Abstract
The major cell wall polysaccharide of mycobacteria is a branched-chain arabinogalactan in which arabinan chains are attached to the 5 carbon of some of the 6-linked galactofuranose residues; these arabinan chains are composed exclusively of d-arabinofuranose (Araf) residues. The immediate precursor of the polymerized Araf is decaprenylphosphoryl-d-Araf, which is derived from 5-phosphoribose 1-diphosphate (pRpp) in an undefined manner. On the basis of time course, feedback, and chemical reduction experiment results we propose that decaprenylphosphoryl-Araf is synthesized by the following sequence of events. (i) pRpp is transferred to a decaprenyl-phosphate molecule to form decaprenylphosphoryl-β-d-5-phosphoribose. (ii) Decaprenylphosphoryl-β-d-5-phosphoribose is dephosphorylated to form decaprenylphosphoryl-β-d-ribose. (iii) The hydroxyl group at the 2 position of the ribose is oxidized and is likely to form decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose. (iv) Decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose is reduced to form decaprenylphosphoryl-β-d-Araf. Thus, the epimerization of the ribosyl to an arabinosyl residue occurs at the lipid-linked level; this is the first report of an epimerase that utilizes a lipid-linked sugar as a substrate. On the basis of similarity to proteins implicated in the arabinosylation of the Azorhizobium caulidans nodulation factor, two genes were cloned from the Mycobacterium tuberculosis genome and expressed in a heterologous host, and the protein was purified. Together, these proteins (Rv3790 and Rv3791) are able to catalyze the epimerization, although neither protein individually is sufficient to support the activity.
The cell envelope of Mycobacterium tuberculosis is made up of three major components: a plasma membrane, a covalently linked mycolic acid, arabinogalactan, and peptidoglycan complex, and polysaccharide-rich capsule-like material. The mycolic acid, arabinogalactan, and peptidoglycan complex is composed of a cross-linked peptidoglycan, which is covalently linked to arabinogalactan chains via phosphoryl-N-acetylglucosaminosyl-rhamnosyl linkage units (3). The arabinogalactan polymer is unique in that both the arabinose (Ara) and galactose (Gal) are in the furanose configuration (9). Decaprenylphosphoryl-d-Araf (DPA) is the only known precursor of polymerized Araf residues in mycobacteria (21). In general, prenylphosphoryl sugars are synthesized through transfer of the sugar from nucleotide sugar precursors, as is the case for mycobacterial decaprenylphosphoryl-d-mannose (18) and decaprenylphosphoryl-d-glucose (16). Surprisingly, DPA does not appear to be synthesized from a sugar nucleotide since concerted attempts to identify a nucleotide diphosphate arabinose in mycobacteria were unsuccessful (authors' unpublished data and reference 8). However, feeding various labeled sugars to growing cultures of M. smegmatis suggested that the d-Araf residues derive from the pentose phosphate shunt (8, 14). In keeping with this observation, it was demonstrated that DPA is, in fact, formed from 5-phosphoribose diphosphate (pRpp) (15).
Thus, the synthesis and activation of d-Ara in mycobacteria apparently follow a series of unique biosynthetic steps unlike any of those described for other organisms. Three reactions are expected: transfer of a 5-phosphopentose to decaprenyl phosphate, removal of the 5′ phosphate, and epimerization. However, the order of these steps and the identity of the enzymes catalyzing them have remained unknown. Structures of likely intermediates in the formation of DPA and three possibilities for when the epimerization occurs are illustrated in Fig. 1. We had previously speculated that the epimerization occurred at the pRpp or decaprenylphosphoryl 5-phosphoribose (DPPR) level (15). Other groups have speculated that decaprenylphosphoryl ribose (DPR) is the precursor of DPA (8, 20). The data presented here, combined with insight provided by Azorhizobium genetics, allowed definition of the pathway from pRpp to DPA in mycobacteria, including the order of events and identification of a cofactor, an unexpected intermediate, and the enzymes responsible for the epimerization.
FIG. 1.
Potential intermediates in the biosynthetic pathway from pRpp to DPA in mycobacteria. The isoprenoid moiety is drawn to conform to the structure of decaprenyl phosphate identified in mycobacteria (21). Epimerization could occur between the compounds indicated by the horizontal arrows.
METHODS AND MATERIALS
Preparation of enzymatically active membrane and cell wall-enriched fractions.
