Abstract
The formation of attaching and effacing (A/E) lesions on intestinal epithelial cells is an essential step in the pathogenesis of human enteropathogenic and enterohemorrhagic Escherichia coli and of the mouse pathogen Citrobacter rodentium. The genes required for the development of the A/E phenotype are located within a pathogenicity island known as the locus of enterocyte effacement (LEE). The LEE-encoded transcriptional regulators Ler, an H-NS-like protein, and GrlA, a member of a novel family of transcriptional activators, positively control the expression of the genes located in the LEE and their corresponding virulence. In this study, we used C. rodentium as a model to study the mechanisms controlling the expression of Ler and GrlA. By deletion analysis of the ler and grlRA regulatory regions and complementation experiments, negative and positive cis-acting regulatory motifs were identified that are essential for the regulation of both genes. This analysis confirmed that GrlA is required for the activation of ler, but it also showed that Ler is required for the expression of grlRA, revealing a novel regulatory loop controlling the optimal expression of virulence genes in A/E pathogens. Furthermore, our results indicate that Ler and GrlA induce the expression of each other by, at least in part, counteracting the repression mediated by H-NS. However, whereas GrlA is still required for the optimal expression of ler even in the absence of H-NS, Ler is not needed for the expression of grlRA in the absence of H-NS. This type of transcriptional positive regulatory loop represents a novel mechanism in pathogenic bacteria that is likely required to maintain an appropriate spatiotemporal transcriptional response during infection.
Enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and Citrobacter rodentium belong to a family of bacterial pathogens causing a destructive lesion of the intestinal enterocyte, called the attaching and effacing (A/E) lesion, as well as gastrointestinal disorders in infected hosts (reviewed in references 28 and 33). EPEC is an important etiological agent of childhood diarrhea in developing countries, whereas EHEC is the cause of frequent outbreaks of food and water poisoning in the developed world. In addition to causing diarrhea, an EHEC infection can result in severe complications, such as hemorrhagic colitis and hemolytic-uremic syndrome (reviewed in reference 33). Due to the specificity of EPEC and EHEC for human hosts, a corresponding small-animal infection model does not exist. Thus, most of the current models to explain EHEC and EPEC pathogen-host interactions, such as those for A/E lesion formation, have been developed based on in vitro studies performed with infected cultured epithelial cells. In recent years, C. rodentium has become accepted as a representative infection system to study the mechanisms leading to the production of the A/E lesion and A/E-associated pathogenesis (12, 13, 47).
The A/E lesion is characterized by a localized loss of microvilli from the surfaces of epithelial cells and important cytoskeleton rearrangements beneath the adherent bacteria, leading to the formation of actin-rich cup-like structures and intimate bacterium-host cell interactions. Intimate adherence is mediated by the interaction between Tir (translocated intimin receptor), a bacterial protein that is translocated and inserted into the host cell membrane, and intimin, a bacterial outer membrane adhesin (reviewed in reference 7). The genes required for the formation of the A/E lesion in EPEC, EHEC, and C. rodentium are located within a pathogenicity island known as the locus of enterocyte effacement (LEE), where they are organized in five polycistronic operons (LEE1-LEE5), two putative bicistronic operons, and four monocistronic units (8). The LEE1 to LEE3 operons encode mostly structural components of a type III secretion system (Esc and Sep), the LEE4 operon encodes proteins involved in protein translocation (EspA, B, and D and SepL), and the LEE5 operon encodes the proteins required for intimate attachment (intimin and Tir). The genes encoding effector proteins, chaperones, and transcriptional regulators are scattered along the LEE (reviewed in references 7 and 8). During A/E lesion formation, several LEE-encoded proteins (Tir, Map, EspF, EspG, EspH, and EspZ), as well as non-LEE-encoded proteins (NleA/EspI, EspFu/TccP, EspJ, and Cif), are translocated by the type III secretion apparatus into the host epithelial cells, where they affect different signaling processes (reviewed in references 10 and 20).
Several studies have shown that Ler (LEE-encoded regulator), a 15-kDa protein encoded by the first gene of the LEE1 operon, is a central positive regulator needed for the expression of the LEE genes (5, 16, 19, 31) as well as the non-LEE-carried gene espC (32). Ler belongs to the H-NS family of nucleoid-associated proteins, exhibiting high amino acid identity with the carboxy termini of these proteins, which contain the DNA binding domain (16). The global regulator H-NS (14) represses the expression of several LEE genes, and Ler induces the expression of these genes by counteracting the H-NS-mediated repression (5, 24, 46). Thus, Ler is primarily an antirepressor needed to conduct gene expression (5, 24, 46).
Different studies of EPEC and EHEC have shown that ler expression is regulated by a complex assortment of global and A/E-specific regulators. The global regulator integration host factor (IHF), which directly binds to a DNA region upstream of the ler promoter, is essential for ler activation (19). ler is also positively regulated by other global regulators, such as BipA, a member of the ribosome-binding GTPase superfamily (23); Fis (factor for inversion stimulation), a bacterial nucleoid-associated protein (21); and QseA (quorum-sensing E. coli regulator A), a factor involved in regulation via quorum sensing (42). H-NS and Hha play a negative role in ler expression, with both binding directly to its regulatory region (40, 46). In addition, specific regulators such as PerC, the product of the third gene of the per locus located in the EPEC adherence factor plasmid, can directly activate the expression of ler (5, 31, 35, 36). PerC-like proteins have also been identified in EHEC and are involved in ler expression (25). GadX regulates the expression of the perABC operon and thus indirectly regulates the expression of ler (41). It has been reported that Ler binds to its own regulatory region and autorepresses its transcription in a concentration-dependent manner (2). The negative regulation of LEE gene expression is also mediated by YhiE and YhiF (44) as well as by EtrA (E. coli type III secretion system 2 regulator A) and EivF (49) by mechanisms that remain to be defined. We have recently identified two novel LEE-encoded regulators, GrlA (global regulator of LEE activator; formerly called Orf11) and GrlR (Grl repressor; formerly called Orf10), which are highly conserved in all A/E pathogens (12). These proteins are encoded by the putative grlRA operon located between the rorf3 gene and the LEE2 operon in the LEE. GrlA is a positive regulator of ler expression (12). The closest GrlA homologue is the putative product of an uncharacterized gene found in different Salmonella enterica serotypes. In addition, GrlA is 23% identical to CaiF, a regulatory protein responsible for the carnitine-dependent induction of the cai and fix E. coli operons under anaerobic conditions and the best-characterized member of this novel family of transcriptional regulators (15). A motif search of GrlA has also revealed the presence of a putative helix-turn-helix DNA binding motif at its N-terminal domain, where most of the similarity with CaiF and the Salmonella GrlA homologue (Sgh) is found (12). GrlR has a significant negative effect on LEE gene expression, probably acting as a negative regulator of ler (12, 26, 27), although its mechanism of action remains to be defined. PSI-BLAST searches have identified only one other GrlR homologue, located next to a GrlA homologue in Salmonella bongori (34). For the present study, we used C. rodentium as a model to study the mechanisms controlling the expression of the genes encoding the positive regulators Ler and GrlA. Although C. rodentium has been used as a model organism to study EPEC and EHEC, there is little known about the regulation of its LEE gene expression. Here we characterize the regulatory regions of the C. rodentium ler and grlRA genes in detail. Furthermore, we demonstrate that Ler and GrlA regulate each other, forming a transcriptional positive regulatory loop that, to our knowledge, represents a novel mechanism controlling gene expression in bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are described in Table 1. Luria-Bertani (LB) broth (37) or Dulbecco's modified Eagle's medium (DMEM) containing glucose (0.45% [wt/vol]) and l-glutamine (584 mg liter−1), but not sodium pyruvate (Gibco BRL Life Technologies), was used for static cultures at 37°C in a 5% CO2 atmosphere. When required, antibiotics were added at the following concentrations for LB cultures: ampicillin (Amp), 100 μg ml−1; carbenicillin (Cb), 100 μg ml−1; kanamycin (Km), 25 μg ml−1; tetracycline (Tc), 12 μg ml−1; and streptomycin (Stp), 100 μg ml−1. The following antibiotic concentrations were used for DMEM cultures, when required: Amp, 50 μg ml−1; Cb, 50 μg ml−1; and Tc, 5 μg ml−1. Test cultures were inoculated as described before (13). Culture samples to determine chloramphenicol acetyltransferase (CAT) activity were collected at 6 h. At this time point, all strains reached similar optical densities. Each experiment was done independently in duplicate at least three times.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| C. rodentium strains | ||
