Abstract
Streptomyces sp. strain HK803 produces six analogues of phoslactomycin (Plm A through Plm F). With the exception of Plm B, these analogues contain a C-18 hydroxyl substituent esterified with a range of short-alkyl-chain carboxylic acids. Deletion of the plmS2 open reading frame (ORF), showing high sequence similarity to bacterial cytochrome P450 monooxygenases (CYPs), from the Plm biosynthetic gene cluster has previously resulted in an NP1 mutant producing only Plm B (N. Palaniappan, B. S. Kim, Y. Sekiyama, H. Osada, and K. A. Reynolds, J. Biol. Chem. 278:35552-35557, 2003). Herein, we report that a complementation experiment with an NP1 derivative (NP2), using a recombinant conjugative plasmid carrying the plmS2 ORF downstream of the ermE* constitutive promoter (pMSG1), restored production of Plm A and Plm C through Plm F. The 1.2-kbp plmS2 ORF was also expressed efficiently as an N-terminal polyhistidine-tagged protein in Streptomyces coelicolor. The recombinant PlmS2 converted Plm B to C-18-hydroxy Plm B (Plm G). PlmS2 was highly specific for Plm B and unable to process a series of derivatives in which either the lactone ring was hydrolyzed or the C-9 phosphate ester was converted to C-9/C-11 phosphorinane. This biochemical analysis and complementation experiment are consistent with a proposed Plm biosynthetic pathway in which the penultimate step is hydroxylation of the cyclohexanecarboxylic acid-derived side chain of Plm B by PlmS2 (the resulting Plm G is then esterified to provide Plm A and Plm C through Plm F). Kinetic parameters for Plm B hydroxylation by PlmS2 (Km of 45.3 ± 9.0 μM and kcat of 0.27 ± 0.04 s−1) are consistent with this step being a rate-limiting step in the biosynthetic pathway. The penultimate pathway intermediate Plm G has less antifungal activity than Plm A through Plm F and is not observed in fermentations of either the wild-type strain or NP2/pMSG1.
Bacterial cytochrome P450 monooxygenases (CYPs) are a major class of heme-thiolate-containing enzymes involved in the oxidative (detoxification of xenobiotic compounds) as well as in the biosynthetic reactions yielding secondary metabolites, steroids, and some vitamins (27). About 40% of the known bacterial CYPs are found in various species of the industrially important streptomycetes (18). The CYPs located in macrolide antibiotic biosynthetic gene clusters catalyze stereospecific and regiospecific oxidation of postpolyketide precursors, leading to both significant increase in antibiotic potency and structural diversity within these molecules (11, 19). An understanding of the Streptomyces CYP superfamily, particularly with respect to their substrate specificity, is an important step towards determining their versatility for developing novel secondary metabolites through combinatorial biosynthesis (19).
A number of enzymes of this family have already been overproduced, purified, and characterized, especially with respect to their substrate specificity. The erythromycin hydroxylases EryF and EryK from Saccharopolyspora erythrae, catalyzing hydroxylation of a 14-membered macrocyclic lactone ring at two different positions, have been shown to possess a strong substrate specificity (1, 12). The methymycin, neomethymycin, and pikromycin hydroxylase PikC from Streptomyces venezuelae is able to catalyze hydroxylation of both 12-membered and 14-membered macrolide rings with two different regioselectivities (2, 7, 28). More recently, S. coelicolor CYP154C1, not associated with any natural product biosynthetic gene cluster, has been characterized, and its structure has been determined (19). The enzyme was also shown to be able to catalyze the same hydroxylation reactions as PikC. Other examples of biochemical characterization of CYPs involved in polyketide biosynthesis include DoxA, catalyzing multiple steps in doxorubicin biosynthesis from Streptomyces sp. strain 5 (4, 5, 25), P-450sca-2 from Streptomyces carbophilus, involved in the production of pravastatin (26), and PimD from Streptomyces natalensis, which converts de-epoxypimaricin to pimaricin (14). Overall these studies have revealed that while some CYPS involved in biosynthetic processes can often catalyze multiple steps and appear to tolerate a variety of structurally related compounds, others have a more strict requirement for the natural substrate.