M. smegmatis mc2155 cells were grown in nutrient broth (EM Science) to midlog phase, harvested, washed, and stored at −70°C until required (11). Cells (10 g) were suspended in 40 ml of buffer A (50 mM morpholinepropanesulfonic acid [MOPS] [pH 7.9], 5 mM 2-mercaptoethanol, 10 mM MgCl2), subjected to probe sonication (11), and centrifuged at 23,000 × g for 20 min at 4°C. The pellet was resuspended in buffer A, and Percoll (Amersham Pharmacia Biotech) was added to achieve a 60% suspension, which was centrifuged at 23,000 × g for 60 min at 4°C. The white upper band, containing a particulate cell wall-enriched fraction, was isolated and Percoll was removed by repeated rounds of suspension in buffer A and centrifugation. The cell wall-enriched fraction was resuspended in buffer A to a protein concentration of 8 to 10 mg/ml for use. A membrane-enriched fraction (membrane protein) was obtained by centrifuging the 23,000 × g supernatant at 100,000 × g for 75 min at 4°C; the resulting supernatant was discarded, and the washed pellet was suspended in buffer A at a protein concentration of 15 to 20 mg/ml.
Reaction mixtures, incubation conditions, and fractionation of reaction products.
Typical reaction mixtures for assessing p[14C]Rpp (synthesized as previously described [15] from uniformly labeled d-[14C]glucose [American Radiolabeled Chemicals, Inc.] [100 mCi/mmol]) incorporation into the lipids typically contained 1 mg of membrane protein, 60 μM ATP, 200,000 dpm of p[14C]Rpp, and buffer A in a final volume of 160 μl. Occasionally, the reaction mixture was enriched with 0.5 to 0.7 mg of the cell wall fraction; when recombinant enzymes were added 5 μg of protein was used. Reaction mixtures were incubated for the indicated times and stopped by the addition of 3 ml of CHCl3-CH3OH (2:1), and the mixture was centrifuged. The supernatant (CHCl3-CH3OH phase) was removed from the pellet, and 340 μl of buffer A was added to form two phases (4). After thorough mixing and a brief centrifugation the upper, aqueous phase was removed and discarded. The bottom, organic phase was backwashed with CHCl3-CH3OH-H2O (3:47:48) (4). The sample was then dried under a stream of N2 at room temperature, and radiolabeled material was dissolved in 200 μl of CHCl3-CH3OH-H2O-NH4OH (65:25:3.6:0.5; solvent I) prior to liquid scintillation spectrometry and thin-layer chromatography (TLC) analysis.
In some cases, radiolabeled products (DPA, DPR, DPPR, and decaprenylphosphoryl-X [DPX]) were purified by preparative TLC or by anion exchange chromatography as described in the analytical procedures paragraph (see below) and used as substrates in subsequent experiments.
Reaction mixtures in which purified radiolabeled DPR and DPPR were used as substrates contained buffer A, 2 mg of membrane protein, 60 μM ATP, and DPR (10,000 dpm) or DPPR (10,000 dpm) in a final volume of 320 μl, and the reaction mixtures were incubated at 37°C for 2 h. Reactions were stopped with 6 ml of CHCl3-CH3OH (2:1), and the products were extracted as described above.
Cloning, expression, and purification of His-tagged Rv3790 and Rv3791.
Primers for PCR amplification of Rv3790 and Rv3791 were designed to include an NdeI restriction site (underlined) in both forward primers. A BamHI restriction site (underlined) as well as a stop codon were engineered into the reverse primers as follows: for Rv3790/Forward, CATATGTTGAGCGTGGGAGCTACC; for Rv3790/Reverse, GGATCCCTACAGCAGCTCCAAGCGTCGGGC; for Rv3791/Forward, CATATGGTTCTTGATGCCGTAGG; and for Rv3791/Reverse, GGATCCTCAGATGGGCAGCTTGCGGAAG.
Vent DNA polymerase (Biolab, Inc.) was used to amplify M. tuberculosis H37Rv genomic DNA. PCR products were checked for size by agarose gel electrophoresis and purified using QIAEX II agarose gel extraction kits (QIAGEN). The purified DNA fragments were ligated to pSTBlue-1 by use of Perfectly Blunt cloning kits (Novagen) to form Rv3790:pSTBlue-1 and Rv3791:pSTBlue-1. These plasmids were used to transform Escherichia coli DH5α. Single colonies were isolated; subsequently, plasmids were isolated and sequenced (Macromolecular Resources, Colorado State University). Rv3790:pSTBlue-1 and Rv3791:pSTBlue-1 were then digested with NdeI and BamHI, and the genes of interest were ligated to pET16b that had also been digested with the restriction enzymes to yield Rv3790c:pET16b and Rv3791c:pET16b. BL21(DE3) containing pKJE7 (Takara Mirus Bio Inc.) was used as the expression strain; transformed bacteria were treated with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at room temperature for 4 h, harvested, resuspended in buffer A, and sonicated. The sonicate was centrifuged at 20,000 × g, the supernatant was applied to an Ni-nitrilotriacetic acid agarose (Sigma-Aldrich) column, and the tagged recombinant proteins were purified.