| DBS100 | Wild type (ATCC 51459) | 39 |
| Δler | DBS100 carrying an in-frame deletion of ler | 12 |
| Δorf11/grlA | DBS100 carrying an in-frame deletion of grlA | 12 |
| E. coli strains | ||
| MC4100 | F′ araD139Δ (argF-lac)U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR | 6 |
| JPMC1 | MC4100 Δhns::Km | This study |
| BL21/pLys21 | F−ompT (lon) hsdSB(rB− mB−) gal dcm (λDE3) | Invitrogen |
| N99 | E. coli K12 F−galK2 rpsLl | 22 |
| K5185 | N99 ΔhimA82 | 18 |
| Plasmids | ||
| pKD46 | Red recombinase system under araB promoter; Apr | 9 |
| pKD4 | Template plasmid containing the Km cassette for lambda Red recombination | 9 |
| pMPM-T3 | Low-copy-number cloning vector; p15A derivative; Tcr | 30 |
| pTCRLer4 | pMPM-T3 derivative carrying the ler structural gene and ribosome binding site under the control of the lac promoter | This study |
| pTCRGrlA1 | pMPM-T3 derivative carrying the grlA structural gene and ribosome binding site under the control of the lac promoter | This study |
| pMPM-T6 | Cloning vector containing an arabinose-inducible promoter; p15A derivative; Tcr | 30 |
| pT6HNS | pMPM-T6 derivative expressing H-NS-His6 under the control of the arabinose-inducible promoter | Unpublished |
| pT6Ler | pMPM-T6 derivative expressing Ler-His6 under the control of the arabinose-inducible promoter | Unpublished |
| pKK232-8 | pBR322 derivative containing a promoterless chloramphenicol acetyltransferase (cat) gene | Pharmacia LKB Biotechnology |
| pLEE2-CAT | pKK232-8 derivative carrying C. rodentium LEE2-cat transcriptional fusion from nucleotides −375 to +121 | 12 |
| PCRler-260 | pKK232-8 derivative carrying C. rodentium ler-cat transcriptional fusion from nucleotides −265 to +216 | This study |
| pCRler-200 | CRler-cat transcriptional fusion from nucleotides −197 to +216 | This study |
| pCRler-160 | CRler-cat transcriptional fusion from nucleotides −163 to +216 (pLEE1-CAT) | 12 |
| pCRler-120 | CRler-cat transcriptional fusion from nucleotides −123 to +216 | This study |
| pCRler-80 | CRler-cat transcriptional fusion from nucleotides −86 to +216 | This study |
| pCRler-40 | CRler-cat transcriptional fusion from nucleotides −44 to +216 | This study |
| pCRgrlRA-1 | pKK232-8 derivative carrying C. rodentium grlRA-cat transcriptional fusion from nucleotides −420 to + 152 | This study |
| pCRgrlRA-2 | CRgrlRA-cat transcriptional fusion from nucleotides −420 to +397 | This study |
| pCRgrlRA-3 | CRgrlRA-cat transcriptional fusion from nucleotides −420 to +565 | This study |
| pCRgrlRA-4 | CRgrlRA-cat transcriptional fusion from nucleotides +212 to +565 | This study |
| pCRgrlRA-5 | CRgrlRA-cat transcriptional fusion from nucleotides −135 to +152 | This study |
| pCRgrlRA-6 | CRgrlRA-cat transcriptional fusion from nucleotides −135 to +565 | This study |
The coordinates for cat transcriptional fusions are indicated with respect to the ler or grlR transcriptional start site.
DNA manipulations.
Recombinant DNA techniques were performed according to standard protocols (1, 37). Restriction enzymes were obtained from Invitrogen or New England Biolabs and used according to the manufacturer's instructions. The oligonucleotides used for amplification by PCR and for primer extension experiments (Table 2) were synthesized by the oligonucleotide synthesis facility at our institute. PCRs were performed in 100-μl reaction mixtures containing a 1.5:1 mixture of AmpliTaq and Pfu DNA polymerases, using an Eppendorf mastercycler gradient thermocycler.
TABLE 2.
Primers used for this study
| Primer | Sequencea (5′-3′) |
|---|---|
| Orf1-H3-R | gctctatAagctTaatgtatg |
| CRler-260 | gaaaaatggAtCcgttacgt |
| CRler-200 | cctggaTCCttgatctga |
| CRler-160 | caatacggAtcCggcgagccg |
| CRler-120 | attaatggaTCCacaata |
| CRler-80 | actagctGGatcCttataat |
| CRler-40 | tttttaattggGatCCtttt |
| CR-ORF10-HIII-A | cccacaggaaGcttcattac |
| CR-ORF10-HIII-B | ctgacataaGcTtcaacaaataac |
| CR-ORF11-H3 | tatacagaAgctTaccattgtaa |
| CR-ORF10-BHI | tgcacccacggGatcccacg |
| CR-ORF11-BHI | atttcctctgtgGatcCggggg |
| CR-RORF3-BH | aaacaatcagaagGatCCcaaaagttagtg |
| Cler-RBS-F | catgtaaggatCCgcttgttaa |
| ClerOrf1-R | gttcagttaaGCTtatcattta |
| CROrf11Xho | cagatttCtcgaGccgttaattat |
| EpCiorf11R-H3 | tactaagaAagcttcgtctaactctcc |
| ompAPE | tttgcgcctcgttatcatccaa |
| hnsH1P1 | cacccaatataagtttgagattactacaatgagcgaagctgtaggctggagctgcttcg |
| hnsH2P2 | gattttaagcaagtgcaatctacaaaagattattgcttcatatgaatatcctcctt |
| hnsM | tgcgagctcatcggtgtaa |
| hnsG | ttgctggcaaaaacccctccg |
Capital letters indicate changes in the oligonucleotide sequence with respect to the wild-type sequence, designed to introduce restriction enzyme sites.
Construction of ler-cat and grlRA-cat transcriptional fusions.
Oligonucleotides were designed for PCR amplification of different fragments spanning the ler regulatory region and the rorf3-grlRA region (Table 2). PCRs were performed using these oligonucleotides, with C. rodentium DBS100 chromosomal DNA as the template. The PCR fragments were double digested with BamHI and HindIII and ligated into pKK232-8 (Pharmacia LKB Biotechnology), which contains a promoterless cat gene, digested with the same enzymes. Combination of the forward primers CRler-260, CRler-200, CRler-160, CRler-120, CRler-80, and CRler-40 with the reverse primer Orf1-H3-R was used for the construction of the fusions pCRler-260, -200, -160, -120, -80, and -40, respectively. Fusions pCRgrlRA-1, -2, and -3 were constructed using the forward primer CR-ORF10-BHI in combination with the reverse primers CR-ORF10-HIII-A, CR-ORF10-HIII-B, and CR-ORF11-H3, respectively. pCRgrlRA-4 was constructed using primers CR-ORF11-BHI and CR-ORF11-H3. The forward primer CR-RORF3-BH and the reverse primers CR-ORF10-HIII-A and CR-ORF11-H3 were used to construct pCRgrlRA-5 and -6, respectively. The nucleotide sequences of the ler-cat and grlRA-cat fusions were determined in the sequencing facility at our institute.
Construction of E. coli MC4100 Δhns::Km mutant.
Deletion of the hns gene from E. coli MC4100 was performed by the one-step mutagenesis procedure for bacterial genes described by Datsenko and Wanner (9). The deletion eliminated 131 codons out of the 137 codons of the hns gene, which were replaced with a Km resistance marker. Primers hnsH1P1 and hnsH2P2 and DNA of plasmid pKD4 were used to generate the deletion cassette. The replacement of hns by the Km resistance marker was confirmed by PCR using primers hnsM and hnsG. The resulting strain was designated JPMC1 (Table 1).
PCR cloning of ler and grlA.
The primer pairs Cler-RBS-F (BamHI)/ClerOrf1-R (HindIII) and CROrf11Xho/EpCiorf11R-H3 were used to amplify the C. rodentium ler and grlA genes, respectively. The resulting PCR products were digested with the BamHI-HindIII and XhoI-HindIII restriction enzymes, respectively, and ligated into pMPM-T3 (30) digested with the same enzyme combinations, generating plasmids pTCRLer4 and pTCRGrlA1 (Table 1). The identity of the inserts was confirmed by DNA sequencing. The plasmids contain the promoterless ler or grlA gene plus the putative ribosome-binding sites and are expressed from the vector lac promoter.
CAT assay.
CAT assays and protein quantification to calculate CAT specific activities were performed as described previously (29).
RNA isolation and primer extension analysis.
