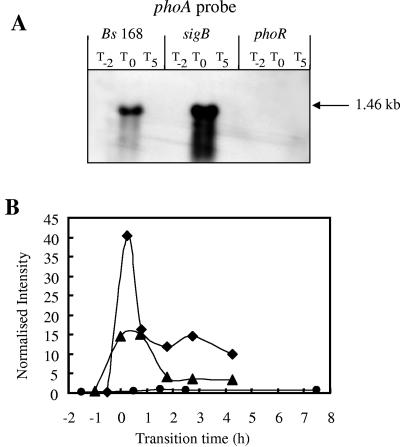
FIG. 2.
Northern blot analyses and transcriptional profiles for the phoA gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB-null and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to a phoA-specific riboprobe. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴) and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex.). Normalization and quantification were performed as described in Materials and Methods.
A number of genes that were induced in the sigB mutant during the transition to phosphate limitation were not induced in the wild type. Four such genes are involved in the synthesis of antimicrobial compounds: srfAA and ppsA are involved in the synthesis of the bioactive lipopeptides surfactin (16) and plipastatin (78), respectively; skfA encodes an antimicrobial peptide (25); and pksE is involved in the synthesis of an antimicrobial polyketide. bpr, pel, and yhcR, encoding bacillopeptidase F, pectate lyase, and a putative 5′ nucleotidase, respectively, are macromolecular hydrolases. asnH, metC, and thrC are involved in the biosynthesis of amino acids. rapA, spoIIAA, and spoIIB are involved in sporulation or sporulation-associated events. yjdB, a gene of unknown function, was previously identified by Ogura and coworkers (54) as being a potential member of the PhoP regulon. Our data tend to confirm this assignment, since the yjdB promoter region contains putative Pho box-like sequences and this gene is induced in the sigB-null mutant but not in the phoR-null mutant. The remaining 11 genes (Table 2) are of unknown function.
Clustering of genes based on expression patterns.
The analysis of the macroarray data was extended using K-means clustering (33). K-means clustering partitions the data into a predetermined number of clusters on the basis of the similarity of their expression profiles. The analysis involves the iterative reallocation of the cluster members to minimize intracluster scattering. Using the normalized data, genes with little or no expression were excluded by using the “filter on expression” script in the GeneSpring software package, arbitrarily setting the minimum expression to 1 for at least three out of the six time points. Genes not conforming to this requirement were excluded, leaving 2,162 genes in the analyzed data set. We selected the number of clusters to be 15 with “standard correlation” as the similarity measure. The analysis was performed on the sigB-null data set, since members of the PhoP regulon generally show enhanced levels of expression in this background (see Fig. S1 in the supplemental material). Cluster 13, containing 139 genes, included 20 genes currently identified as members of the PhoP regulon (see Fig. S2 in the supplemental material).
Comparison of the cluster analyses for the wild-type and phoR-null mutant data sets (see Fig. S3 in the supplemental material) show, as expected, little or no expression of the PhoP regulon genes in the phoR-null mutants. This method also identified vpr, yurI, yfkN, and bpr as potential additional members of the PhoP regulon.
Analysis of promoter regions for PhoP-like consensus sequences.
To identify potential PhoP binding sequences of Pho member candidates, the B. subtilis genome was interrogated using the Pattern Match function of SubtiList (http://genolist.pasteur.fr/SubtiList/) (47). The PhoP consensus TTHACA3-7TTHACA (H = A/C/T) (11) can be repeated as few as twice, as in the case of the resA promoter that additionally requires ResD for induction, or as many as eight times in the case of the tuaA promoter. In all cases there is always an even numbers of repeats. Only targets which were upstream of gene boundaries and within 150 bp of the start codon were reported, and in addition, genes were reported only if they were transcribed monocistronically or were the first gene in a polycistronic operon. A deviation of 1 bp from the consensus was allowed in each part of the search sequence. The pattern match gave good results with known PhoP promoters, correctly predicting 8 out of 11 PhoP-regulated promoters (Table 2). PhoP-repressed promoters, such as tagAB and tagDEF, were not considered, as their consensus sequence usually comprise two repeats on the noncoding strand. Of the promoters not identified by this method, phoP has a very weak consensus sequence (60), and yttP and glpQ would be included if a 2-bp deviation from the consensus sequence were used.
Analyses of new potential members of the PhoP regulon.
The outlier analysis identified 29 genes that were induced significantly in the wild type or sigB-null mutant in response to phosphate starvation but crucially not in the phoR-null mutant. Five of these genes, namely, bpr, rapA, yfkN, yukJ, and yxnB, were induced in the wild type and sigB-null mutant and had not previously been identified as members of the PhoP regulon; they were therefore considered to be good candidates for inclusion in the regulon. Additionally, yjdB was chosen for further analysis, as Ogura and coworkers (54) identified it as a potential member of the PhoP regulon member in experiments in which phoP was overexpressed in the absence of the phoPR operon. Our analysis showed that yjdB was induced in the sigB-null mutant and had Pho box-like repeats in its promoter region. rapA was not analyzed further, since its expression in response to growth arrest has been the subject for previous studies (73).