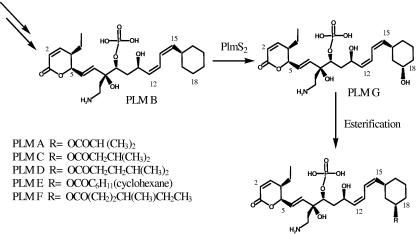
CYPs have also been proposed to be involved in the biosynthesis of phoslactomycins (Plms), potent protein phosphatase 2A (PP2A) inhibitors with both antifungal and antitumor activity (6, 16). Streptomyces sp. strain HK803 produces six analogues of Plms, namely Plm A through Plm F (Fig. 1) (24). With the exception of Plm B, these analogues contain a C-18 hydroxyl substituent esterified with a range of short-alkyl-chain carboxylic acids. The entire 75-kbp Plm biosynthetic gene cluster has been cloned, sequenced, and analyzed previously in our laboratory (17). The analysis revealed two open reading frames (ORFs), plmT4 and plmS2, both encoding proteins with high sequence similarity to each other as well as to several bacterial CYPs, including those involved in the biosynthesis of macrolide antibiotics. Insertional inactivation of plmS2 resulted in a mutant (NP1) producing only Plm B. This NP1-blocked-mutant study suggested (i) a Plm biosynthetic pathway in which the final two steps are a PlmS2-catalyzed C-18 hydroxylation of Plm B followed by esterification (Fig. 1) and (ii) that the other two proposed hydroxylation steps in Plm biosynthesis occur earlier in the pathway and are catalyzed by PlmT4 (17).
FIG. 1.
Proposed final steps of the Plm biosynthetic pathway and the role of PlmS2. The NP1 and NP2 mutants produce only Plm B. The NP2/pMSG1 mutant produces Plm A through Plm F with the exception of Plm B, while the wild-type strain produces Plm A through Plm F.
Here we report further characterization of the plmS2 ORF using both a genetic and biochemical approach. Complementation of NP2 (a derivative of the Plm B-producing NP1 mutant) with the plmS2 ORF under control of the ermE* promoter using pMSG1 led to restoration of production of Plm A as well as Plm C through Plm F. No Plm B was observed in the NP2/pMSG1 strain, in contrast to both the NP1 and NP2 mutants and the Streptomyces sp. HK803 wild-type strain. The plmS2 ORF was expressed as an N-terminal polyhistidine-tagged protein in Streptomyces coelicolor M511. The affinity-purified PlmS2 selectively converts Plm B to C-18-hydroxy Plm B (Plm G), the proposed final intermediate in the pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The NP2 mutant was generated from the NP1 mutant of Streptomyces sp. strain HK803 by deletion of the apramycin resistance gene using the standard method of PCR targeting (8). The plmS2 deletion mutants NP1 and NP2 were maintained on SY agar (1% soluble starch, 0.1% yeast extract, 0.1% N-Z amine type A), and Plm B was produced as described previously (17). Escherichia coli TG2 and ET12567 were grown at 37°C in Luria-Bertani medium supplemented with either ampicillin (100 μg/ml) or apramycin (50 μg/ml) when necessary (22). R2YE medium was used for regeneration of S. coelicolor M511 protoplasts after transformation, whereas MS agar (2% mannitol, 2% soy flour, 2% agar, made in tap water) was used for selecting potential NP2/pMSG1 exconjugants (10). Recombinant S. coelicolor M511 was grown in 50 ml YEME medium containing thiostrepton (10 μg/ml) at 30°C for 96 h. δ-Aminolevulinic acid (Sigma) (1 mM) and FeCl3 (0.5 mM), precursors for the heme prosthetic group, were added as described previously (9).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Streptomyces sp. strain HK803 | Wild type | Riken, Japan |
| NP1 mutant | plmS2 replaced by Amr | 17 |
| NP2 mutant | plmS2-deleted mutant; Ams | Unpublished |
| S. coelicolor M115 | 15 | |
| E. coli TG2 | supE hsd5 thi (lac-proAB) (srl-recA) 306::Tn10(tetr) F[traD36 proAB+laclq lac ZM15] | 22 |
| E. coli ET12567 | Methylation-deficient E. coli host | John Innes Center |
| E. coli ET12567/pUZ8002 | Nonmethylating E. coli harboring nontransmissible pUZ8002 (plasmid capable of mobilizing oriT-containing vectors) | John Innes Center |
| Plasmids | ||
| pET15b | E. coli expression vector, Apr | Novagen |
| pMSG | pET15b with 1.2-kbp NdeI-BamHI plmS2 ORF | This work |
| pGF200 | pSET152 with 2.2-kbp EcoRI S. collinus ccr fragment from pHL18 | 13 |
| pMSG1 | pGF200 with 0.7-kbp NdeI-BglI fragment from ccr replaced by 1.2-kbp NdeI-BamHI plmS2 | This work |
| pSE34 | pWHM3 with ermE* promoter | Pfizer Inc. |
| pMSG3 | pSE34 with 1.6-kbp XbaI-HindIII fragment from pMSG | This work |
Amr, apramycin resistance; Ams, apramycin sensitivity; Apr, ampicillin resistance.