Total RNAs were isolated from samples of cultures grown for 6 h in DMEM at 37°C in a 5% CO2 atmosphere without agitation, using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. The RNA concentration and quality were determined by measuring the A260-to-A280 ratio and by gel electrophoresis. Primer extension reactions were performed as described previously (29). Briefly, oligonucleotides complementary to the grlR (CR-ORF10-HIII-A) or ompA (ompAPE) (Table 2) coding region were end labeled with [γ-32P]dATP, using T4 polynucleotide kinase, and annealed with 8 μg (for grlR) or 0.8 μg (for ompA) of total RNA in 0.37 M NaCl-0.035 M Tris-HCl (pH 7.5) by heating for 3 min at 90°C and then cooling slowly to 50°C. Reverse transcription reactions were performed at 42°C for 2 h with 10 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) in avian myeloblastosis virus buffer containing 1 mM dithiothreitol, a 0.3 mM concentration of each deoxynucleoside triphosphate, and 50 U of RNase inhibitor (Invitrogen). The reverse transcription products were cleaned and concentrated using a Microcon YM-30 microconcentrator (Amicon) according to the specifications of the manufacturer, denatured by heating to 95°C for 5 min in loading buffer, and resolved by electrophoresis through an 8% polyacrylamide-7 M urea-Tris-borate-EDTA sequencing gel. The gel was analyzed using a PhosphorImager scanner (Molecular Dynamics). The transcriptional start site was determined by comparison with a DNA ladder obtained by sequencing plasmid pCRgrlRA-3 (Table 1), using primer CR-ORF10-HIII-A (Table 2).
Expression and purification of His-tagged H-NS and Ler proteins.
E. coli BL21/pLys21 harboring the pT6HNS or pT6Ler plasmid (Table 1), expressing H-NS-His6 or Ler-His6, respectively, was grown to mid-logarithmic phase at 37°C. l-(+)-Arabinose (Sigma-Aldrich) was added to a final concentration of 0.1%, and the bacteria were further incubated for 4 h at 30°C and 250 rpm. Cells were then pelleted by centrifugation at 4°C, resuspended in urea buffer (pH 8.0) (8 M urea, 20 mM NaH2PO4, and 2 M Tris-HCl), and disrupted by sonication. The suspension was centrifuged at 4°C, and the supernatant was filtered through a 0.22-μm membrane (Millipore) and applied to a HiTrap Ni2+-chelating column, which was loaded with 100 mM NiSO4 and connected to a minichromatographer ÄKTA prime system (Amersham Pharmacia Biotech). Proteins were eluted with a pH gradient (pH 8.0 to 4.5) of urea buffer (8 M urea, 20 mM NaH2PO4, and 2 M Tris-HCl). Fractions containing purified H-NS-His6 or Ler-His6 were selected based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. The selected fractions were loaded into a Slyde-A-Lyzer 10K cassette (Pierce) and gradually dialyzed at 4°C in a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 20% glycerol, 0.5 M NaCl, 0.1% Triton X-100, and various amounts of urea (4, 1, and 0.2 M), which was changed every hour. The final dialysis was done in storage buffer containing 30 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 20% glycerol, 240 mM NaCl, 0.1% Triton X-100, and 3 mM EDTA, and aliquots of the purified proteins were stored at −70°C. Protein concentrations were determined by the Bradford procedure.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as follows. Approximately 100-ng samples of PCR-generated DNA fragments corresponding to the inserts carried by the grlRA-cat fusions were mixed with increasing concentrations of purified Ler-His6 or H-NS-His6 protein in a buffer containing 11.7 mM Tris-HCl, pH 7.5, 0.975 mM EDTA, 78 mM NaCl, 9.75 mM 2-mercaptoethanol, 0.975 mM dithiothreitol, and 6.5% glycerol. The reactions were incubated for 30 min at room temperature and then separated by electrophoresis in 4% polyacrylamide gels in 0.45× Tris-borate-EDTA buffer at room temperature. The DNA bands were stained with ethidium bromide and visualized with an Alpha-Imager UV transilluminator (Alpha Innotech Corp.). A fragment containing the ler structural gene of EPEC was used as a negative control when evaluating H-NS-DNA interactions, as previously described (17).
RESULTS
cis-acting elements involved in transcriptional regulation of C. rodentium ler.
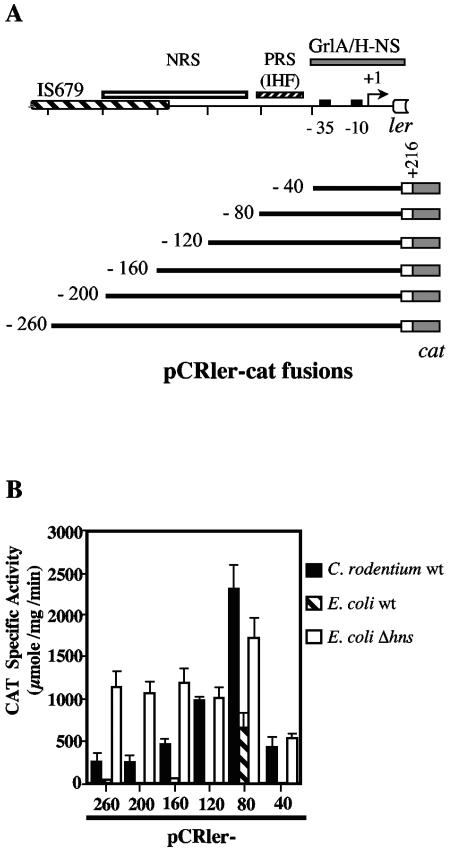
We constructed a series of transcriptional fusions to the cat reporter gene in plasmid pKK232-8, encompassing different lengths of the ler 5′ upstream regulatory region, to determine the cis-acting elements controlling its expression (Fig. 1A). The promoterless cat reporter gene has proven to be a reliable system for analyzing gene expression in A/E pathogens (5, 29, 38). The ler-cat fusions were called pCRler-260, -200, -160, -120, -80, and -40 according to the positions of their 5′ ends with respect to the transcriptional start site (12). All of the ler-cat fusions contained a common 3′ end at position +216 with respect to the transcriptional start site (Fig. 1A). The plasmids containing the fusions were transformed into C. rodentium DBS100, the prototype wild-type strain (Table 1), and the CAT specific activity was determined from bacterial cultures grown under inducing conditions for the expression of C. rodentium LEE genes, as described in Materials and Methods. Fusions pCRler-260 and pCRler-200 expressed similar levels of CAT (Fig. 1B), whereas pCRler-160, -120, and -80 showed a gradual increase in CAT activity with respect to the expression shown by pCRler-200 (Fig. 1B). These results indicate that the region between positions −200 and −80 contains cis-acting elements that negatively control ler expression. The fusion pCRler-40, which still contains the ler promoter, showed an approximately sixfold reduced activity with respect to the expression shown by pCRler-80 (Fig. 1B), indicating the presence of positive regulatory cis-acting elements between positions −80 and −40. This region is equivalent to the one containing the IHF binding site previously found to be essential for ler expression in EPEC (19), suggesting that IHF plays a similar role in the expression of C. rodentium ler. In agreement with this hypothesis, the pCRler-80 fusion, which renders significant levels of expression in an E. coli K-12 strain (Fig. 1B), was no longer active in an isogenic E. coli ihf mutant (data not shown).
FIG. 1.
Expression of C. rodentium ler is regulated by global and specific regulators. (A) Schematic representation of the ler regulatory region. The bent arrow indicates the previously reported transcriptional start site (+1) (12). −35 and −10 consensus sequences are shown as black boxes. A large hatched box represents the insertion sequence element (IS679) localized at the 5′ end of the C. rodentium LEE (11). Open and hatched boxes indicate the approximate positions of negative and positive regulatory sequences (NRS and PRS), respectively, revealed by expression analysis of ler-cat transcriptional fusions. The PRS contains the putative IHF binding site. A gray box indicates a region required for GrlA and H-NS-mediated regulation of ler. Schematic representations of the ler-cat transcriptional fusions are shown below the diagram of the ler regulatory region. The ler-cat fusions were named pCRler and numbered according to the position of the 5′ end of the ler region contained in each fusion with respect to the transcriptional start site. (B) Expression of the ler-cat fusions was monitored in C. rodentium DBS100, E. coli MC4100, and E. coli MC4100 Δhns. The CAT specific activity was determined from samples collected from bacterial cultures grown for 6 h in DMEM at 37°C without agitation in a 5% CO2 atmosphere. The values are the means of at least three independent experiments performed in duplicate. Standard errors are shown with error bars.