Analytical procedures.
TLC analyses of enzymatically radiolabeled lipid-linked products were done using aluminum-backed silica gel 60 (F254) plates (Merck) developed in CHCl3-CH3OH-1 M CH3COONH4-concentrated (conc.) NH4OH-H2O (180:140:9:9:23; solvent II). Radiolabeled compounds were visualized by autoradiography (BioMax MR film [Kodak]). Preparative TLC of radiolabeled lipids was carried out under the same conditions. Radioactive bands of the individual lipids were visualized by autoradiography and were extracted with 1 ml of CHCl3-CH3OH (2:1) containing 0.05% NH4OH. Extraction of DPPR required a more polar solvent made up of water-ethanol-diethyl ether-pyridine-conc. ammonium hydroxide (15:15:5:1:0.017) (2). Extracts were dried under a stream of N2 at room temperature and dissolved in solvent I. In some cases chromatography on DEAE cellulose (acetate form) was used to purify radiolabeled lipids. DEAE cellulose was packed in a Pasteur pipette and equilibrated with CHCl3-CH3OH (2:1); lipid extracts were dried under N2, dissolved in CHCl3-CH3OH (2:1), and loaded on the column. The material bound to the column was eluted stepwise with CHCl3-CH3OH (2:1), CH3OH, and CH3OH containing 10, 50, and 70 mM ammonium acetate. The partially purified lipids were desalted by phase partitioning (4).
Mild acid hydrolysis of the radiolabeled lipids was conducted in 50 μl of 1-propanol-100 μl 0.02 N HCl at 60°C for 30 min, the solution was neutralized with 10 μl of 0.2 N NaOH, and 600 μl of CHCl3-CH3OH (2:1) was added to form two phases. Radioactive material in both phases was subjected to liquid scintillation spectrometry and analyzed by TLC.
Mild base hydrolysis of the radiolabeled lipids was conducted in 200 μl of CHCl3-CH3OH (1:1) containing 0.1 M NaOH. The mixture was incubated at 37°C for 20 min and neutralized with glacial acetic acid. CHCl3 (100 μl) and 60 μl of H2O were added, the solution was mixed, and the resulting biphase mixture was centrifuged. The upper (aqueous) phase was removed, and the lower (organic) phase was evaporated under a stream of N2. The radiolabeled lipids were dissolved in solvent I, and aliquots were subjected to liquid scintillation spectrometry and TLC analysis.
For chemical reduction of DPX, TLC-purified material was dried under N2 and treated with 100 μl of 1 M NH4OH in 50% ethanol containing 10 mg/ml of NaBH4 overnight at room temperature. Subsequently, 3 ml of CHCl3-CH3OH (2:1) and 0.5 ml of H2O were added, which resulted in formation of two phases. The lower (organic) phase was removed, dried under a stream of N2, and analyzed by TLC. Sugar analysis of the reduced material was performed after complete acid hydrolysis as described below.
In order to identify radiolabeled sugars, enzymatically labeled material was hydrolyzed in 2 M CF3COOH at 120°C for 1 h, dried under conditions of N2, washed twice with methanol, and partitioned between hexane and water. The hexane was removed and discarded, and the aqueous phase was dried on a rotary vacuum evaporator (Savant). Repeated washes of the hydrolyzed samples with deionized water removed residual CF3COOH. The hydrolysates were subjected to TLC analysis on silica gel 60 plates (F254; Merck) developed twice in pyridine-ethyl acetate-glacial acetic acid-water (5:5:1:3). TLC plates were subjected to autoradiography as described above. Unlabeled standards were visualized by charring after spraying the plates with a solution of 10% cupric sulfate in 8% phosphoric acid. In some cases sugars were analyzed by anion exchange high-performance liquid chromatography on a PA-1 column as previously described (15).
Bacterial alkaline phosphatase (Invitrogen) was used to remove the phosphate from the 5 position of ribose 5-phosphate released from the DPPR by acid hydrolysis in buffer containing 10 mM TRIS-HCl (pH 8.0).