Analysis of the transcriptional profiles by K-means clustering identified four putative new members of the PhoP regulon, namely, bpr, yfkN, vpr, and yurI.
We selectively analyzed expression of seven of these putative new members of the PhoP regulon using strains carrying transcriptional reporter gene fusions generated during the Bacillus Functional Analyses (BFA) project. In BFA mutants, the target gene was inactivated using the pMUTIN integration vector that simultaneously fuses its transcription to a lacZ reporter gene (79). Mutants were grown in LPM and HPM, and the production of β-galactosidase and APase was monitored throughout growth. β-Galactosidase assays were performed on a minimum of three independent cultures. Additionally, selective Northern blot analyses were performed on RNA extracted from the wild type and phoR- and sigB-null mutants during exponential phase and phosphate starvation-induced stationary phase. The hybridization of a riboprobe against a well-established member of the PhoP regulon, namely, phoA, was used as a control. These analyses confirmed that little or no phoA mRNA transcripts were detected at T−2, confirming the lack of induction of this gene prior to phosphate starvation. As expected, a single prominent band of ∼1.45 kb was detected at T0 (Fig. 2A), disappearing at T5, confirming the transient expression of the PhoP regulon (28) in response to phosphate starvation. A more intense band was seen at T0 in the sigB mutant than in the wild type, while, as expected, no transcript was seen in the phoR mutant. The quantitative transcription profiles for phoA in the wild type and phoR- and sigB-null mutants obtained from the DNA macroarrays are shown in Fig. 2B.
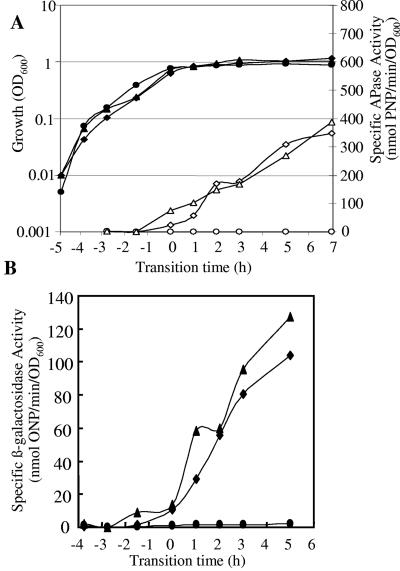
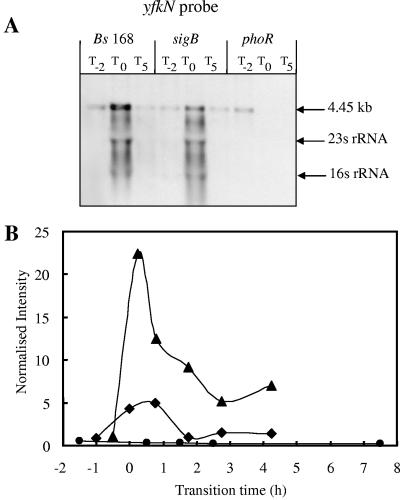
yfkN, encoding an extracellular 2′,3′ cyclic nucleotide 2′-phosphodiesterase (13), is a good candidate for inclusion in the PhoP regulon, since it is induced in response to phosphate starvation in a PhoR-dependent manner and has a Pho box-like sequence (Table 2) in the vicinity of its putative σA promoter 38 bases upstream of the GTG start codon. Growth of a BFA yfkN mutant (YFKNd-PR) in LPM showed the concomitant induction of β-galactosidase and APase activities at T0 (Fig. 3). The introduction of a sigB-null mutation into this strain had very little influence on the induction of yfkN (∼100 nmol o-nitrophenyl [ONP]/min/OD unit at T0), while the introduction of the phoP-null mutation reduced it 100-fold (∼1 nmol ONP/min/OD unit at T0). No β-galactosidase activity was detected in HPM (data not shown), indicating that transcription of yfkN was induced only in response to phosphate starvation. Northern blot analyses confirmed the induction of the yfkN transcript in the wild type and sigB mutant and the absence of this transcript in phosphate-replete conditions (Pi >0.1 mM) (Fig. 4A). The size of the primary transcript (4.45 kb) was consistent with the predicted length of yfkN, indicating that this gene comprises a monocistronic operon. Discrete smaller bands are likely to be processed products (3). The Northern blot analyses confirmed the data obtained in the reporter gene experiments and transcriptional profiling (Fig. 4B), namely, that yfkN is induced at T0 in the wild type and sigB mutant but not in the phoR mutant. yfkN showed fourfold hyperinduction at T0 in the sigB-null mutant using DNA macroarray, and twofold hyperinduction was detected using β-galactosidase reporter fusion (Fig. 3 and 4) at T1. On the basis of this evidence, we have putatively assigned yfkN as a new member of the PhoP regulon.