H-NS negatively regulates the expression of C. rodentium ler.
It has previously been reported that H-NS represses the expression of ler in EPEC (46). To further characterize the role of H-NS and the elements controlling the expression of C. rodentium ler, the plasmids containing the ler-cat fusions were transformed into E. coli K-12 and its isogenic hns mutant, and the CAT activity was measured after the strains were grown under inducing conditions. Expression in E. coli K-12 was close to the background level for all fusions except for pCRler-80 (Fig. 1B), further supporting the notion that C. rodentium contains specific positive regulatory factors for ler expression that are not present in E. coli K-12. In contrast, all fusions were expressed in the E. coli hns mutant at a similar or even higher level than that in C. rodentium (Fig. 1B), confirming the role of H-NS as a repressor of ler expression. Interestingly, the fusion pCRler-40, containing only the promoter, was still partially expressed in C. rodentium and in the E. coli hns mutant, but not in wild-type E. coli K-12 (Fig. 1B). This indicates that cis-acting elements required for positive regulation by a C. rodentium factor and for H-NS-mediated repression are present in the region between positions −40 and +216 of ler.
GrlA positively regulates the expression of C. rodentium ler.
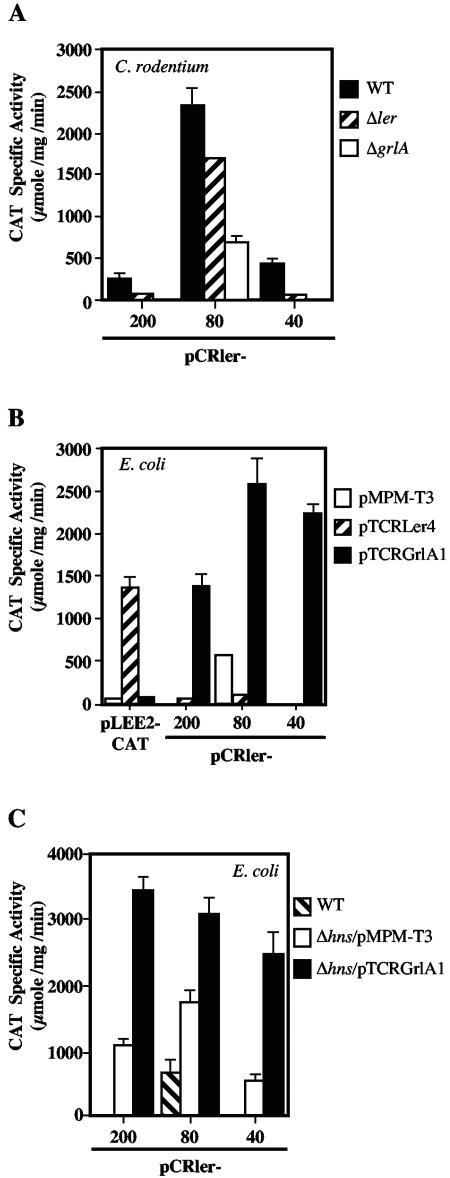
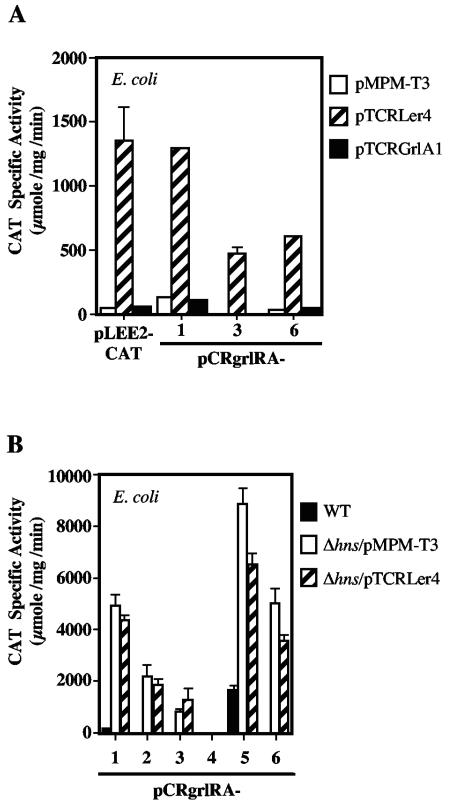
In addition to the set of global regulators currently known to regulate ler expression in A/E pathogens, we have recently reported that the expression of ler, and thus of the LEE genes involved in the development of the A/E lesion, requires a second LEE-encoded regulator called GrlA (12). To define the regulatory region required for the GrlA-mediated activation of ler, we analyzed the CAT activity driven from three representative ler fusions (pCRler-200, -80, and -40) in wild-type C. rodentium and its ΔgrlA derivative. According to the results shown in Fig. 1, pCRler-200 contains all of the regulatory elements involved in ler regulation, pCRler-80 lacks putative negative regulatory elements located upstream of the putative IHF binding site and showed a 10-fold increase in activity with respect to the longest fusions in the wild-type strain, and pCRler-40 contains the promoter and downstream elements involved in positive and negative regulation. In the grlA mutant, the transcriptional activity of pCRler-200 was reduced to background levels, confirming the requirement of GrlA for ler expression (Fig. 2A). The activity of pCRler-80 showed a threefold decrease in the grlA mutant compared to that in the wild-type strain, indicating that even in the absence of negative cis-acting regulatory elements, GrlA was still needed for full ler promoter activation. Interestingly, the expression of pCRler-40 was also abolished in the absence of GrlA (Fig. 2A). To further confirm the direct positive role of GrlA on ler expression, the CAT activities of these three fusions were determined in the nonpermissive E. coli K-12 strain in the presence of a plasmid carrying grlA (pTCRGrlA1) expressed from the lac promoter on the vector. As shown in Fig. 2B, GrlA activated high levels of expression of fusions pCRler-200 and pCRler-40 and further increased (approximately fivefold) the activity of pCRler-80, while no changes were observed with the vector alone. Together, these results strongly suggest that GrlA is directly involved in ler activation, probably interacting with cis-acting elements located between positions −40 and +216 (Fig. 1A). In addition, these results indicated that sequences located upstream of position −40, including the putative IHF binding site, are not required for the GrlA-mediated activation of the ler promoter. Nonetheless, the presence of the sequence up to position −80 enhances the GrlA-dependent expression of the ler promoter as well as the level of GrlA-independent ler expression.
FIG. 2.
GrlA is required for expression of C. rodentium ler. The expression of representative ler-cat fusions was monitored in C. rodentium DBS100, C. rodentium Δler, and C. rodentium ΔgrlA (A) or in E. coli MC4100 containing the pMPM-T3 vector or its derivative pTCRLer4 or pTCRGrlA1, expressing Ler or GrlA, respectively. As a control, the expression of a LEE2-cat fusion (pLEE2-CAT) was analyzed in the same strains (B). The expression of representative ler-cat fusions was monitored in E. coli MC4100 and its isogenic hns mutant containing plasmid pMPM-T3 or pTCRGrlA1 (C). The CAT specific activity was determined as described for Fig. 1. The values are the means of at least three independent experiments performed in duplicate. Standard errors are shown with error bars.
Autoregulation of C. rodentium ler.
The autoregulation of ler expression was examined by performing a similar analysis of the pCRler-cat fusions in the C. rodentium ler mutant strain as well as in E. coli K-12 carrying a plasmid expressing Ler. The expression of pCRler-200, pCRler-80, and pCRler-40 showed a 4-, 1.4-, and 10-fold reduction, respectively, in the ler mutant compared with the expression in the wild-type strain (Fig. 2A). The high levels of expression of pCRler-80 in the Δler strain were roughly the same in the wild-type strain, supporting the proposal that this fusion lacks a negative regulatory motif that is required for repression of the ler promoter (see above). The results obtained with pCRler-200 and pCRler-40 suggested that ler expression could be directly autoregulated by its own product or indirectly regulated through an additional regulator encoded by a Ler-regulated gene. To discriminate between these two possibilities, we measured the expression of fusions pCRler-200, -80, and -40 in E. coli K-12 containing a plasmid carrying the ler gene (pTCRLer4). In contrast to the strong GrlA-mediated activation of ler expression in the nonpermissive E. coli background, the presence of Ler did not increase ler-cat expression (Fig. 2B). Conversely, the GrlA-independent expression of the ler promoter in pCRler-80 was reduced sevenfold in the presence of a plasmid expressing Ler (Fig. 2B), supporting the notion that Ler may negatively autoregulate its own expression to optimize its cellular levels, preventing the uncontrolled expression of LEE genes, as recently proposed (2). As a control, the expression of a transcriptional fusion to the LEE2 promoter (pLEE2-cat), whose expression is Ler dependent, was measured. As expected, this fusion was not active in the presence of plasmid-encoded GrlA, while as previously shown (5), its expression was increased significantly in the presence of plasmid-encoded Ler (Fig. 2B).