Cloning of the plmS2 gene.
General DNA manipulations were performed following standard protocols (22). The plmS2 ORF was amplified using forward primer (5′-GGC GTC CTC CAT ATG GTG ACC GTC GAC C-3′), introducing a unique NdeI site at the 5′ end of the gene, and the reverse primer (5′-AGC GTG CGG GAT CCC TCA CCA GCG CA-3′), introducing a unique BamHI site downstream to the TGA translational stop codon. Cosmid clone 3A11 from the Streptomyces sp. strain HK803 genomic library was used as a template (17). The 1.2-kbp amplified DNA fragment was first cloned into the pCR2.1 TOPOTA vector (Invitrogen). The 1.2-kbp NdeI-BamHI insert was further subcloned into pET15b to generate pMSG. This plasmid was digested with XbaI and HindIII, excising the plmS2 gene, along with the pET15b sequence encoding an N-terminal polyhistidine tag and the T7 gene 10 ribosome binding site (RBS) and was cloned into the corresponding sites of the Streptomyces expression vector (pSE34) 3′ of the ermE* promoter. The resulting recombinant plasmid, pMSG3, was sequenced to confirm the insert. The plasmid pMSG1 was generated by cloning the 1.2-kbp NdeI-BamHI plmS2 fragment from pMSG into NdeI-BglI-digested pGF200 (a derivative of the Streptomyces conjugative vector pSET152) (13) such that it was also under the control of the constitutive ermE* promoter.
Transformations.
Preparation and transformation of competent E. coli cells as well as that of Streptomyces protoplasts was performed by standard methods (10, 22). Conjugative transfer of pMSG1 into the NP2 mutant was also performed by standard procedures (10).
Expression and purification of N-terminal hexahistidine-PlmS2.
Recombinant S. coelicolor M511/pMSG3 was grown in YEME medium as described above. After 96 h of incubation the cells were harvested, washed with 10.3% sucrose solution, and resuspended in 10 ml conversion buffer (50 mM potassium phosphate buffer, pH 7.5, 0.5 mM EDTA, 0.1 mM dithiothreitol, 10% glycerol). Lysozyme (1 mg/ml) and phenylmethylsulfonyl fluoride (1 mM) were added, and the tube was incubated on ice for 30 min. Cells were further ruptured by sonication, cell debris was removed by centrifugation, and the soluble fraction was applied to Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) equilibrated with conversion buffer. The column was washed with 20 ml of conversion buffer, and the protein was eluted with 300 mM imidazole. Fractions of 500 μl were collected, and aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to check the presence of PlmS2. Fractions containing the PlmS2 protein were pooled and subjected to desalting using a DG-10 column (Bio-Rad). Protein elution was monitored at 280 nm. Protein concentration was determined by the Bio-Rad protein assay dye reagent concentrate, using bovine serum albumin as a standard. Typical yields of purified PlmS2 were 1.5 mg/liter. The spectrum of the purified PlmS2 in conversion buffer was recorded from 250 nm to 650 nm.
Western blot analysis.
Cell extracts of S. coelicolor M511/pMSG3 were run on SDS-PAGE gels and transferred to polyvinylidene difluoride (Invitrogen) membranes by standard techniques. The membrane was blocked by incubation in 5% nonfat dry milk in Tris-buffered saline (TBS) at room temperature for 1 h. The blot was further incubated with anti-polyhistidine antibodies (Sigma) conjugated with alkaline phosphatase (AP) for 2 h. A triple wash with TBS containing 0.05% Tween-20 was performed between incubations. At the end the membrane was treated with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate mix (Roche) in AP-specific buffer. Color development was stopped by washing the membrane with tap water thoroughly.
Plm production and analysis.
Production, purification, and analysis of Plm B and other Plm analogues using high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) was carried out as described previously (17). Plm B was stored as a concentrated solution (10 mM) in methanol.
Conversion of Plm B to Plm G in vitro.
Enzymatic conversion of Plm B to Plm G was monitored by HPLC assay. The purified PlmS2 (0.3 μM) was incubated in conversion buffer with Plm B (100 μM), spinach ferredoxin (50 μg), ferredoxin-NADP reductase (0.1 U), NADPH (1 μmol), NADP+ (1 μmol), glucose-6-phosphate (10 μmol), and glucose-6-phosphate dehydrogenase (1 U) in a reaction volume of 1 ml at 30°C for 10 min. NADP+, glucose-6-phosphate, and glucose-6-phosphate dehydrogenase were added as an “NADPH regenerating system” as described elsewhere (25). Simultaneous enzyme and substrate controls were also run. An aliquot of reaction mixture (500 μl) was injected on HPLC to detect Plm G formation under conditions described previously (17). The identity of Plm G was assessed by LC-MS. One unit of enzyme activity was defined as the amount of enzyme hydroxylating 1 μM of Plm B per min under these assay conditions.