FIG. 3.
Growth and reporter activity of B. subtilis yfkN-lacZ fusion mutants grown in LPM. (A) OD600 values of lacZ fusion yfkN-lacZ (♦), ΔsigB yfkN-lacZ (▴), and ΔphoR yfkN-lacZ (•) mutants are shown with closed symbols. APase activities of yfkN-lacZ (⋄), ΔsigB yfkN-lacZ (▵), and ΔphoR yfkN-lacZ (○) strains are shown with open symbols. PNP, p-nitrophenyl. (B) Specific β-galactosidase activities of yfkN-lacZ (♦), ΔsigB yfkN-lacZ (▴), and ΔphoR yfkN-lacZ (•) strains are shown with closed symbols.
FIG. 4.
Northern blot analyses and transcriptional profiles for the yfkN gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB- and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to yfkN-specific riboprobes. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). The arrows labeled 16S rRNA and 23S rRNA indicate the locations of these rRNA species that are known to trap smaller RNA species (1). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴), and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex). Normalization and quantification were performed as described in Materials and Methods.
bpr, encoding bacillopeptidase (71), was also a good candidate for inclusion in the PhoP regulon, since it is induced in response to phosphate starvation in a PhoR-dependent manner and has a Pho box-like sequence (Table 2) in the vicinity of a SigA promoter and 88 bases upstream of its ATG start codon. Northern blot analyses, using a bpr-specific riboprobe, confirmed the induction of the bpr transcript at T0, but this induction was seen in all strains. Since its expression is both PhoR and SigB independent, we conclude that bpr is induced in response to growth arrest by an as-yet identified regulator (data not shown).
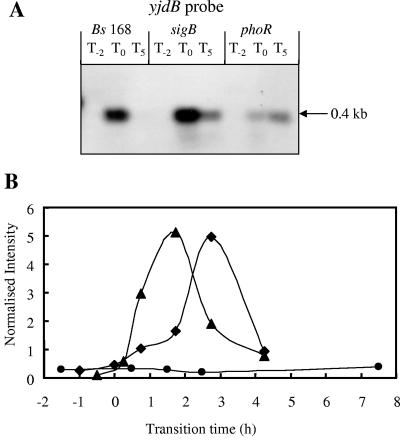
yjdB was previously identified by Ogura and coworkers (54) as being a potential member of the PhoP regulon. The data shown here support its inclusion in the PhoP regulon, as it is induced in response to phosphate starvation in a PhoR-dependent manner, and the promoter region contains Pho box-like sequences (Table 2) 63 bases upstream of the ATG start codon. Northern blot analyses using a riboprobe specific to the monocistronic yjdB gene showed induction of a single strong band of ∼0.4 kb at T0 which was much reduced at a later time point (T5). The band intensity was highest in the sigB mutant and lowest in the phoR mutant (Fig. 5A), and this was confirmed by the DNA array data (Table 2). The strength of the transcriptional profiles of yjdB in the wild type and sigB mutant were similar, whereas little induction was seen with the phoR mutant (Fig. 5B). On the basis of this evidence and that of supporting reporter gene data (data not shown), we have putatively assigned yjdB as a new member of the PhoP regulon.
FIG. 5.
Northern blot analyses and transcriptional profiles for the yjdB gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB-null and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to yjdB-specific riboprobes. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴) and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex.). Normalization and quantification were performed as described in Materials and Methods.
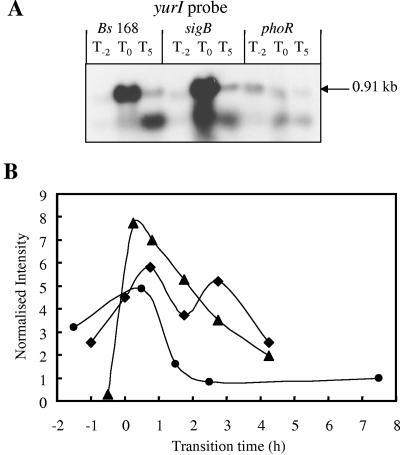
yurI was identified as a candidate for inclusion in the PhoP regulon by K-means clustering. yurI encodes an RNase which shows 78% amino acid identity to the RNase Bsn. Bsn has no apparent sequence specificity and can hydrolyze RNA endonucleolytically to yield 5′-phosphorylated oligonucleotides (50). Growth of a BFA yurI mutant, BFA1234, in LPM showed the concomitant induction of β-galactosidase and APase at T0 (Fig. 6). The introduction of a sigB-null mutation into this strain had little influence on the induction of yurI (∼300 nmol ONP/min/OD unit at T0), while the introduction of the phoR-null mutation reduced it 300-fold (∼1 nmol ONP/min/OD unit at T0). No transcription activity was detected in HPM (data not shown), confirming the phosphate starvation-specific induction of yurI. Northern blot analyses confirmed the induction of the yurI transcript in the wild type and sigB-null mutant, with a much lower level of transcript present in phosphate-replete conditions (Pi concentration of >0.1 mM) (Fig. 7A). The size of the primary transcript (∼0.9 kb) was consistent with the predicted length of yurI. A second band of ∼0.5 kb that was observed at T5 in the wild type and the sigB-null mutant and to a lesser extent in the phoR-null mutant may indicate posttranscriptional processing or multiple promoters. The Northern blot analyses confirmed the data obtained in the reporter gene experiments and transcriptional profiling (Fig. 6 and 7), namely, that yurI was induced at T0 in the wild type and sigB mutant but not in the phoR mutant. On the basis of this evidence, we have putatively assigned yurI as a new member of the PhoP regulon. In order to determine whether the product of yurI was responsible for the nonspecific degradation of RNA in the environment as a source of Pi, the wild type and a yurI-null mutant were grown on LPM agar supplemented with RNA. Plate tests confirmed the absence of a secreted RNase in the yurI mutant (data not shown).