Taken together, these results rule out a direct positive autoregulation of ler expression by Ler itself, at least in the absence of other A/E-specific factors, and suggest that Ler could be involved in regulating a positive regulatory loop by reciprocally controlling GrlA expression (see below).
Effect of GrlA on ler expression in the absence of H-NS.
The expression of pCRler-40, which lacks the sequences upstream of the ler promoter, was abolished in a C. rodentium grlA mutant and restored in E. coli K-12 by a plasmid expressing GrlA (Fig. 2A and B). In addition, this fusion was active in the absence of H-NS (Fig. 1B) but did not reach the levels seen in wild-type E. coli K-12 carrying the plasmid expressing GrlA (Fig. 2B). These results led us to believe that both regulators (GrlA and H-NS) perform their function by interacting with elements located downstream of position −40 and that GrlA, although it can in part counteract H-NS-mediated repression, is essential for the efficient activation of the ler promoter, even in the absence of H-NS. In order to investigate this hypothesis, the expression of fusions pCRler-200, -80, and -40 in E. coli K-12 Δhns containing plasmid pTCRGrlA1 was determined. As shown in Fig. 2C, the presence of GrlA further increased the expression of pCRler-200, -80, and -40 approximately three-, two-, and fivefold, respectively, compared to the activity observed in the E. coli K-12 hns mutant strain carrying the vector. Although other scenarios cannot be excluded at this point, two possibilities may explain this result. In addition to H-NS, another factor could also partially repress ler expression, and thus GrlA could counteract the total repression exerted by more than one negative regulator. Alternatively, GrlA may counteract the H-NS-mediated repression but also promote the interaction of the RNA polymerase with the ler promoter.
To further define the mechanism by which GrlA induces the expression of ler, GrlA fused to a six-His or maltose binding protein (MBP) tag was purified. Both fusion proteins restored protein secretion in the C. rodentium grlA mutant when expressed in trans (data not shown). However, when using the purified proteins, we were unable to detect GrlA binding to DNA fragments containing the regulatory region of ler by EMSA, even with protein concentrations as high as 25 μM (data not shown).
Identification of cis-acting elements involved in the regulation of grlRA expression.
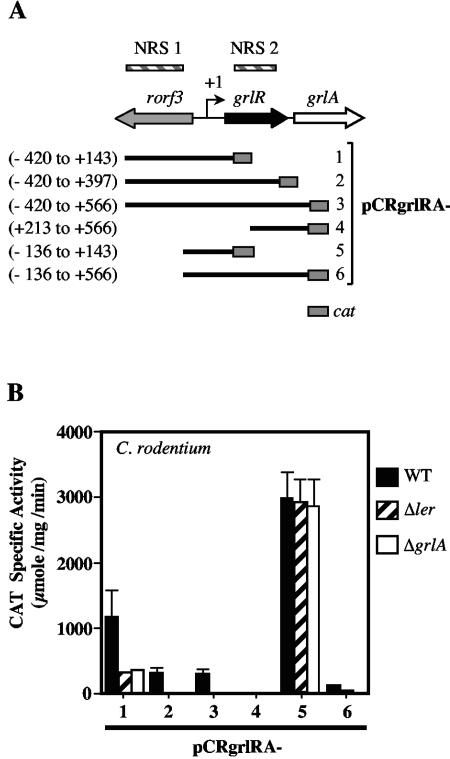
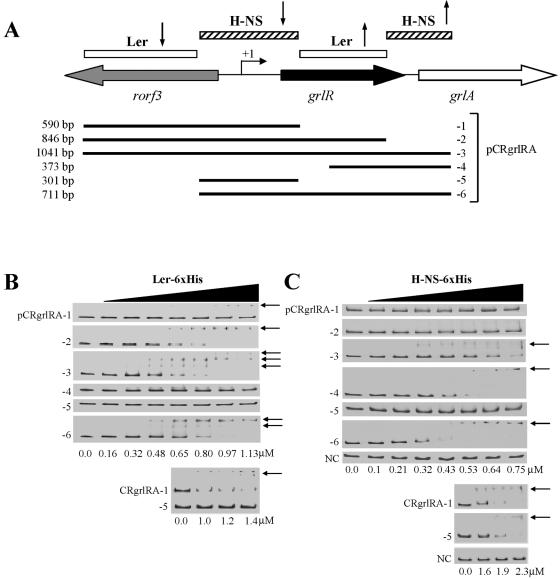
As described above, Ler does not directly regulates its own expression, but could indirectly autoregulate it in a positive manner by reciprocally regulating GrlA expression. In order to test this hypothesis, the regulation of the grlR and grlA genes was studied using a series of transcriptional fusions containing different segments of the 5′ upstream region of grlR and grlA fused to the cat reporter gene (Fig. 3A). Expression was measured in wild-type C. rodentium and its isogenic ler and grlA mutants. The tandem organization of the grlR and grlA genes suggested that they were transcribed as an operon from a promoter located upstream of grlR. In support of this notion, a transcriptional fusion between the grlR-grlA intergenic region and the cat reporter gene (pCRgrlRA-4) was inactive in all three strains tested, while a fusion carrying the intergenic region between grlR and the divergently transcribed rorf3 gene (pCRgrlRA-5) was highly active in the wild-type strain (Fig. 3B). In addition, the expression of pCRgrlRA-5 was Ler and GrlA independent, as it was equally active in the wild-type and mutant strains (Fig. 3B). The presence of further upstream sequences in fusion pCRgrlRA-1 with respect to pCRgrlRA-5 decreased the expression of the grlRA promoter about 2.5-fold in the wild-type strain. In addition, the activity of this fusion was further decreased in the ler and grlA mutants, suggesting that the region from −420 to −136 with respect to the transcriptional start site (see below) contains a negative cis-acting element, which we named NRS1 (negative regulatory sequence 1), and a putative Ler binding region. The presence of further downstream elements in fusions pCRgrlRA-2 (down to the end of grlR) and pCRgrlRA-3 (down to the 5′ end of grlA) with respect to pCRgrlRA-1 (Fig. 3A) reduced their transcriptional activity about fourfold in the wild-type strain, but they were still Ler and GrlA dependent, as their expression was abolished in the mutant strains (Fig. 3B). Since the activities of pCRgrlRA-2 and pCRgrlRA-3 were very similar, these results suggested the presence of a second negative regulatory element (NRS2) between positions +143 and +397 with respect to the grlR transcriptional start site. In agreement with these observations, fusion pCRgrlRA-6, which contains the rorf3-grlRA intergenic region carried by pCRgrlRA-5 plus the NRS2 motif, was 36-fold less active in the wild-type strain than was pCRgrlRA-5 (Fig. 3B).
FIG. 3.
Ler is required for C. rodentium grlRA expression. (A) Schematic representation of the rorf3-grlRA region. Hatched boxes indicate negative regulatory sequences (NRS) revealed by expression analysis of the grlRA-cat transcriptional fusions. The bent arrow indicates the transcriptional start site (+1) for grlRA determined in this study. Schematic representations of the grlRA-cat transcriptional fusions are shown below the diagram of the rorf3-grlRA region. The positions for the 5′ and 3′ ends of the rorf3-grlRA region contained in each fusion, with respect to the transcriptional start site of grlRA, are shown to the left of the fusions. The grlRA-cat fusions were named pCRgrlRA and numbered consecutively as shown at the right of the diagram. (B) Expression of the grlRA-cat fusions was monitored in C. rodentium DBS100 and its isogenic ler and grlA mutants. The CAT specific activity was determined as described for Fig. 1. The values are the means of at least three independent experiments performed in duplicate. Standard errors are shown with error bars.
Taken together, this analysis demonstrated that grlR and grlA form an operon under the control of a promoter located upstream of grlR. In addition, it revealed that sequences flanking the grlRA operon promoter, named NRS1 and NRS2 in this study, are involved in its negative regulation as well as its Ler- and GrlA-dependent activation. In the absence of these elements, grlRA expression becomes constitutive, resembling the regulation of other Ler-dependent promoters (5, 24, 38).