Other procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis was performed using 12% gels; proteins were visualized with Coomassie brilliant blue R250. Polyvinylidene difluoride membranes were used for Western blot analysis, and monoclonal antipolyhistidine (mouse immunoglobulin G2a isotype; Sigma-Aldrich) and anti-mouse immunoglobulin G-AP conjugate (Sigma-Aldrich) were used as primary and secondary antibodies. Protein concentrations were estimated using a Pierce BCA kit. Restriction digests, ligations, and electroporations were done as described by Sambrook and Russell (13) unless otherwise noted. BLAST searches were done on the National Center for Biotechnology Information website and the Mycobacterium tuberculosis Structural Genomics Consortium website by use of standard protein-protein BLAST (blastp). Alignments were done using multiple sequence alignments with hierarchical clustering using the “Multalin” interface at the Institut National dela Recherche Agronomique (Toulouse, France) website.
RESULTS
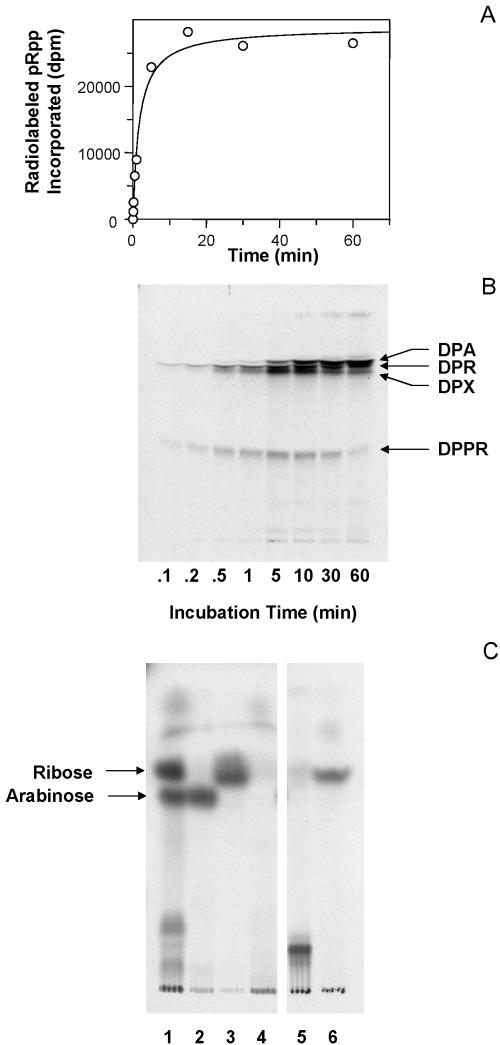
As shown previously (15), when p[14C]Rpp is incubated with a particulate fraction prepared from M. smegmatis, radioactivity is rapidly incorporated into CHCl3-CH3OH (2:1) soluble material. The incorporation of radioactivity into lipids increased linearly with time for approximately 5 min and then reached a plateau (Fig. 2A). Analysis by TLC indicated that the radioactivity was primarily incorporated into four compounds, the relative amounts of which differed over time (Fig. 2B). Three of these compounds were identified as DPPR, DPR, and DPA on the basis of previous studies (15), sensitivity to mild acid hydrolysis, and sugar analysis (Fig. 2C). A fourth compound, which ran as a somewhat diffuse band just below DPR, was stable to mild base and sensitive to mild acid hydrolysis. Thus, it had the properties of a prenyl phosphate-linked compound and was designated decaprenylphosphoryl-X.
FIG. 2.
Enzymatic conversion of p[14C]Rpp into [14C]DPA. (A) Incorporation of radioactivity from p[14C]Rpp into CHCl3-CH3OH (2:1) soluble material. (B) TLC analysis of enzymatically radiolabeled chloroform-methanol (2:1) soluble material. Enzymatically radiolabeled material was applied to aluminum-backed silica gel 60 F254 TLC plates (10 by 10 cm), which were developed in CHCl3-CH3OH-1 M CH3COONH4-conc. NH4OH-H2O (180:140:9:9:23). Radiolabeled compounds were visualized by autoradiography. (C) TLC analysis of sugar residues released by treatment with 2 M TFA. Lane 1, entire lipid extract; lane 2, purified DPA; lane 3, purified DPR; lane 4, purified DPX; lane 5, purified DPPR; lane 6, purified DPPR that was treated with alkaline phosphatase after acid hydrolysis. Enzymatically radiolabeled material was applied to aluminum-backed silica gel 60 F254 TLC plates (10 by 10 cm), which were developed twice in pyridine-ethyl acetate-glacial acetic acid-water (5:5:1:3). The arrows indicate the positions of authentic ribose and arabinose. Radiolabeled compounds were visualized by autoradiography, and unlabeled standards were visualized by charring. In all cases reaction mixtures contained 1 mg of membrane-enriched protein, 0.6 mg of cell wall protein, 60 μM ATP, 200,000 dpm of p[14C]Rpp, and buffer A in a final volume of 160 μl. Reactions were stopped by addition of CHCl3-CH3OH (2:1). Extracts were subsequently treated as described in Methods and Materials prior to liquid scintillation spectrometry or TLC analysis.