FIG. 6.
Growth and reporter activity of B. subtilis yurI-lacZ fusion mutants grown in LPM. (A) OD600 values of lacZ fusion mutant yurI-lacZ (♦), ΔsigB yurL-lacZ (▴), and ΔphoR yurL-lacZ (•) strains are shown with closed symbols. APase activities of yurI-lacZ (⋄), ΔsigB yurL-lacZ (▵), and ΔphoR yurL-lacZ (○) strains are shown with open symbols. PNP, p-nitrophenyl. (B) Specific β-galactosidase activities of yurI-lacZ (♦), ΔsigB yurL-lacZ (▴), and ΔphoR yurL-lacZ (•) strains are shown.
FIG. 7.
Northern blot analyses and transcriptional profiles for the yurI gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB- and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to yurI-specific riboprobes. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴) and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex.). Normalization and quantification were performed as described in Materials and Methods.
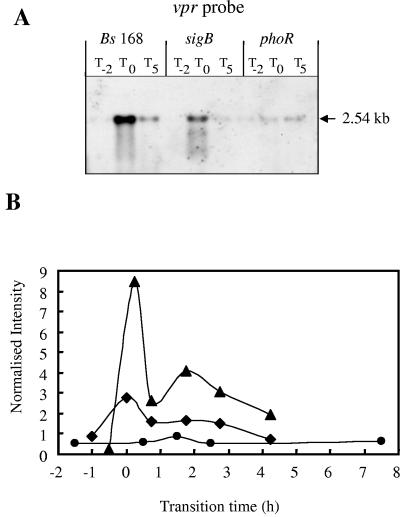
vpr was identified as a candidate for inclusion in the PhoP regulon by K-means clustering. Since no pMUTIN constructs were available, we used Northern blot analysis and vpr-specific riboprobes to analyze the transcription of vpr. This confirmed the transient induction of the vpr transcript in response to phosphate starvation in the wild type and sigB-null mutant but not in the phoR mutant (Fig. 8). The size of the primary transcript (∼2.5 kb) was consistent with the predicted length of vpr, indicating that this gene is transcribed as part of a monocistronic operon. The DNA macroarray data showed that the transcription levels of vpr were induced threefold in the wild type and eightfold in the sigB mutant at T0 compared to the transcription levels in the phoR mutant. On the basis of this evidence and the fact that the promoter has PhoP box-like consensus sequences, we have putatively assigned vpr as a new member of the PhoP regulon.
FIG. 8.
Northern blot analyses and transcriptional profiles for the vpr gene. (A) RNA was isolated from wild-type B. subtilis strain 168 (Bs 168) and sigB-null and phoR-null mutants. Bacteria were grown in LPM, and samples were taken 2 h before (T−2) and 0 h (T0) and 5 h (T5) after entry into the stationary growth phase, which was provoked by phosphate starvation. Five micrograms of RNA was applied per lane; after the filters were capillary blotted, they were hybridized to vpr-specific riboprobes. Transcript size was determined by comparison with DIG-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). (B) RNA was isolated from wild-type B. subtilis (168) (♦) and sigB-null (▴) and phoR-null (•) mutants grown in LPM. Samples were taken before, during, and after entry into the stationary growth phase, which was provoked by phosphate starvation. Total RNA was isolated and used as the template for reverse transcriptase incorporating radiolabeled [33P]dATP, which was hybridized to a whole-genome macroarray (Sigma-Genosys, The Woodlands, Tex.). Normalization and quantification were performed as described in Materials and Methods.
yxnB and yukJ were identified as potential PhoR- and SigB-independent members of the Pho stimulon, and Northern blot analyses confirmed their induction at T0 in the wild type and sigB- and phoR-null mutants (data not shown). To determine whether yxnB was induced in response to phosphate starvation, strains containing a reporter gene fusion to yxnB were grown in LPM and HPM. The data indicated that yxnB was induced at T0 in both high- and low-phosphate media, indicating that this gene is induced in response to growth arrest, independently of phosphate concentration (data not shown).