The kinetics of Plm B hydroxylation was determined using 0.3 μM purified His-tagged PlmS2 with varying substrate concentrations (2 to 200 μM). The data from lower substrate concentration reactions were used to fit the Michaelis-Menten equation to determine the enzymatic kinetic parameters.
The substrate specificity of PlmS2 was probed by incubating Plm B (100 μM) in 100 mM potassium chloride-hydrochloride buffer, pH 2.0, for 12 h. A similar incubation was carried out under basic conditions using 100 mM carbonate-bicarbonate buffer at pH 10.0 for 24 h. The pH of both of these solutions was then neutralized, and the resulting mixture was incubated for 1 h with PlmS2 under standard assay conditions. Control incubations without PlmS2 were also carried out.
RESULTS
Genetic complementation of plmS2 deletion mutant NP2.
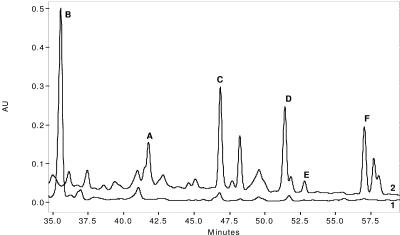
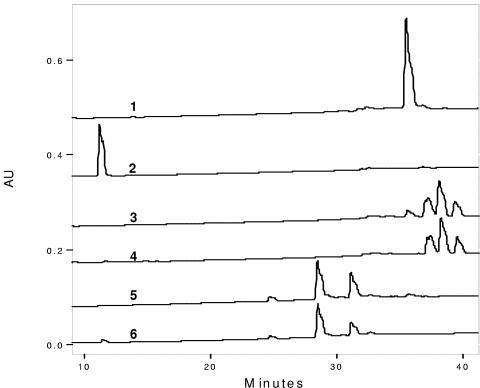
The apramycin-sensitive NP2 derivative of the NP1 mutant (ΔplmS2) strain produced only Plm B (Fig. 2). Complementation of the NP2 mutant by recombinant conjugative plasmid pMSG1 resulted in a strain producing five Plm products, namely A, C, D, E, and F (Fig. 2). The production of these Plms was confirmed by coinjections with purified authentic standards and by mass spectrometric analysis. The results of this complementation experiment are consistent with the proposed role for the plmS2 gene product in the hydroxylation of Plm B. No Plm B was detected in this strain, whereas Plm B typically represents about 13% of the total Plm pool produced by the wild-type strain. More complete conversion of Plm B in the NP2/pMSG1 mutant is likely due to higher levels of PlmS2, and these in turn are a result of the difference in the promoters and RBS used for expression of the corresponding gene.
FIG. 2.
Complementation of NP2 with plmS2 restores production of Plm A and Plm C through Plm F. HPLC C-18 column chromatogram of an Amberlitel XAD 4 extract of fermentation broth of (line 1) NP2 (producer of Plm B) and (line 2) NP2/pMSG1 (producer of Plm A and Plm C through Plm F). AU, arbitrary units.
Homologous expression of PlmS2.
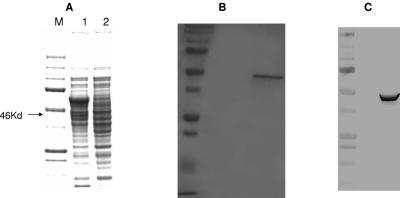
To analyze PlmS2-catalyzed monooxygenation in vitro, the plmS2 gene was cloned into pSE34 under the control of the ermE* promoter and pET T7 gene 10 RBS to give pMSG3. PlmS2 was expressed efficiently in S. coelicolor M511/pMSG3 as an N-terminal hexahistidine-tagged protein. The fermentation media were supplemented with δ-aminolevulinic acid and ferric chloride, heme biosynthetic precursors. SDS-PAGE analysis revealed the presence of a significant quantity of a soluble protein of 46 kDa, which was consistent with the predicted molecular mass of PlmS2 (44 kDa) and the hexahistidine tag (2 kDa) (Fig. 3A). This band was absent in a control culture of S. coelicolor M511/pSE34. PlmS2 was purified as a His-tagged protein by affinity chromatography on a Ni-NTA agarose column. The purified protein was electrophoretically homogeneous (Fig. 3B) and reacted strongly with anti-His-tag antibodies in a Western blot analysis (Fig. 3C). The UV-visible absorption spectrum of the purified protein showed the characteristic 390- to 450-nm CYP peak with an absorption maximum at 415.5 nm.
