Abstract
We report that the cold shock protein CspA of Staphylococcus aureus is required for maximal production of pigment. Results from transcriptional studies revealed that loss of CspA resulted in decreased expression of genes needed for the biosynthesis of 4,4′-diaponeurosporene and the alternative sigma factor SigB.
The yellowish-orange pigment produced by most clinical isolates of Staphylococcus aureus (6) has been associated with enhanced bacterial survival in harsh environments and increased staphylococcal pathogenicity (5, 7, 9, 11, 19, 20). Recently, pigment production by S. aureus has been tied to bacterial virulence by the finding that it impairs the antimicrobial action of neutrophils (20). Accordingly, an understanding of the molecular mechanisms that control pigment production in S. aureus is important.
The orange and yellow pigments that produce the “golden” colonies of S. aureus are the products of a C30 triterpenoid biosynthetic pathway (22, 23). The yellow carotenoid pigment is produced through 4,4′-diaponeurosporene, which is then converted to an orange end product, staphyloxanthin. The protein products of the proposed crtMN operon (29) are responsible for the first stage of carotenoid pigment production. CrtM, a dehydrosqualene synthase, combines two molecules of farnesyl pyrophosphate to form 4,4′-diapophytoene. The 4,4′-diapophytoene then undergoes three rounds of dehydrogenation, directed by CrtN, a 4,4′-diapophytoene desaturase, to produce the intermediate yellow pigment 4,4′-diaponeurosporene (26). The dehydrogenation and conversion of this pigment to staphyloxanthin are thought to be carried out by one or more of the protein products of the four-gene open reading frame orf1-orf4 (GenBank accession number X97985) (28).
Production of pigment is influenced by the rsbUVWsigB system (9, 18, 24, 25). Specifically, the alternative sigma factor SigB (31) positively regulates expression of the crtMN operon (2). In experiments using a genetic derivative of S. aureus strain 8325-4 carrying Tn551, we determined that the susceptibility of S. aureus to an antimicrobial peptide of human lysosomal cathepsin G (CG117-136) is associated with expression of cspA (16), which has been annotated (www.tigr.org) as a cold shock gene in S. aureus. We subsequently found that transfer of a cspA null mutation to the highly pigmented strain COL confirmed a relationship between cspA and staphylococcal susceptibility to CG117-136. Surprisingly, however, cspA-negative mutants of strain COL lost the capacity to produce pigment. We now report that CspA regulates pigment production in S.aureus through a SigB-dependent mechanism.
Bacterial strains, plasmids used, growth conditions, and analysis of pigment.
The strains of Escherichia coli and S. aureus and the plasmids used in this investigation are listed in Table 1. To determine if pigment production is temperature dependent, two 250-ml flasks containing 50 ml tryptic soy broth (TSB) plus the appropriate antibiotic were inoculated (1:100 dilution) from an overnight culture of a specific strain and incubated at 30°C or 15°C. Cultures were sampled every 24 to 30 h until pigmentation was visible. A 20-ml sample of each culture was centrifuged at 6,000 × g for 10 min. To determine the concentration of carotenoid pigment present in each strain, the methanol extraction procedure of Morikawa et al. (24) was employed using S. aureus strains grown at 30°C for 30 h in 50 ml of TSB.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80 ΔlacZΔM15 Δ(lacZA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Stratagene |
| S. aureus | ||
| RN4220 | rsbU res mutant strain used for electroporation with E. coli-replicated plasmids | J. Iandolo (17) |
| COL | Wild type, Mecr with Tetr carried on plasmid | M. Bischoff (29) |
| Newman | Clinical isolate with high level of clumping factor rsbU+ | M. Bischoff (6) |
| SH1000 | rsbU+ variant of strain 8325-4, plasmid free | K. Bayles (12) |
| SKC31 | Wild-type COL with a kanamycin resistance cassette deletion-insertion of the cspA coding region | 16 |
| SKC31(pBCSA) | SKC31 with pBCSA, cspA complementation construct | 16 |
| SKC31(pBT2) | SKC31 with pBT2 vector | 16 |
| IK183 | COL ΔrsbUVWsigB | M. Bischoff (9,18) |
| SKC32 | As IK183 but cspA::Km | This study |
| Plasmids | ||
| pBT2 | Low-copy-number shuttle vector with Ampr in E. coli and temperature-sensitive replication with Cmr in S. aureus | A. Peschel (4) |
| pBCSA | pBT2 vector with 528-bp cspA complementation sequence | 16 |
| pBTCSKOKM | pBT2 vector with 850-bp nonpolar kanamycin resistance cassette ligated into SmaI site between 750-bp region upstream of the cspA start codon and 466-bp region downstream of the cspA stop codon | This study |
Genetic and molecular procedures.
Protocols for the isolation of both chromosomal and plasmid DNA have been previously described (16). The construction of plasmids containing wild-type or mutated cspA coding sequences and transformation by electroporation into S. aureus strain RN4220 and other staphylococcal strains was performed essentially as described (1, 4, 13, 16). A derivative of strain COL bearing a deletion in the rsbUVWsigB operon (strain IK183; Table 1) (9, 18) was transduced with phage 80α (17, 27) that carried the cspA::Km cassette from SKC31, and a transductant (strain SKC32) was identified (Table 1).