DISCUSSION
Soil-dwelling bacteria, such as B. subtilis, have evolved physiological responses to overcome environmental stress. One of the most commonly encountered stresses in soil is phosphate starvation (28). B. subtilis responds to phosphate starvation via the transient induction of a variety of genes with diverse functions. In this study we have characterized the phosphate starvation stimulon of B. subtilis at the whole-genome level using DNA macroarrays. Although the main aim was to obtain a global perspective of the B. subtilis cell to the phosphate starvation response, comparison of the data with existing information obtained using complementary technologies has provided information on the relative advantages and disadvantages of DNA array data. To this end, we have characterized the phosphate starvation response in wild-type B. subtilis and sigB- and phoR-null mutants in which the general and specific responses to phosphate starvation are nonfunctional, respectively. The data shown here and previously (7, 22, 60) indicate that the phosphate starvation stimulon consists, at least, of the phosphate starvation-specific PhoP regulon and the σB general stress regulons. The SigB-dependent stress proteins provide a nonspecific, prospective stress resistance (26), while the PhoP regulon provides proteins with functions specific for making alternative sources of phosphate available.
The response of B. subtilis to phosphate starvation has been extensively studied during the last 15 years. Prominent among the cell's response is the induction of APases and a high-affinity phosphate-specific transporter which cooperatively recover phosphate from organic sources and transport it into the cell. Important marker genes involved, namely, glpQ, phoB, phoD, and pstS (7), were all identified as significantly induced, confirming the validity of the methodology. However, it should be borne in mind that different transcriptomic approaches (e.g., DNA arrays, Northern blotting, and reporter gene technology) do not detect and measure the same elements: although they usually show the same trends, factors such as mRNA stability and processing can influence their congruence.
The outlier analysis identified 24 genes in the wild type which were significantly induced in response to phosphate starvation, 15 of which were previously known members of the PhoP regulon. In contrast, only one of these genes (ydbD) was a previously known member of the SigB regulon (58). This method provided a stringent method of analysis which reduced the identification of false-positive results, but probably at the expense of excluding genes that are only weakly induced in response to phosphate starvation.
Hulett and coworkers showed that the resABCDE operon was induced under phosphate starvation conditions (10) and that the ResDE two-component system modulates this activity, as mutations in these genes lead to decreased PhoP regulon expression (74). However, our experiments provided no evidence for the induction of the res operon in response to phosphate starvation. Discrepancies between our results and those of Hulett and colleagues may be due to differences between in the growth media or strain genotypes.
The majority of PhoP-regulated genes are expressed at T0, the most notable exceptions being yhaX and yhbH, which are induced at T3 (62). The timing of their expression is compatible with the observation that they are expressed from SigE-dependent promoters. However, we have shown previously that PhoP does not bind in the region to the yhaX promoter (60), indicating either that PhoP regulates yhaX indirectly or that a product of the Pho regulon is required for its transcription. On the basis of this evidence, we do not currently include yhaX as a member of the PhoP regulon. Additionally, because of the similarity of its induction kinetics, we currently also exclude yhbH.
Twenty-nine genes were induced significantly in the phoR-null mutant in response to phosphate starvation. As expected, none of the currently recognized PhoP regulon genes are included, but a large number of these genes belong to the SigB regulon. They include SigB-dependent genes, such as gsiB and dps, that were shown previously to be induced in response to phosphate starvation (7). Dps is required for oxidative stress resistance and is thought to bind DNA to prevent damage (6). Six other SigB-dependent genes were significantly induced in response to phosphate starvation: yheK, ykzA, ysnF, and csbD were identified by screening BFA mutants (62), and yjbC and ytxH were identified in a two-dimensional gel electrophoresis study of the phosphate starvation proteome (7).
The questions arise as to why the analysis did not include all members of the PhoP regulon and why no members of the SigB regulon were significantly induced. One explanation of the weak response of the SigB regulon may be that its activation by the RsbP phosphatase on the energy side of the signal transduction pathway is not as strong as the induction by environmental stress that is mediated via RsbU (Fig. 9). Hyperinduction of the SigB regulon can occur if a mild physical stress is applied during energy depletion (80).
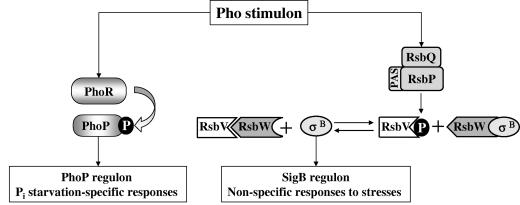
FIG. 9.