FIG. 3.
Expression and purification of PlmS2 from recombinant Streptomyces coelicolor. A. SDS-PAGE of an intracellular extract of S.coelicolor carrying pMSG3 (lane 1) and a control plasmid, pSE34 (lane 2). B. SDS-PAGE of Ni-NTA column-purified polyhistidine-tagged PlmS2. C. Immunoblot of N-terminal polyhistidine-tagged PlmS2 with anti-polyhistidine antibodies.
Conversion of Plm B to Plm G in vitro.
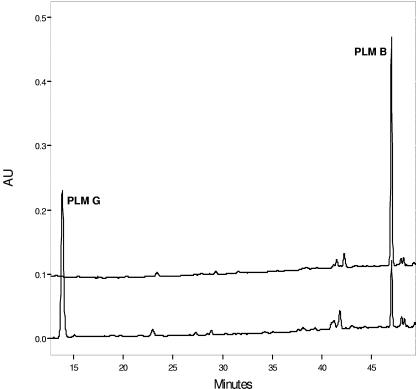
The enzymatic activity of PlmS2 was shown to readily convert the proposed Plm B substrate to Plm G in an HPLC assay (Fig. 4). In a standard 1-ml assay, almost a complete conversion of Plm B (100 μM) with a retention time of 47 min to a new peak with a retention time of 14 min by PlmS2 was observed in 1 h. LC-MS analysis revealed that the new product had a molecular mass of 529 Da, consistent with the addition of one oxygen atom to Plm B (molecular mass of 513 Da).
FIG. 4.
In vitro conversion of Plm B to Plm G. HPLC chromatogram of Plm B hydroxylation by PlmS2. AU, arbitrary units.
Substrate specificity of PlmS2.
The substrate specificity of PlmS2 was probed using Plm B preincubated under both acid and base conditions as a substrate. Under basic conditions three major degradation products are obtained, involving either direct hydrolysis of the lactone ring or hydration of the C2-C3 double bond of the lactone ring followed by either hydrolysis or methanolysis (3). These compounds have an earlier retention time than Plm B under the standard HPLC conditions. Under acidic conditions more than three compounds are formed, and all of these are dehydration products (in all cases the lactone ring remains intact). One of the acid degradation products has been identified as a C9-C11 phosphorinane derivative, while the other compounds appear to be mixtures of products containing an additional double bond in the C7-C11 region of the molecule. In all products the cyclohexane ring remains unchanged, and the structural changes caused by both acid and base occur either in the central core or the distal end of the molecule. No detectable hydroxylation of these products over 1 h of incubation was observed (Fig. 5, traces 4 and 6), as indicated by unchanged levels of the starting materials. The Plm B in these samples, however, was completely converted. Under the same conditions, PlmS2 catalyzed a complete conversion of Plm B to Plm G (Fig. 5, traces 1 and 3). The activity of PlmS2 towards Plm B is at least 20-fold (the value is limited by sensitivity of the assay) higher than that with any of these Plm B-related compounds, demonstrating that the enzyme has significant substrate specificity.
FIG. 5.
Substrate specificity of PlmS2. HPLC chromatograms 1, 3, and 5, Plm B control as well as acid- and alkali-degraded Plm B, respectively, incubated without PlmS2. Chromatograms 2, 4, and 6, corresponding reaction mixtures after 1 h of incubation with PlmS2. AU, arbitrary units.
Kinetic parameters of the reaction catalyzed by N-terminal hexahistidine PlmS2.
The rate of Plm B hydroxylation by PlmS2 (0.3 μM) under the assay conditions was determined at varying concentrations of substrate (2 to 200 μM) by measuring the area of unconsumed Plm B peak in an HPLC chromatogram. High-substrate concentrations (>200 μM) led to marked inhibition of the enzyme activity. At lower substrate concentrations, where this inhibition is negligible, the data could be fit to the Michaelis-Menten equation, resulting in the values for Km of 45.3 ± 9.0 μM and Vmax of 4.8 ± 0.8 μM/min. Assuming that all the enzyme was in active confirmation, this would correspond to a kcat of 0.27 ± 0.04 s−1 and a kcat/Km of 0.01 ± 0.001 μM−1 s−1. These kinetic values are comparable to those previously observed for PikC (kcat, 0.6 ± 0.05 s−1; kcat/Km, 0.029 ± 0.008 μM−1 s−1) and PimD (kcat, 0.78 ± 0.04 s−1; kcat/Km, 0.024 ± 0.003 μM−1 s−1) CYPs towards their substrates (28, 14).