Experiments conducted over very short time periods indicated that radioactivity derived from p[14C]Rpp is rapidly incorporated into DPPR and DPR. However, even when incubation for a period as short as 0.1 min was used it was not possible to determine which molecule was labeled first by this method. However, logic dictates the formation of DPPR was followed by dephosphorylation to DPR, a hypothesis supported by the recent identification of a DPPR synthase (7). Clearly, the compound designated DPX became labeled after 0.5 min, and significant labeling was observed in DPA after 1 min of incubation. Incorporation of radioactivity continued for periods of up to 60 min, but the amount of radioactivity in DPPR, DPR, and DPX began to decrease after 30 min. The data shown in Fig. 2 suggest that DPA derives from either DPPR or DPR and that DPX could be an intermediate.
In contrast to an earlier report (15), the radiolabeled material identified as DPPR did not contain any decaprenylphosphoryl phosphoarabinose. This was shown by analysis of products after acid hydrolysis and alkaline phosphatase treatment. TLC analysis of the radiolabeled material after acid hydrolysis indicated the presence of a compound that comigrated with various glycosyl phosphates (Fig. 2C, lane 5; pentose phosphates do not separate on the TLC system used). However, when this material was subsequently treated with alkaline phosphatase, a compound that comigrated with ribose was liberated with no indication of the presence of Ara (Fig. 2C, lane 6), suggesting that pRpp is not epimerized to pApp. Sugar analysis of the bands identified as DPR and DPA indicated that these compounds contained only ribose and Ara, respectively. Interestingly, acid hydrolysis of DPX did not release radioactivity that could be associated with a glycose residue by TLC analysis (Fig. 2C, lane 4).
In order to examine the possibility that epimerization occurs at the level of DPR (Fig. 1) and the precursor-product relationship of DPPR and DPR, both compounds were purified by anion exchange chromatography on DEAE cellulose and subsequently incubated with a membrane preparation from M. smegmatis. Figure 3A shows that DPPR can be enzymatically converted to DPR and DPA. In addition, there is a shadow just below the DPR band, suggesting that DPX may also have been formed. Figure 3B shows that the addition of purified DPR can also be enzymatically converted to DPA and perhaps DPX. However, there was no DPPR formed from DPR. Thus, the data clearly indicate that DPPR is a precursor of DPR and importantly, in contrast to our earlier speculation (15), that DPR is a precursor of DPA.
FIG. 3.
Identification of intermediates in DPA synthesis. Purified DPPR and DPR (lane 1 of panels A and B, respectively) were incubated with the particulate fraction prepared from M. smegmatis for 120 min (lane 2 of panels A and B). Reaction mixtures contained buffer A, 2 mg of membrane-enriched protein, 60 μM ATP, and DPR (10,000 dpm) or DPPR (10,000 dpm). Reactions were stopped with 6 ml of CHCl3-CH3OH (2:1). Extracts were subsequently treated as described in Methods and Materials prior to TLC analysis. Radiolabeled material was applied to aluminum-backed silica gel 60 F254 TLC plates (10 by 10 cm), which were developed in CHCl3-CH3OH-1 M CH3COONH4-conc. NH4OH-H2O (180:140:9:9:23). Radiolabeled compounds were visualized by autoradiography.
In keeping with the fact that epimerization often involves oxidation and reduction, the addition of nicotinamide adenine dinucleotides to the reaction mixture resulted in an increase in the formation of DPA (Fig. 4A). Unexpectedly, there was little specificity for either the oxidation state of the nicotinamide adenine dinucleotides or the presence of a phosphate. That is, addition of NADPH and addition of NADH appeared to be equally effective, as did addition of NAD and addition of NADP (data not shown). Despite this lack of specificity, these results indicated that epimerization occurred via a sequential oxidation-reduction mechanism and suggested that DPX may be an oxidized intermediate such as a pentose with a keto function at C-2 or C-3. In support of this hypothesis, reduction of DPX with sodium borohydride resulted in the formation of a compound chromatographically identical to DPA (Fig. 4B). Subsequent acid hydrolysis of the chemically reduced material released a compound that is chromatographically identical to Ara (Fig. 4C), indicating that DPX is not only an intermediate in the formation of DPA but is also likely the immediate precursor of DPA.