Pho stimulon of B. subtilis. Model for the activation of the PhoP and SigB (σB) regulons in response to phosphate starvation (energy) stress. The activation of PhoP via phosphorylation by PhoR leads to the induction of genes involved in the recovery and acquisition of Pi (28). The energy-sensing pathway of the SigB regulon is mediated via the Per-Arnt-Sim (PAS) domain of the RsbP phosphatase and RsbQ. Activated RsbP removes the serine phosphate (P) from RsbV∼P, which in turn sequestrates anti-σB factor RsbW. Released from its anti-σ factor, SigB is now free to interact with the core RNA polymerase to induce the nonspecific general stress genes.
The overexpression of genes belonging to the SigB regulon in a phoR-null mutant has been described previously by Prágai and Harwood (62), who observed a two- to fourfold increase in expression of SigB-dependent genes. The data obtained from the DNA arrays show that expression of 124 SigB genes (64) in response to phosphate starvation was higher in the phoR mutant than in the wild-type strain (Table 3). This increased expression seems to indicate that the SigB response is proportional to the level of the stress. This might occur if an active PhoP regulon reduces the extent to which the cellular concentration of ATP falls in response to phosphate starvation. In the absence of an active PhoP regulon, the cellular ATP concentration may be lower, triggering a greater response from the energy-sensing pathway of the SigB regulon.
Putative new members of the PhoP regulon.
Two methods of analysis were used to identify new candidate members of the PhoP regulon: outlier analysis and K-means cluster analysis. Genes bpr, skfA, yfkN, yjdB, yukJ, yurI, yxnB, and vpr were identified as being induced in response to phosphate starvation and as potential members of the PhoP regulon. Subsequent Northern blot analysis of these candidate genes identified that bpr, yxnB, and yukJ were induced at T0 in a PhoR- and SigB-independent manner and are therefore not members of either the PhoP or SigB regulon. These data confirm the importance of secondary analysis of candidate genes identified solely by DNA array data, and a combination of Northern blotting and reporter gene analyses was used to confirm genes yfkN, yurI, yjdB, and vpr as putative new members of the PhoP regulon.
Northern blot analysis showed that the yfkN, yurI, yjdB, and vpr genes were induced transiently at T0 and that this signal was absent from RNA isolated from a phoR-null strain. Northern blot analysis confirmed that each of these genes was transcribed from monocistronic operons: yfkN is transcribed as an 4.45-kb transcript; yurI as transcripts of 0.9 kb and 0.5 kb, indicating that it may be posttranscriptionally processed; yjdB as a 0.4-kb transcript and vpr as a 2.5-kb transcript. In the case of yfkN and yurI, the introduction of a phoR mutation into their respective BFA strains resulted in the loss of phosphate starvation-induced β-galactosidase activity.
Interestingly, many of the previously identified PhoP regulon members and all of the putative new members described here are known or predicted to be secreted. These include alkaline phosphatases, phosphodiesterases, glycerophosphoryl diester phosphodiesterases, and now, a protease, RNase, and 2′,3′ cyclic nucleotide phosphodiesterase, 2′ (or 3′) nucleotidase, and 5′ nucleotidase. These proteins all have the potential for salvaging Pi from organic sources in the environment. Ultimately, the recovered Pi is transported into the cell using the pit (low-affinity) and the pst (high-affinity) phosphate-specific-transporters.
Vpr is an extracellular serine protease (71) and may help recover phosphate from phosphoproteins and has been identified as a protease capable of processing subtilin (15).
YurI is predicted to be a secreted protein (76) with nonspecific hydrolytic activity on extracellular RNA to generate 3′ or 5′ phosphonucleotides. These phosphonucleotides provide substrates for APases (e.g., PhoA) and YfkN, which exhibits 2′,3′ cyclic nucleotide phosphodiesterase, 2′ (or 3′) nucleotidase, and 5′ nucleotidase activities (13) to provide a complete pathway for the recovery of Pi from extracellular RNA.
In conclusion, our data show the value of the global transcriptomic analysis that is possible using DNA macroarrays, and this has enabled us to identify five putative new members of the PhoP regulon. However, despite using the rigorous outlier technique, the technology still identified a number of putative PhoP regulon genes that were rejected by subsequent analysis by Northern blotting or reporter gene fusion technology. The global transcriptome analysis revealed the relatively weak induction of the SigB regulon during phosphate starvation and the hyperinduction of this regulon in the absence of the specific, PhoPR-mediated response.
Supplementary Material
Acknowledgments
We thank J. Sekiguchi (Shinshu University, Japan) for the gift of strain YFKNd and members of the European Bacillus Functional Analysis program for other pMUTIN-based strains.