DISCUSSION
The sequence of the Plm biosynthetic pathway and the subsequent generation of the plmS2-deleted mutant (17) led to a proposal for the last steps of phoslactomycin biosynthesis and the role of the PlmS2 protein (Fig. 1). A genetic complementation experiment with this mutant and biochemical characterization of PlmS2, reported here, have proven entirely consistent with this initial hypothesis.
The appearance of Plm A and Plm C through Plm F (and the absence of Plm B) in the NP2/pMSG1 mutant of Streptomyces sp. strain HK803 is consistent with higher overall levels of PlmS2 compared to those of the wild-type strain (which does produce some Plm B). The pMSG1 integrative plasmid used in this experiment is a pSET152 derivative (13) with the ermE* constitutive promoter. The complementation experiment provided the first evidence that this promoter works in Streptomyces sp. strain HK803 and its derivatives. This integrative plasmid represents a convenient vector to deliver genes into the Streptomyces sp. strain HK803 chromosome for stable expression and an important tool for further genetic studies probing the role of individual plm genes in the biosynthetic process.
To the best of our knowledge, previous biochemical characterization of streptomycetes CYPs has involved heterologous expression in E. coli (2, 7, 14, 28). The only exception is DoxA from the doxorubicin biosynthesis pathway, which was heterologously expressed in Streptomyces lividans without an affinity tag and purified using multiple chromatographic steps (25). All attempts to obtain heterologous expression of PlmS2 in E. coli using a battery of standard techniques were unsuccessful. In contrast, heterologous expression of PlmS2 with a hexahistidine tag as a His-tagged protein from S. coelicolor M511 cell extract was successful and afforded the pure protein in single-step affinity chromatography. This strategy has proven successful for expression of a number of Streptomyces proteins where problems with heterologous expression in E. coli have been encountered (unpublished data).
As noted above, Plm G cannot be detected in the Streptomyces sp. HK803 wild-type strain or any of the existing mutants. In principle, a mutant producing Plm G might be obtained once the gene encoding the enzyme responsible for esterification of Plm G is identified. The availability of pure Plm B (from either the NP1 or NP2 mutant strain) and purified PlmS2 permits production of Plm G. Chemical or enzymatic modification of the C-18-hydroxyl of Plm G can permit access to production of specific known Plms or structurally novel Plms. Such compounds may be expected to have biological activity (Plm A through Plm F all have activity [6, 16]) and likely greater antifungal activity than Plm G. The molecular basis for the antifungal activity of the Plms and related compounds is unknown, and it is curious that Plm G has such a marked decrease in activity relative to Plm A to F. Previously, Shibata et al. (23) showed that leustroducsin H (LSN H) was 100 times less effective than LSN B (a potent cytokine inducer from Streptomyces platensis SANK 60191) in vitro but was found to show comparative thrombopoetic effects in vivo. LSNs differ from Plms only in the nature of the C-18 acyl substituent. LSN H was prepared from a mixture of LSNs by enzymatic hydrolysis of this group and would appear to be structurally identical to Plm G. Acylation of the C-18 hydroxyl group clearly plays a significant role in the biological activity of Plm G (LSN H).
The deduced amino acid sequence and the characteristic UV-VIS spectrum, as well as the monomeric and soluble nature, suggest PlmS2 to be a class I type bacterial P450 in which ferredoxin acts as an electron donor. In the enzyme assays PlmS2 was able to use the spinach ferredoxin. The Plm biosynthetic gene cluster (17) contains plmT6, whose predicted protein product has high sequence identity to known ferredoxin proteins and presumably functions as the electron donor for both PlmS2 and PlmT4 (the other proposed CYP). It has been shown that, in cases where an antibiotic biosynthetic process requires hydroxylation at more than one position and the corresponding biosynthetic gene cluster encodes only one CYP, this enzyme has relaxed substrate specificity (for example, PikC and DoxA) (5, 28). There are three proposed CYP-dependent hydroxylations (C-8, C-18, and the C-18 ethyl substituents) in Plm B biosynthesis, proposed to be carried out by two enzymes (PlmS2 and PlmT4). The formation of Plm B in the NP1 and NP2 mutants implicates PlmT4 as being responsible for two steps and thus also having relaxed substrate specificity (characterization of the role of plmT4 is under way). In contrast, PlmS2 is responsible for just the C-18 hydroxylation step and has greater substrate specificity, reacting only with Plm B (the enzyme was unable to hydroxylate related compounds bearing the same 4-cyclohexyl-1,3-butadiene moiety). Podust et al. (20), from their comparative studies on Streptomyces coelicolor A3 (2) CYPs and several other CYPs from antibiotic biosynthetic pathways, have predicted that regions located outside the substrate binding site influence the catalytic specificity of these enzymes.