FIG. 4.
Reduction of DPX. (A) Effect of addition of NAD on DPA formation. Reactions were done under standard conditions (lane 1) or with the addition of 50 μM NADH (lane 2). Standard reaction mixtures contained 1 mg of membrane-enriched protein, 60 μM ATP, 200,000 dpm of p[14C]Rpp, and buffer A in a final volume of 160 μl. Reactions were incubated for 1 h and stopped by addition of CHCl3-CH3OH (2:1). Extracts were subsequently treated as described in Methods and Materials prior to TLC analysis. Radiolabeled material was applied to aluminum-backed silica gel 60 F254 TLC plates (10 by 10 cm), which were developed in CHCl3-CH3OH-1 M CH3COONH4-conc. NH4OH-H2O (180:140:9:9:23). Radiolabeled compounds were visualized by autoradiography. (B) Effect of sodium borohydride reduction on DPX. Purified DPX was treated overnight at room temperature with 100 μl of 1 M NH4OH in 50% ethanol containing 10 mg/ml of NaBH4. Samples were phase partitioned as described in Methods and Materials and analyzed by TLC. Lane 1 contains unreduced DPX, and lane 2 contains reduced DPX. (C) Sugar analysis of the reduced material was performed after complete acid hydrolysis as described in Methods and Materials. Lane 1 contains hydrolysate from DPX, and lane 2 contains hydolysate from reduced DPX. The arrows indicate the position of authentic unlabeled ribose and arabinose. Radiolabeled compounds were visualized by autoradiography, and unlabeled standards were visualized by charring.
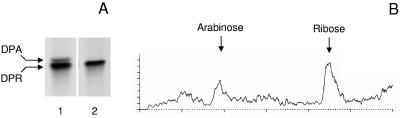
Based on similarity to the Noe proteins of Azorhizobium caulinodans it was hypothesized that Rv3790 and Rv3791 of M. tuberculosis could be involved in the epimerization of ribose to Araf. Rv3790 is annotated as a probable oxidoreductase and has a flavin adenine dinucleotide (FAD) binding domain, while Rv3791 is annotated as a probable short-chain dehydrogenase that contains a FAD/NAD(P)-binding Rossmann fold (TB Structural Genomics Consortium website). Therefore, the genes encoding these proteins were cloned, expressed in E. coli, and subsequently purified to near homogeneity on immobilized metal affinity columns (data not shown). When both proteins were included in a reaction mixture containing FAD, NAD+, and NADPH, DPR was epimerized to DPA (Fig. 5A, lane 1), but no epimerization was observed when the reaction mixture contained only the enzymes, DPR, and FAD (Fig. 5A, lane 2). In the first case, formation of Ara was confirmed by sugar analysis (Fig. 5B). No conversion to DPA was observed when the enzymes were added to the reaction mixture individually.
FIG. 5.
Purified, recombinant Rv3790 and Rv3971 catalyze the conversion of DPR to DPA. (A) Pure, radiolabeled DPR was incubated with Rv3790 and Rv3791 for 2 h, and the radioactive material was analyzed by TLC. Reaction mixtures contained DPR (4,000 dpm), 5 μg Rv3790, 5 μg Rv3791, 1 mM NAD+, 1 mM FAD, and 1 mM NADH in buffer A (lane 1) or DPR (4,000 dpm), 5 μg Rv3790, 5 μg Rv3791, and 1 mM FAD+ in buffer A (lane 2). The final volume of the reaction mixtures was 60 μl. (B) The radiolabeled material shown in panel A, lane 1, was hydrolyzed with 2 M TFA and subjected to high-performance liquid chromatography analysis as described in Methods and Materials.
DISCUSSION
The data presented here support the hypothesis that in mycobacteria, DPA is formed from pRpp as follows. First, pRpp is converted to DPPR (via a 5-phospho-α-d-ribose-1-diphosphate-decaprenyl-phosphate 5-phosphoribosyltransferase) (7), which is then dephosphorylated to form DPR. DPR is subsequently oxidized to form DPX followed by reduction to form DPA.
If this hypothesis is correct, it is highly probable that DPX is the product of oxidation at C-2 of DPR, specifically decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose. This conclusion is supported by the observation that chemical reduction of DPX yields DPA. One might expect the reduction to yield both DPA and DPR, but the results suggest that the trans configuration is highly favored over the cis.