This work was funded by the European Commission (QLG2-CT-1999-01455) and the United Kingdom Biotechnology and Biological Sciences Research Council (13/PRES/12179). N.E.E.A. was in receipt of a BBSRC studentship.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdel-Fattah, W. R., Y. Chen, A. Eldakak, and F. M. Hulett. 2005. Bacillus subtilis phosphorylated PhoP: direct activation of the EσA- and repression of the EσE-responsive phoB-PS+V promoters during Pho response. J. Bacteriol. 187:5166-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., S. Y. Lee, S. A. Boylan, and C. W. Price. 1999. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor σB. Microbiology 145:1069-1078. [DOI] [PubMed] [Google Scholar]
- 3.Allenby, N. E., N. O'Connor, Z. Pragai, N. M. Carter, M. Miethke, S. Engelmann, M. Hecker, A. Wipat, A. C. Ward, and C. R. Harwood. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 150:2619-2628. [DOI] [PubMed] [Google Scholar]
- 4.Anagnastopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antelmann, H., J. Bernhardt, R. Schmid, and M. Hecker. 1995. A gene at 333 degrees on the Bacillus subtilis chromosome encodes the newly identified σB-dependent general stress protein GspA. J. Bacteriol. 177:3540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 179:7251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asai, K., S. H. Baik, Y. Kasahara, S. Moriya, and N. Ogasawara. 2000. Regulation of the transport system for C4-dicarboxylic acids in Bacillus subtilis. Microbiology 146:263-271. [DOI] [PubMed] [Google Scholar]
- 9.Auger, S., W. H. Yuen, A. Danchin, and I. Martin-Verstraete. 2002. The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination. Microbiology 148:507-518. [DOI] [PubMed] [Google Scholar]
- 10.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 11.Bookstein, C., C. W. Edwards, N. V. Kapp, and F. M. Hulett. 1990. The Bacillus subtilis 168 alkaline phosphatase III gene: impact of a phoAIII mutation on total alkaline phosphatase synthesis. J. Bacteriol. 172:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambert, R., Y. Pereira, and M. F. Petit-Glatron. 2003. Purification and characterization of YfkN, a trifunctional nucleotide phosphoesterase secreted by Bacillus subtilis. J. Biochem. (Tokyo) 134:655-660. [DOI] [PubMed] [Google Scholar]
- 14.Chesnut, R. S., C. Bookstein, and F. M. Hulett. 1991. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol. Microbiol. 5:2181-2190. [DOI] [PubMed] [Google Scholar]
- 15.Corvey, C., T. Stein, S. Dusterhus, M. Karas, and K. D. Entian. 2003. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem. Biophys. Res. Commun. 304:48-54. [DOI] [PubMed] [Google Scholar]
- 16.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 17.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of σB and σH. J. Bacteriol. 180:6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eder, S., L. Shi, K. Jensen, K. Yamane, and F. M. Hulett. 1996. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology 142:2041-2047. [DOI] [PubMed] [Google Scholar]
- 20.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 21.Engelmann, S., and M. Hecker. 1996. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63-69. [DOI] [PubMed] [Google Scholar]
- 22.Eymann, C., H. Mach, C. R. Harwood, and M. Hecker. 1996. Phosphate-starvation-inducible proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 142:3163-3170. [DOI] [PubMed] [Google Scholar]
- 23.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez, M., and S. M. Cutting. 1997. Identification of a new σB-controlled gene, csbX, in Bacillus subtilis. Gene 188:29-33. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 26.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 27.Huisman, G. W., D. A. Siegele, M. M. Zambrano, and R. Kolter. 1996. Morphological and physiological changes during stationary phase, p. 1672-1682. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 28.Hulett, F. M. 2002. The Pho regulon, p. 193-203. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 29.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 30.Hulett, F. M., C. Bookstein, and K. Jensen. 1990. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J. Bacteriol. 172:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulett, F. M., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. F. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igo, M., M. Lampe, C. Ray, W. Schafer, C. P. Moran, Jr., and R. Losick. 1987. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J. Bacteriol. 169:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain, A. K., and R. C. Dubes. 1988. Algorithms for clustering data. Prentice-Hall, Englewood Cliffs, N.J.