The natural substrate Plm B inhibited PlmS2 at higher concentrations. Substrate inhibition has previously been observed in the hydroxylations catalyzed by EryK (12), PikC (28), and PimD (23) toward erythromycin D, YC-17, and de-epoxypimaricin, respectively. A combination of this inhibitory effect and the observed low kcat and kcat/Km values obtained for PlmS2 at lower Plm B concentrations presumably contribute to the conversion to Plm G being a rate-limiting step in Plm biosynthesis in the wild-type strain under typical fermentation conditions. This rate-limiting step (resulting in observation of Plm B intermediate) is presumably overcome as a result of increased levels of PlmS2 in the NP2/pMSG1 mutant (see above).
Efficient manipulation of Streptomyces in generating novel and potentially useful polyketide antibiotics by combinatorial-based or semisynthetic strategies can be achieved through modification of post-polyketide synthase tailoring steps, including those catalyzed by CYP monooxygenases (20, 21). Identification and/or engineering of the monooxygenases such as PlmS2 with activities towards a diverse array of chemical substrates and particularly unusual natural products expands the set of genetic tools available in this endeavor. Nonetheless, the analyses of PlmS2 provide a cautionary example that for some CYPs the substrate specificity of the native form of the enzyme may preclude the most facile direct application in generating these “unnatural” or “hybrid” natural products.
Acknowledgments
This work was supported by grant AI51629 from the National Institutes of Health.
The NP2 mutant was generated by Nadaraj Palaniappan, and the original Streptomyces HK803 strain was kindly provided by Hiroyuki Osada (Riken, Japan). Conditions for Plm B degradation and characterization of the major products were determined by Suparna DasChouduri and have been published separately (3).
REFERENCES
- 1.Anderson, J. F., K. Tatsuta, H. Gunji, T. Ishiyama, and C. R. Hutchinson. 1993. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry 32:1905-1913. [DOI] [PubMed] [Google Scholar]
- 2.Betlach, M. C., J. T. Kealey, M. C. Betlach, G. W. Ashley, and R. McDaniel. 1998. Characterization of the macrolide P-450 hydroxylase from Streptomyces venezuelae which converts narbomycin to picromycin. Biochemistry 37:14937-14942. [DOI] [PubMed] [Google Scholar]
- 3.Das Choudhuri, S., S. Ayers, W. H. Soine, and K. A. Reynolds. A pH-stability study of phoslactomycin B and analysis of the acid and base degradation products. J. Antibiot., in press. [DOI] [PubMed]
- 4.Dickens, M. L., and W. R. Strohl. 1996. Isolation and characterization of a gene from Streptomyces sp. strain C5 that confers the ability to convert daunomycin to doxorubicin on Streptomyces lividans TK24. J. Bacteriol. 178:3389-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickens, M. L., N. D. Priestley, and W. R. Strohl. 1997. In vivo and in vitro bioconversion of ε-rhodomycinone glycoside to doxorubicin: functions of DauP, DauK, and DoxA. J. Bacteriol. 179:2641-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fushimi, S., S. Nishikawa, A. Shimazu, and H. Seto. 1989. Studies on new phosphate ester antifungal antibiotics phoslactomycins. I. Taxonomy, fermentation, purification and biological activities. J. Antibiot. 42:1019-1025. [DOI] [PubMed] [Google Scholar]
- 7.Graziani, E. I., D. E. Cane, M. C. Betlach, J. T. Kealey, and R. McDaniel. 1998. Macrolide biosynthesis: a single cytochrome P450, PicK, is responsible for the hydroxylations that generate methymycin, neomethymycin, and picromycin in Streptomyces venezuelae. Bioorg. Med. Chem. Lett. 8:3117-3120. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., T. Kieser, and K. F. Chater. 2002. REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. John Innes Center, Norwich, United Kingdom.