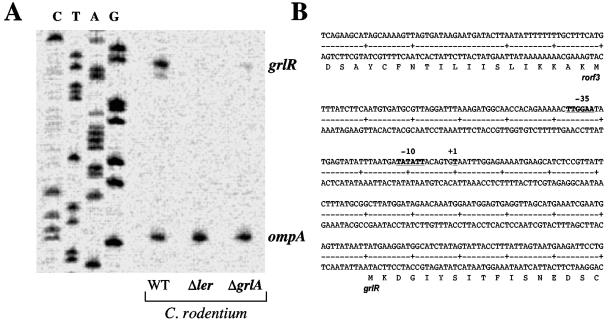
To further support the role of Ler and GrlA in the regulation of the grlRA promoter in C. rodentium and to map the promoter, primer extension analysis was performed using total RNAs purified from the wild-type strain and the ler and grlA mutants. A predominant primer extension product was detected for the wild-type strain (Fig. 4A), revealing that the transcriptional start site of the grlRA promoter corresponds to the T residue located 102 bp upstream of the grlR start codon (Fig. 4B). Putative promoter sequences which show identity to the consensus −10 (five of six [TATATT]) and −35 (four of six [TTGGAA]) sequences of sigma 70-dependent promoters were found upstream of the grlRA transcriptional start site (Fig. 4B). This promoter closely matches the promoter previously reported for EPEC orf10/grlR (31).
FIG. 4.
Primer extension analysis of the C. rodentium grlRA promoter region. (A) Total RNAs were obtained from culture samples of C. rodentium wild-type (WT), Δler, and ΔgrlA strains grown for 6 h in DMEM at 37°C without agitation in a 5% CO2 atmosphere. Primer extension assays were performed with purified total RNA and a primer specific for the grlR structural gene or a primer specific for ompA, which was used as a control. (B) Sequence of the intergenic region between rorf3 and grlRA. The transcriptional start site (+1) and the −10 and −35 promoter sequences for grlRA are shown with bold underlined letters.
In contrast, a primer extension product was not detected in the ler mutant, in agreement with a previous report showing that the expression of the orf10 (grlR) transcript in EPEC was reduced in the absence of Ler (16). Similarly, grlRA transcription was reduced in the grlA mutant (Fig. 4A). To control the RNA load size and integrity, primer extension was performed in parallel to detect the expression of ompA, a constitutively expressed gene coding for an outer membrane protein (17). As shown in Fig. 4A, the ompA transcript was detected at similar levels in the wild-type strain and the ler and grlA mutants.
Ler directly regulates the expression of the grlRA operon.
To further confirm the role of Ler on grlRA regulation, the expression of fusions pCRgrlRA-1, -3, and -6 was analyzed in the nonpermissive E. coli K-12 strain in the presence of a plasmid expressing Ler (pTCRLer4) or GrlA (pTCRGrlA1). The expression levels of these fusions were slightly above the background in the presence of only the vector or the plasmid expressing GrlA (Fig. 5A). In contrast, significant levels of expression were obtained in the presence of Ler (Fig. 5A). This pattern of expression resembles the regulation of the LEE2-cat control fusion (Fig. 5A), which is directly regulated by Ler (5).
FIG. 5.
Ler activates and H-NS represses the expression of grlRA-cat fusions in an E. coli K-12 strain. (A) The expression of representative grlRA-cat fusions was monitored in E. coli MC4100 containing plasmid pMPM-T3 (vector), pTCRLer4, or pTCRGrlA1. As a control, the expression of pLEE2-CAT was analyzed in the same strains. (B) H-NS mediates repression of grlRA expression. The expression of grlRA-cat fusions was monitored in E. coli MC4100 and its isogenic hns mutant containing plasmid pMPM-T3 (vector) or pTCRLer4 (ler). The CAT specific activity was determined as described for Fig. 1. The values are the means of at least three independent experiments performed in duplicate. Standard errors are shown with error bars.
Taken together, these results highlight the existence of a novel positive regulatory loop where GrlA and Ler reciprocally regulate each other to modulate the expression of LEE genes.
H-NS is a negative regulator of grlRA expression.
Previous reports indicated that Ler induces LEE gene expression by counteracting the repression exerted by H-NS on their promoters (5, 24, 46). The results described above indicated that grlRA is positively regulated by Ler and subjected to negative regulation resembling that of other Ler-regulated genes. In order to evaluate whether H-NS is involved in the negative regulation of grlRA, we measured the expression of the grlRA-cat fusions (Fig. 3A) in the E. coli K-12 strain and its isogenic hns mutant. Increased CAT activity was observed for all fusions in the hns mutant, except for pCRgrlRA-4 (which lacks the grlRA promoter), indicating that H-NS negatively regulates grlRA expression (Fig. 5B). However, the fact that the grlRA-catfusions were expressed at different levels in the hns mutant suggested that, in addition to H-NS, other regulators could be involved in repressing grlRA expression. In this regard, compared to pCRgrlRA-1, the pCRgrlRA-2 and pCRgrlRA-3 fusions were between three- and sixfold less active in the hns mutant. This difference could be due to the presence of the grlR gene in these fusions, either because the structural sequence contains cis-acting negative regulatory motifs or because the expression of GrlR, which has been shown to act as a repressor of LEE gene expression (12, 26, 27), has a negative effect on the expression of its own promoter. However, further studies are needed to distinguish between these possibilities.
Fusion pCRgrlRA-5 was also expressed in E. coli K-12, further supporting the notion that it lacks negative cis-acting regulatory elements; however, its expression was further increased (approximately fivefold) in the hns mutant (Fig. 5B). This observation suggests that H-NS negatively controls grlRA expression by interacting with the rorf3-grlRA intergenic region in the vicinity of the promoter between positions −136 and +143. The presence of Ler did not further increase the expression of the grlRA-cat fusions in the E. coli hns mutant (Fig. 5B), strongly suggesting that Ler induces grlRA expression by mainly counteracting the H-NS-mediated repression of this promoter.
Since different attempts to delete or interrupt the C. rodentium hns gene have so far been unsuccessful (despite our success in the generation of deletion mutants in C. rodentium [12]), the experiments described above were performed with E. coli strains. The C. rodentium hns gene, as provided by the Wellcome Trust Sanger Institute, codes for a protein sharing 96% identity with E. coli H-NS, with six amino acid changes located outside functional domains (data not shown). This high degree of conservation suggests that the two proteins are functionally equivalent. In order to confirm the role of H-NS in the transcriptional repression of the grlRA promoter in C. rodentium, we took advantage of the dominant-negative effect shown by E. coli H-NS mutants that are defective in the ability to repress transcription but not in the ability to interact with other H-NS monomers (45). Plasmids expressing E. coli H-NS and the H-NS R12C and G113D mutants under the control of an arabinose-inducible promoter (4, 5) were introduced into C. rodentium Δler carrying the fusion plasmid pCRgrlRA-1 to determine the CAT activity in the presence or absence of arabinose. The expression of the grlRA promoter in the Δler strain was further repressed when wild-type H-NS was induced in C. rodentium Δler. In contrast, when the R12C or G113D H-NS mutant was induced, a dominant-negative effect that allowed the expression of the grlRA promoter was observed (data not shown). These results are in agreement with those obtained using E. coli strains (Fig. 5B).
Ler and H-NS bind to different motifs in the rorf3-grlRA region.
In order to identify the DNA binding sites of Ler and H-NS in the grlRA region, EMSAs with purified Ler-His6 and H-NS-His6 proteins and PCR products corresponding to the fragments contained in the grlRA-cat fusions were performed (Fig. 6A). These experiments demonstrated that Ler binds to DNA fragments corresponding to those present in pCRgrlRA-2, pCRgrlRA-3, and pCRgrlRA-6, starting at a concentration of 480 nM, whereas no binding was detected to pCRgrlRA-4 or -5 fragments, even at a concentration of 1.4 μM (Fig. 6B). The common region between pCRgrlRA-2, -3, and -6 which is not present in pCRgrlRA-4 and -5 is located within the grlR structural sequence between positions +143 and +213 (Fig. 6A), indicating that this region contains sequences recognized by Ler.
FIG. 6.
Binding of Ler and H-NS to the rorf3-grlRA region. (A) Schematic representation of the rorf3-grlRA region. The bent arrow indicates the grlRA transcriptional start site determined in this study. The binding regions for H-NS and Ler revealed by EMSAs are represented by hatched and open boxes, respectively. The DNA fragments used in EMSAs are represented below the diagram of the rorf3-grlRA region. The sizes of the fragments are indicated to the left, and the corresponding fusion numbers are indicated to the right. Increasing concentrations of purified Ler-His6 (B) or H-NS-His6 (C) protein were incubated with the PCR-generated DNA fragments represented in panel A, resolved in 4% polyacrylamide gels, and stained with ethidium bromide. The fragments correspond to the pCRglrRA transcriptional fusions, as indicated to the left of the gels. The EPEC ler structural gene was used as a negative control (NC) for the EMSAs. Arrows indicate DNA-protein complexes.