- 34.Jongbloed, J. D., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Muller. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350-41357. [DOI] [PubMed] [Google Scholar]
- 35.Jurgen, B., T. Schweder, and M. Hecker. 1998. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol. Gen. Genet. 258:538-545. [DOI] [PubMed] [Google Scholar]
- 36.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 37.Lahooti, M., and C. R. Harwood. 1999. Transcriptional analysis of the Bacillus subtilis teichuronic acid operon. Microbiology 145:3409-3417. [DOI] [PubMed] [Google Scholar]
- 38.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 39.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP∼P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, W., and F. M. Hulett. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 41.Loos, A., C. Glanemann, L. B. Willis, X. M. O'Brien, P. A. Lessard, R. Gerstmeir, S. Guillouet, and A. J. Sinskey. 2001. Development and validation of Corynebacterium DNA microarrays. Appl. Environ. Microbiol. 67:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 43.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauel, C., M. Young, A. Monsutti-Grecescu, S. A. Marriott, and D. Karamata. 1994. Analysis of Bacillus subtilis tag gene expression using transcriptional fusions. Microbiology 140:2279-2288. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moszer, I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28-36. [DOI] [PubMed] [Google Scholar]
- 48.Moszer, I., P. Glaser, and A. Danchin. 1995. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology 141:261-268. [DOI] [PubMed] [Google Scholar]
- 49.Muller, J. P., Z. An, T. Merad, I. C. Hancock, and C. R. Harwood. 1997. Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143:947-956. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura, A., Y. Koide, H. Miyazaki, A. Kitamura, H. Masaki, T. Beppu, and T. Uozumi. 1992. Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem. 209:121-127. [DOI] [PubMed] [Google Scholar]
- 51.Nakano, M. M., Y. Zhu, M. Lacelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 37:1198-1207. [DOI] [PubMed] [Google Scholar]
- 52.Nakatani, Y., W. L. Nicholson, K. D. Neitzke, P. Setlow, and E. Freese. 1989. Sigma-G RNA polymerase controls forespore-specific expression of the glucose dehydrogenase operon in Bacillus subtilis. Nucleic Acids Res. 17:999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasser, W., A. C. Awade, S. Reverchon, and J. Robert-Baudouy. 1993. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett. 335:319-326. [DOI] [PubMed] [Google Scholar]
- 54.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsot, C., and G. N. Cohen. 1988. Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogenase. Structural and evolutionary relationships with Escherichia coli aspartokinases-homoserine dehydrogenases I and II. J. Biol. Chem. 263:14654-14660. [PubMed] [Google Scholar]
- 56.Paul, S., S. Birkey, W. Liu, and F. M. Hulett. 2004. Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EσA- and EσE-responsive promoters by phosphorylated PhoP. J. Bacteriol. 186:4262-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perego, M., and J. A. Hoch. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:1549-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petersohn, A., H. Antelmann, U. Gerth, and M. Hecker. 1999. Identification and transcriptional analysis of new members of the σB regulon in Bacillus subtilis. Microbiology 145:869-880. [DOI] [PubMed] [Google Scholar]
- 60.Pragai, Z., N. E. Allenby, N. O'Connor, S. Dubrac, G. Rapoport, T. Msadek, and C. R. Harwood. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pragai, Z., C. Eschevins, S. Bron, and C. R. Harwood. 2001. Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J. Bacteriol. 183:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prágai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 63.Prágai, Z., and C. R. Harwood. 2000. Screening for mutants affected in their response to phosphate, p.245-249. In W. Schumann, S. D. Ehrlich, and N. Ogasawara (ed.), Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 64.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 65.Qi, Y., and F. M. Hulett. 1998. PhoP-P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 66.Qi, Y., and F. M. Hulett. 1998. Role of Pho∼P in transcriptional regulation of genes involved in cell wall anionic polymer biosynthesis in Bacillus subtilis. J. Bacteriol. 180:4007-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reischl, S., S. Thake, G. Homuth, and W. Schumann. 2001. Transcriptional analysis of three Bacillus subtilis genes coding for proteins with the alpha-crystallin domain characteristic of small heat shock proteins. FEMS Microbiol. Lett. 194:99-103. [DOI] [PubMed] [Google Scholar]
- 69.Robichon, D., M. Arnaud, R. Gardan, Z. Pragai, M. O'Reilly, G. Rapoport, and M. Debarbouille. 2000. Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J. Bacteriol. 182:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, and R. Losick. 1990. Control of developmental transcription factor σF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sloma, A., G. A. Rufo, Jr., K. A. Theriault, M. Dwyer, S. W. Wilson, and J. Pero. 1991. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J. Bacteriol. 173:6889-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soldo, B., V. Lazarevic, M. Pagni, and D. Karamata. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol. Microbiol. 31:795-805. [DOI] [PubMed] [Google Scholar]
- 73.Stephenson, S., C. Mueller, M. Jiang, and M. Perego. 2003. Molecular analysis of Phr peptide processing in Bacillus subtilis. J. Bacteriol. 185:4861-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 75.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tosato, V., A. M. Albertini, M. Zotti, S. Sonda, and C. V. Bruschi. 1997. Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology 143:3443-3450. [DOI] [PubMed] [Google Scholar]
- 78.Tsuge, K., T. Ano, M. Hirai, Y. Nakamura, and M. Shoda. 1999. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 43:2183-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 80.Voelker, U., A. Dufour, and W. G. Haldenwang. 1995. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of σB. J. Bacteriol. 177:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Volker, U., K. K. Andersen, H. Antelmann, K. M. Devine, and M. Hecker. 1998. One of two osmC homologs in Bacillus subtilis is part of the σB-dependent general stress regulon. J. Bacteriol. 180:4212-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.