A reasonable alternative is that DPR could be oxidized to form decaprenylphosphoryl-3-keto-β-d-xylo-pentofuranose. Such a compound could support epimerization at C-2; epimerization α to carbonyl groups is well known, as demonstrated in the dTDP-rhamnose biosynthesis pathway (5, 6, 17). After epimerization the resulting decaprenylphosphoryl-3-keto-β-d-threo-pentofuranose could then be reduced to DPA. However, the expected products of chemical reduction of the xylo-pentofuranose would be DPR and/or decaprenylphosphoryl xylose, which were not observed. To date it has not been possible to isolate sufficient quantities of the rather unstable DPX to be able to do an unambiguous structural determination of this molecule; these studies are ongoing.
Given the probable identification of DPX as decaprenylphosphoryl-2-keto-β-d-erythro-pentofuranose it is interesting to compare the epimerization of DPR to DPA with that of UDP-glucose to UDP-galactose, which is catalyzed by GalE. The most significant difference between the mechanism proposed here and that of GalE is that the 2-keto pentofuranose is not a transient intermediate that remains bound to the enzyme; hence, it seemed likely that two separate enzymes for oxidation and reduction are required, as opposed to the single enzyme represented by GalE. The reaction catalyzed by GalE is complicated, as it requires nonstereospecificity of hydride return from the B side of the nicotinamide ring of NADH to the 4′ ketopyranose intermediate; this occurs via rotation of the intermediate in the active site of the enzyme (1, 19). Active site rotation of a 2-keto pentofuranose may be more difficult than that of a 4-keto hexopyranose, as the keto group would likely move further from the site of oxidation. However, if two separate enzymes are utilized rotation in the active site is obviously not required.
The identification of these enzymes was aided by related work on d-arabinofuranosylation of the nodulation factor in A. caulinodans. In what began as discrete studies, two of us (M.H. and W.D.) studying d-arabinofuranosylation of nodulation factor in A. caulinodans reported that inactivation of noeC or downstream genes in A. caulinodans prevents d-arabinofuranosylation of the nodulation factor (10). Subsequently, disruption of the downstream genes noeH, noeO, and noeP was also shown to disrupt d-arabinofuranosylation of the nodulation factor (data to be published elsewhere). Orthologs of noeC, noeH, and noeO (Rv3806c, Rv3790, and Rv3791, respectively) can be found in the M. tuberculosis genome. Of these, Rv3806c was shown to encode a DPPR synthase (7). Rv3790 and Rv3791 are both annotated as encoding oxidoreductases, thus fitting well with the hypothesis that two enzymes were involved in the epimerization of DPR to DPA.
Results presented here strongly suggest that Rv3790 and Rv3791 work in concert to catalyze the conversion of DPR to DPA. However, neither protein alone appears to be sufficient to catalyze the reaction individually (data not shown). Thus, it is likely that one of these enzymes oxidizes DPR to the keto intermediate and the other reduces the intermediate to DPA. Thus far, experiments indicate that little DPX accumulates when only one recombinant enzyme is present in reaction mixtures, making it difficult to unambiguously determine which protein catalyzes the initial oxidation.
A particularly exciting aspect of the formation of DPA in mycobacteria is that the epimerization occurs when the sugar is linked to a lipid and not a nucleotide, as exemplified by enzymes such as UDP-galactose 4-epimerase and ADP-l-glycero-d-mannoheptose 6-epimerase or as free sugars as in the case of d-ribulose-5-phosphate 3-epimerase and galactose mutarotase (see reference 1 for a review). It has previously been hypothesized that glycose residues could be epimerized at the lipid-linked level (8, 12, 20, 21), but this report appears to be the first unambiguous demonstration of epimerization of sugar attached to a prenylphosphate.
.