Binding of Ler to the fragment contained in pCRgrlRA-1 at higher concentrations (Fig. 6B, bottom panel) revealed the presence of an additional lower-affinity binding site. In agreement with this observation, this fusion was still regulated by Ler (Fig. 3B and 5A). The lack of binding to the fragment contained in pCRgrlRA-5 at the same protein concentrations (Fig. 6B, bottom panel) suggested that this putative Ler binding site is located between positions −420 and −136, within the structural sequence of rorf3. Ler binding to fragment 3 generates at least two distinctive complexes (Fig. 6B), suggesting that the binding of Ler to the higher-affinity binding site precedes subsequent binding to the lower-affinity binding site. The expression analysis of grlRA-cat fusions described above suggested that both Ler binding sites could independently mediate grlRA induction, since the expression of grlRA-cat fusions containing only one of these binding sites (pCRgrlRA-1 or pCRgrlRA-6) was still Ler dependent (Fig. 3B and 5A). More defined deletions and site-directed mutagenesis will be required to further map the Ler binding sites involved in grlRA expression, since footprinting analysis has shown that Ler binds to extended regions, complicating the definition of primary binding sites (2, 24, 43; our unpublished results).
Using the same approach, we showed that H-NS binds to the fragments carried by fusions pCRgrlRA-3, -4, and -6, at concentrations ranging from 430 to 750 nM, but not to fragments contained in fusions pCRgrlA-1, -2, and -5 or to a DNA fragment corresponding to the ler structural gene, which was used as a negative control for Ler and H-NS binding (Fig. 6C and data not shown). Fragments pCRgrlRA-3, -4, and -6 share a common region that is absent in pCRgrlRA-1, -2, and -5, localized between positions +397 and +566 spanning the last codons of grlR and the first codons of grlA, indicating that this region contains sequences recognized by H-NS. However, considering that fusions pCRgrlRA-2 and pCRgrlRA-3 have very similar regulatory patterns (Fig. 3B and 5B), it is likely that this binding site does not play a major role in the negative regulation of grlRA expression.
Since fusions pCRgrlRA-1 and pCRgrlRA-5 are still strongly regulated by H-NS (Fig. 5B), another EMSA was performed using higher concentrations of H-NS to explore the existence of lower-affinity binding sites in the vicinity of the grlRA promoter region. At concentrations between 1.6 and 2.3 μM, H-NS bound to the DNA fragments corresponding to pCRgrlRA-1 and -5, but not to the negative control (Fig. 6C, bottom panel), indicating that the sequence contained in pCRgrlRA-5 spanning positions −136 to +143 is bound by H-NS to repress grlRA expression.
DISCUSSION
Different studies have demonstrated that Ler is the primary positive regulator of virulence gene expression in A/E bacterial pathogens (12, 16, 31). Ler expression is finely regulated by a myriad of regulatory factors, as described in the introduction. In addition to all of the regulatory proteins shown thus far to be involved in ler regulation, it was recently shown that ler expression, and thus the expression of other LEE genes, requires the additional LEE-encoded regulator GrlA (12).
In the present study, we demonstrate that GrlA and Ler positively regulate each other's expression, forming a novel transcriptional positive regulatory loop. This notion is supported by the results showing that ler expression was severely reduced in a C. rodentium grlA mutant and restored by GrlA in the nonpermissive E. coli K-12 background (Fig. 2), while the expression of the grlRA operon was impaired in a C. rodentium ler mutant (Fig. 3 and 4) and restored in E. coli K-12 by Ler (Fig. 5A). The complementation experiments with E. coli K-12 clearly reproduced the reciprocal regulation between GrlA and Ler observed in the experiments performed with C. rodentium mutants.
Our results also indicate that Ler positively regulates the expression of grlRA by counteracting, at least in part, the H-NS-mediated repression of its promoter (Fig. 5B). In this regard, we and other groups have shown that H-NS exerts a global repressing effect on EPEC LEE promoters and that Ler acts as an antirepressor counteracting this negative effect (5, 24, 46). For example, H-NS-mediated repression of the divergently transcribed LEE2 and LEE3 operons involves the binding of H-NS to silencer regulatory sequences 1 and 2 (SRS1 and -2) flanking the LEE2 and LEE3 promoters, which favors the formation of a repressor nucleoprotein complex that is probably stabilized by H-NS-H-NS bridging interactions (4, 5). Specific binding of Ler to SRS1 destabilizes the repressor nucleoprotein complex and releases the expression of the LEE2 and LEE3 operons. The expression of both operons is constitutive and is no longer affected by Ler in the absence of any of the SRSs or of H-NS (4, 5). A similar model has been proposed for the regulation of the LEE5 operon (24). However, overcoming transcriptional repression by H-NS is a common mechanism for inducing virulence gene expression in pathogenic bacteria and involves different families of transcriptional activators (reviewed in reference 14).
In agreement with their role in grlRA regulation, H-NS and Ler bind to nonoverlapping sites in the rorf3-grlRA region. DNA binding assays showed that a higher-affinity H-NS-binding site is located between the 3′ end of grlR and the 5′ end of grlA and a lower-affinity H-NS-binding site is located in the intergenic region between rorf3 and grlRA. In contrast, for Ler a lower-affinity binding site is contained within the rorf3 structural gene and a higher-affinity Ler binding site is located at the beginning of the grlR structural gene flanking the grlRA promoter (Fig. 6). The lower-affinity H-NS-binding site, but not the higher-affinity H-NS-binding site, seems to be the one involved in the repression of the grlRA promoter, as all the grlRA-cat fusions containing the rorf3-grlRA intergenic region were derepressed in the Δhns background (Fig. 5B). In contrast, both Ler binding sites could independently mediate grlRA induction by Ler, as fusions carrying one or the other were still regulated in a Ler-dependent manner (Fig. 3B and 5A). It is likely that the binding of Ler to sequences flanking the grlRA promoter region, where the H-NS-binding site resides, induces structural changes that may destabilize H-NS binding, thus releasing promoter expression. However, H-NS is not fully responsible for the negative regulation, since activation of the different pCRgrlRA fusions showed different degrees of derepression in its absence (Fig. 5B). The fact that derepression was only partial in the presence of one or both NRS elements in the Δhns background suggests that an additional factor or mechanism which is not yet defined is required for a second level of repression. Thus, in contrast to the case for the LEE2 and LEE3 promoters, full strength Ler-independent expression of the grlRA promoter is only achieved in the absence of H-NS and both NRSs. Considering the putative role of GrlR as a repressor of LEE gene expression (12, 27), we cannot rule out the possibility that the presence of the grlR gene in some of the pCRgrlRA fusions has a negative influence on its own expression. H-NS also represses the expression of ler in E. coli K-12, but in contrast to the grlRA and LEE2-LEE3 promoters, the ler promoter does not become fully constitutive (e.g., GrlA independent) in the absence of H-NS or negative cis-acting regulatory elements.
It has been previously reported that H-NS represses ler expression at 27°C, but not at 37°C, as a mechanism controlling thermoregulation (46). Our observations confirm the role of H-NS in ler regulation, but they also show that H-NS can exert its negative effect even at 37°C in the absence of ler-specific activators. They also indicate that both H-NS and GrlA require sequences located in close proximity to the ler promoter to exert their functions.
The results reported here indicate that GrlA is required for promoter activation, probably favoring productive interactions of the RNA polymerase with the ler promoter, as well as for counteracting H-NS repression. Similar double functions have been observed, for example, for the regulator ToxT in the expression of ctx and tcp (48). GrlA contains a putative helix-turn-helix motif potentially involved in DNA binding (12). Mutations of this domain at residues that are conserved in CaiF and the Salmonella GrlA homologue abolish GrlA's ability to activate ler expression (unpublished observations). However, despite all the evidence implicating GrlA in binding to DNA, we have not yet been able to detect GrlA binding to the ler promoter region by EMSAs using purified MBP-GrlA and GrlA-His6 fusion proteins, which fully complement the C. rodentium grlA mutant strain (data not shown). The lack of binding in vitro may be the result of different situations, including the possibility that GrlA may become inactive upon purification or that it requires another factor for DNA binding. Correlating with the second possibility, it has been shown that CaiF, the only characterized homologue of GrlA, binds more efficiently to the intergenic cai-fix regulatory region when CRP is present (3) and also counteracts H-NS repression (15).