- 9.Hussain, H. A., and J. M. Ward. 2003. Enhanced heterologous expression of two Streptomyces griseolous cytochrome P450s and Streptomyces coelicolor ferredoxin reductase as potentially efficient hydroxylation catalysts. Appl. Environ. Microbiol. 69:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 11.Lamb, D. C., H. Ikeda, D. R. Nelson, J. Ishikawa, T. Skaug, C. Jackson, S. Omura, M. R. Waterman, and S. L. Kelly. 2003. Cytochrome P450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem. Biophys. Res. Commun. 307:610-619. [DOI] [PubMed] [Google Scholar]
- 12.Lambalot, R. H., D. E. Cane, J. J. Aparicio, and L. Katz. 1995. Overproduction and characterization of the erythromycin C-12 hydroxylase, EryK. Biochemistry 34:1858-1866. [DOI] [PubMed] [Google Scholar]
- 13.Li, C., G. Florova, K. Akopiants, and K. A. Reynolds. 2004. Crotonyl-coenzyme A reductase provides methylmalonyl-CoA precursors for monensin biosynthesis by Streptomyces cinnamonensis in oil-based extended fermentation. Microbiology 150:3463-3472. [DOI] [PubMed] [Google Scholar]
- 14.Mendes, M. V., N. Anton, J. F. Martin, and J. F. Aparicio. 2005. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 386:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mo, S., B. S. Kim, and K. A. Reynolds. 2005. Production of branched-chain alkylprodiginines in S. coelicolor by replacement of the 3-ketoacyl ACP synthase III initiation enzyme, RedP. Chem. Biol. 12:191-200. [DOI] [PubMed] [Google Scholar]
- 16.Ozasa, T., K. Suzuki, M. Sasamata, K. Tanaka, M. Kobori, S. Kadota, K. Nagai, T. Saito, S. Watanabe, and M. Iwanami. 1989. Novel antitumor antibiotic phospholine. 1. Production, isolation and characterization. J. Antibiot. 42:1331-1338. [DOI] [PubMed] [Google Scholar]
- 17.Palaniappan, N., B. S. Kim, Y. Sekiyama, H. Osada, and K. A. Reynolds. 2003. Enhancement and selective production of phoslactomycin B, a protein phosphatase IIa inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J. Biol. Chem. 278:35552-35557. [DOI] [PubMed] [Google Scholar]
- 18.Parajuli, N., D. B. Basnet, H. C. Lee, J. K. Sohng, and K. Liou. 2004. Genome analyses of Streptomyces peucetius ATCC 27952 for the identification and comparison of cytochrome P450 complement with other Streptomyces. Arch. Biochem. Biophys. 425:233-241. [DOI] [PubMed] [Google Scholar]
- 19.Podust, L. M., Y. Kim, M. Arase, B. A. Neely, B. J. Beck, H. Bach, D. H. Sherman, D. C. Lamb, S. L. Kelly, and M. R. Waterman. 2003. The 1.92-Å structure of Streptomyces coelicolor A3(2) CYP154C1. A new monooxygenase that functionalizes macrolide ring systems. J. Biol. Chem. 278:12214-12221. [DOI] [PubMed] [Google Scholar]
- 20.Podust, L. M., H. Bach, Y. Kim, D. C. Lamb, M. Arase, D. H. Sherman, S. L. Kelly, and M. R. Waterman. 2004. Comparison of the 1.85-Å structure of CYP154A1 from Streptomyces coelicolor A3(2) with the closely related CYP154C1 and CYPs from antibiotic biosynthetic pathways. Protein Sci. 13:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rix, U., C. Fischer, L. L. Remsing, and J. Rohr. 2002. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 19:542-580. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Shibata, T., S. Kurihara, T. Oikawa, N. Ohkawa, N. Shimazaki, K. Sasagawa, T. Kobayashi, T. Kohama, F. Asai, A. Shiraishi, and Y. Sugimura. 1995. Preparation of Leustroducsin H and the structure-activity relationship of its derivatives. J. Antibiot. (Tokyo) 48:1518-1520. [DOI] [PubMed] [Google Scholar]
- 24.Tomiya, T., M. Uramoto, and K. Isono. 1990. Isolation and structure of phosphazomycin C. J. Antibiot. 43:118-121. [DOI] [PubMed] [Google Scholar]
- 25.Walczak, R. J., M. L. Dickens, N. D. Priestley, and W. R. Strohl. 1999. Purification, properties, and characterization of recombinant. Streptomyces sp. strain C5 DoxA, a cytochrome P-450 catalyzing multiple steps in doxorubicin biosynthesis. J. Bacteriol. 181:298-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe, I., F. Nara, and N. Serizawa. 1995. Cloning, characterization and expression of the gene encoding cytochrome P-450sca-2 from Streptomyces carbophilus involved in the production of pravastatin, a specific HMG-CoA reductase inhibitor. Gene 163:81-85. [DOI] [PubMed] [Google Scholar]
- 27.Werck-Reichhart, D., and R. Feyereisen. 2000. Cytochromes P450: a success story. Genome Biol. 1:3003.1-3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue, Y., D. Wilson, L. Zhao, H. Liu, and D. H. Sherman. 1998. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem. Biol. 5:661-667. [DOI] [PubMed] [Google Scholar]