Acknowledgments
This work was supported by grants AI49151 (D.C.C.), AI18357 (P.J.B.), and AI33706 and R03TW200627 (M.R.M.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Allard, S. T. M., M. F. Giraud, and J. H. Naismith. 2001. Epimerases: structure, function and mechanism. Cell. Mol. Life Sci. 58:1650-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, W. W., and R. L. Lester. 1972. Turnover of inositol and phosphorus containing lipids in Saccharomyces cerevisiae—extracellular accumulation of glycerophosphorylinositol derived from phosphatidylinositol. Arch. Biochem. Biophys. 151:483-&. [DOI] [PubMed] [Google Scholar]
- 3.Crick, D. C., S. Mahapatra, and P. J. Brennan. 2001. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11:107R-118R. [DOI] [PubMed] [Google Scholar]
- 4.Folch, J., M. Lees, and G. H. S. Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 5.Giraud, M. F., G. A. Leonard, R. A. Field, C. Berlind, and J. H. Naismith. 2000. RmlC, the third enzyme of dTDP-l-rhamnose pathway, is a new class of epimerase. Nat. Struct. Biol. 7:398-402. [DOI] [PubMed] [Google Scholar]
- 6.Graninger, M., B. Nidetzky, D. E. Heinrichs, C. Whitfield, and P. Messner. 1999. Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar typhimurium LT2. J. Biol. Chem. 274:25069-25077. [DOI] [PubMed] [Google Scholar]
- 7.Huang, H., M. S. Scherman, W. D'Haeze, D. Vereecke, M. Holsters, D. C. Crick, and M. R. McNeil. 2005. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-α-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J. Biol. Chem. 280:24539-24543. [DOI] [PubMed] [Google Scholar]
- 8.Klutts, J. S., K. Hatanaka, Y. T. Pan, and A. D. Elbein. 2002. Biosynthesis of d-arabinose in Mycobacterium smegmatis: specific labeling from d-glucose. Arch. Biochem. Biophys. 398:229-239. [DOI] [PubMed] [Google Scholar]
- 9.McNeil, M., S. J. Wallner, S. W. Hunter, and P. J. Brennan. 1987. Demonstration that the galactosyl and arabinosyl residues in the cell-wall arabinogalactan of Mycobacterium leprae and Mycobacterium tuberculosis are furanoid. Carbohydr. Res. 166:299-308. [DOI] [PubMed] [Google Scholar]
- 10.Mergaert, P., W. D'Haeze, M. Fernandez-Lopez, D. Geelen, K. Goethals, J.-C. Prome, M. Van Montagu, and M. Holsters. 1996. Fucosylation and arabinosylation of Nod factors in Azorhizobium caulinodans: involvement of nolK, nodZ as well as noeC and/or downstream genes. Mol. Microbiol. 21:409-419. [DOI] [PubMed] [Google Scholar]
- 11.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 12.Russomando, G., and M. A. Dankert. 1992. Lipid monophosphate galacturonic acid, a new lipid linked sugar from Rhizobium. An. Asoc. Quim. Argent. 80:175-186. (In Spanish.) [Google Scholar]
- 13.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Scherman, M., A. Weston, K. Duncan, A. Whittington, R. Upton, L. Deng, R. Comber, J. D. Friedrich, and M. McNeil. 1995. Biosynthetic origin of mycobacterial cell wall arabinosyl residues. J. Bacteriol. 177:7125-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherman, M. S., L. Kalbe-Bournonville, D. Bush, Y. Xin, L. Deng, and M. McNeil. 1996. Polyprenylphosphate-pentoses in mycobacteria are synthesized from 5-phosphoribose pyrophosphate. J. Biol. Chem. 271:29652-29658. [DOI] [PubMed] [Google Scholar]
- 16.Schultz, J., and A. D. Elbein. 1974. Biosynthesis of mannosyl-phosphoryl polyprenols and glucosyl-phosphoryl polyprenols in Mycobacterium smegmatis— evidence for oligosaccharide-phosphoryl-polyprenols. Arch. Biochem. Biophys. 160:311-322. [DOI] [PubMed] [Google Scholar]
- 17.Stern, R. J., T. Y. Lee, T. J. Lee, W. Yan, M. S. Scherman, V. D. Vissa, S. K. Kim, B. L. Wanner, and M. R. McNeil. 1999. Conversion of dTDP-4-keto-6-deoxyglucose to free dTDP-4-keto-rhamnose by the rmIC gene products of Escherichia coli and Mycobacterium tuberculosis. Microbiology 145:663-671. [DOI] [PubMed] [Google Scholar]
- 18.Takayama, K., and D. S. Goldman. 1970. Enzymatic synthesis of mannosyl-1-phosphoryl-decaprenol by a cell-free system of Mycobacterium tuberculosis. J. Biol. Chem. 245:6251-6257. [PubMed] [Google Scholar]
- 19.Thoden, J. B., and H. M. Holden. 1998. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 37:11469-11477. [DOI] [PubMed] [Google Scholar]
- 20.Wolucka, B. A., and E. de Hoffmann. 1995. The presence of beta-d-ribosyl-1-monophosphodecaprenol in mycobacteria. J. Biol. Chem. 270:20151-20155. [DOI] [PubMed] [Google Scholar]
- 21.Wolucka, B. A., M. R. McNeil, E. de Hoffmann, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]