Furthermore, IHF has been shown to be essential for ler expression in EPEC (19) and for pCRler-cat fusion expression in E. coli K-12 (unpublished results), making it a candidate for acting synergistically with GrlA to activate ler expression. However, our results suggest that IHF is not necessary for the GrlA-mediated activation of ler, since in the absence of the putative IHF binding site, as for pCRler-40, GrlA was still able to activate ler expression (Fig. 2). Similarly, a transcriptional fusion of the EPEC ler regulatory region lacking the IHF binding sequence was still activated in a GrlA-dependent manner (unpublished results). It is worth noting that pCRler-80 rendered significant levels of GrlA-independent expression of the ler promoter in C. rodentium ΔgrlA and E. coli K-12 (Fig. 1B and 2B). These observations suggest that upstream of position −80, there is a putative negative regulatory motif that negatively modulates ler repression. In support of this notion, it has been shown that Hha negatively regulates ler expression in EHEC and interacts with its regulatory region (40). These results also suggest that binding of IHF to its putative binding site, located between position −80 and the ler promoter, may generate architectural changes that partially counteract the negative regulation mediated by, for example, H-NS and/or facilitate RNA polymerase productive interactions with the ler promoter in the absence of GrlA.
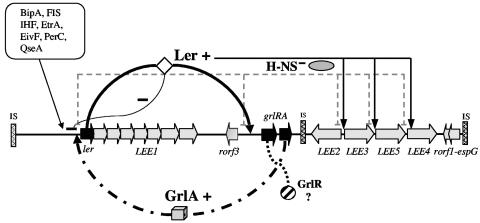
It is not yet possible to determine whether Ler or GrlA is responsible for initiating the feedback regulatory loop. However, it is tempting to suggest that under inducing conditions, preexisting basal levels of Ler and/or GrlA adopt a transcriptionally proficient conformation that allows the reciprocal activation of the grlRA or ler promoter, respectively. Alternatively, or in parallel, the initial increase in ler or grlRA expression could be mediated by DNA structural changes that set the promoters to a more competent transcriptional state or by additional regulatory proteins in response to specific environmental cues. In this way, the active feedback loop will increase the cellular concentration of Ler, which then specifically counteracts the H-NS-mediated repression of several LEE and non-LEE promoters. To prevent the detrimental accumulation of Ler or of the proteins encoded by Ler-regulated genes in the cell, the Ler-GrlA feedback loop could be negatively modulated when Ler reaches the threshold concentration that represses ler transcription, as recently proposed (2). Alternatively, other elements could establish a checkpoint to prevent Ler overexpression. One candidate is GrlR, a protein encoded by the first gene of the grlRA operon that has shown to be involved in the negative regulation of ler expression and thus of Ler-regulated genes (12, 26, 27). Intriguingly, as shown here, grlR is cotranscribed with grlA in a Ler-dependent manner, suggesting that, while the feedback loop is active, GrlR may reach a concentration that down regulates the feedback loop to set it back to the steady-state level. We propose that the Ler-GrlA positive regulatory loop is functionally similar in all A/E pathogens, since the expression of LEE-encoded proteins is also abolished in EPEC and EHEC grlA mutants (unpublished results) and since grlRA (orf10-11) expression is abolished in ler mutants (16; unpublished results). In this way, the concentration of Ler required for the appropriate induction of the LEE genes in A/E pathogens would be maintained by the combined action of positive and negative regulatory loops. A model for the regulation of LEE genes, with emphasis on the positive and negative regulatory loops controlling the expression of Ler, is depicted in Fig. 7.
FIG. 7.
Model for the regulation of LEE genes in A/E pathogens. Ler positively regulates LEE gene expression by counteracting the H-NS-mediated repression of LEE gene promoters. The expression of ler is tightly regulated by specific regulators, such as GrlA, GrlR, and PerC, as well as by global regulators, such as IHF, Fis, BipA, EtrA, EivF, and QseA. Appropriate levels of ler expression are maintained by a positive regulatory loop formed by Ler and GrlA, which could be negatively modulated by GrlR through a mechanism that is still not well understood or by the ability of Ler to negatively autoregulate its own expression (see the text).
In summary, we identified a novel regulatory mechanism involving a reciprocal positive regulatory circuit integrated by the LEE-encoded positive regulatory proteins Ler and GrlA. Although the role of this regulatory loop during infection remains to be elucidated, it is probably required to maintain appropriate levels of different regulatory proteins to achieve a precise and optimal spatiotemporal response to the host environment. This would allow the successful colonization of the preferred niche and prevent the disproportionate production of virulence factors that could potentially jeopardize subsequent stages of the infectious process.
Acknowledgments
We are particularly thankful to R. Oropeza and K. Carrillo for their support with H-NS and Ler purification, to J. A. Ibarra for help with early primer extension experiments, to C. Lara and M. I. Villalba for their help with GrlA purification and binding assays, to J. M. Tellez for his advice, and to A. Vázquez and F. J. Santana for their excellent technical assistance. We also thank E. Calva for helpful discussions and continuous support.
J.B. is supported by a Ph.D. fellowship from CUNACyT. J.L.P. is funded by Dirección General de Asuntos del Personal Académico (DGAPA), Consejo Nacional de Ciencia y Tecnología (CONACyT), and the Howard Hughes Medical Institute (HHMI). B.B.F. is supported by grants from Canadian Institutes of Health Research (CIHR) and the HHMI. J.L.P. and B.B.F. are HHMI International Research Scholars, and B.B.F. is a CIHR Distinguished Scientist and the Peter Wall Distinguished Professor.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 2.Berdichevsky, T., D. Friedberg, C. Nadler, A. Rokney, A. Oppenheim, and I. Rosenshine. 2005. Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J. Bacteriol. 187:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchet, A., W. Nasser, K. Eichler, and M. A. Mandrand-Berthelot. 1999. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol. Microbiol. 34:562-575. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante, V. H., J. A. Ibarra, K. Carrillo, A. Vazquez, and J. L. Puente. 2004. The locus of enterocyte effacement-encoded regulator (Ler) overcomes repression of type III secretion operons by modifying a nucleoprotein complex formed by H-NS. Presented at the 104th General Meeting of the American Society for Microbiology, New Orleans, La., 23 to 27 May 2004.
- 5.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29:83-98. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, S. C., R. D. Haigh, P. P. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8:28-34. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, W., B. A. Vallance, Y. Li, J. L. Puente, and B. B. Finlay. 2003. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol. Microbiol. 48:95-115. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 15.Eichler, K., A. Buchet, R. Lemke, H. P. Kleber, and M. A. Mandrand-Berthelot. 1996. Identification and characterization of the caiF gene encoding a potential transcriptional activator of carnitine metabolism in Escherichia coli. J. Bacteriol. 178:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores-Valdez, M. A., J. L. Puente, and E. Calva. 2003. Negative osmoregulation of the Salmonella ompS1 porin gene independently of OmpR in an hns background. J. Bacteriol. 185:6497-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friden, P., K. Voelkel, R. Sternglanz, and M. Freundlich. 1984. Reduced expression of the isoleucine and valine enzymes in integration host factor mutants of Escherichia coli. J. Mol. Biol. 172:573-579. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 20.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman, M. E., and M. B. Yarmolinsky. 1968. Integration-negative mutants of bacteriophage lambda. J. Mol. Biol. 31:487-505. [DOI] [PubMed] [Google Scholar]
- 23.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 24.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357-2371. [DOI] [PubMed] [Google Scholar]
- 26.Iyoda, S., and H. Watanabe. 2005. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 187:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lio, J. C., and W. J. Syu. 2004. Identification of a negative regulator for the pathogenicity island of enterohemorrhagic Escherichia coli O157:H7. J. Biomed. Sci. 11:855-863. [DOI] [PubMed] [Google Scholar]
- 28.Luperchio, S. A., J. V. Newman, C. A. Dangler, M. D. Schrenzel, D. J. Brenner, A. G. Steigerwalt, and D. B. Schauer. 2000. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38:4343-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 30.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41-46. [DOI] [PubMed] [Google Scholar]
- 31.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 32.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallen, M. J., S. A. Beatson, and C. M. Bailey. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter, M. E., P. Mitchell, A. Free, D. G. Smith, and D. L. Gally. 2005. The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J. Bacteriol. 187:458-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter, M. E., P. Mitchell, A. J. Roe, A. Free, D. G. Smith, and D. L. Gally. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54:1117-1133. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 42.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 44.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263:149-162. [DOI] [PubMed] [Google Scholar]
- 46.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 47.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell. Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 48.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]