Transcription of S. aureus genes was monitored by quantitative real-time reverse transcription-PCR (qRT-PCR), using RNA extracted from 15-h TSB cultures grown at 30°C. RNA was extracted from approximately 5.0 × 109 CFU, resuspended in Tris-EDTA (pH 7.6), that were lysed using the FastProtein blue matrix (Qbiogene) in a FastPrep (FP120) Bio 101 Savant instrument. RNA was separated from the cell lysate using the RNeasy minikit (QIAGEN, Inc.).
The 16S rRNA RT-PCR products were used to normalize the concentration of transcripts from selected genes (data not presented). These genes were the alkaline shock protein, asp23, which is regulated by the secondary sigma factor SigB (8), crtN, and sigB. Specific primers used in qRT-PCR are listed in Table 2. qRT-PCR was performed on an iCycler iQ real-time PCR detection system (Bio-Rad); 0.5 μg of total RNA was used as the template for cDNA synthesis that employed a random hexamer and SuperScript II reverse transcriptase (Invitrogen). iQ SYBR Green Supermix (Bio-Rad) was used in a total reaction volume of 25 μl with 200 nM of 5′ and 3′ primers and twofold-diluted RT reaction mixtures as templates for each PCR.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′ → 3′) |
|---|---|
| 16S5516 | GCCGCGGTAATACGTAGGTG |
| 16S3566 | TTTACGCCCAATAATTCCGG |
| Asp528 | CAAGCATACGACAATCAAACTGG |
| Asp378 | ACGCTCTTCTCTTTCTTTTTCGTTA |
| CrtN517 | TTGGTGCAGGTGTCACAGGA |
| CrtN367 | CTTGAGAAGCAATACGGGCTG |
| SigB5363 | AGTGAGCGATGAACTAACCGC |
| SigB3414 | TCACTGATAGAAGGTGAACGCTCT |
The stability of asp23, crtN, and sigB transcripts and 16S rRNA was determined using RNA extracted from late-log-phase TSB (10 ml) cultures of strains COL and SKC31 grown at 30°C. Prior to the addition of rifampin (200 μg/ml), 1-ml samples were removed for RNA extraction. After a 1-minute incubation period with shaking, 1-ml samples were removed immediately and at time periods corresponding to 2.5, 5, and 10 min after addition of rifampin. The samples were immediately centrifuged at 10,000 × g in a tabletop microcentrifuge, and the pelleted bacteria were rapidly frozen. RNA was extracted by the use of the RNeasy minikit.
Loss of CspA results in decreased pigment production by S. aureus.
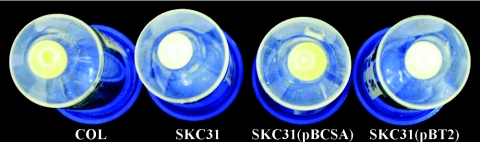
Unlike the parental strain COL, a broth culture of strain SKC31 (isogenic to COL but with cspA::Km) failed to produce the characteristic orange pigment after 30 h of incubation at 30°C (Fig. 1); similar observations were made for broth cultures of strains COL and SKC31 incubated at 15°C or 37°C (data not presented). Pigment (orange) production could, however, be restored in strain SKC31 by placing cspA in trans (strain SKC31/pBCSA) but the strain remained nonpigmented when only the vector (pBT2) was introduced (Fig. 1). Through measurements of the methanol-extractable carotenoid pigment (24) in these strains, we found (Table 3) that strain SKC31 produced substantially less pigment than parental strain COL and that pigment production was restored by expressing cspA in trans (strain SKC31/pBCSA). The low level of the slightly yellow pigment produced by strain SKC31 was likely the intermediate pigment 4,4′-diaponeurosporene (29).
FIG. 1.
Visual comparison of pigment production by the cold shock gene of S. aureus, cspA. Each pellet, at the bottom of a 50-ml conical polystyrene centrifuge tube, represents 20 ml of a 50-ml culture that was harvested after growth at 30°C for 30 h. The strains used were the wild-type S. aureus COL, the cspA::Km insertion-deletion mutant SKC31, SKC31(pBCSA), an SKC31 mutant complemented in trans by a 528-bp insert of the cspA gene in the pBT2 vector, and SKC31(pBT2), an SKC31 mutant control carrying only the pBT2 vector.
TABLE 3.
Comparison of methanol-extracted carotenoid pigments from strains of Staphylococcus aureus
| Strain | Carotenoida |
|---|---|
| COL | 100.0 |
| SKC31b | 32.4 |
| SKC31(pBCSA)b | 107.6 |
| SKC31(pBT2)b | 20.9 |
| IK183b | 15.4 |
| SKC32b | 15.2 |
Values are relative optical density units at 465 nm that have been normalized to and based on strain COL, which was set at 100. In triplicate experiments, the difference between strains COL and SKC31 was significant (P = 0.0006) and the difference between strains SKC31 (pBCSA) and SKC31(pBT2) was also significant (P = 0.004).
All strains are derivatives of strain COL. SKC31 has cspA::Km, while SKC32 has cspA::Km and the rsbUVWsigB deletion mutation in strain IK183; see Table 1.
cspA and gene expression in S. aureus.
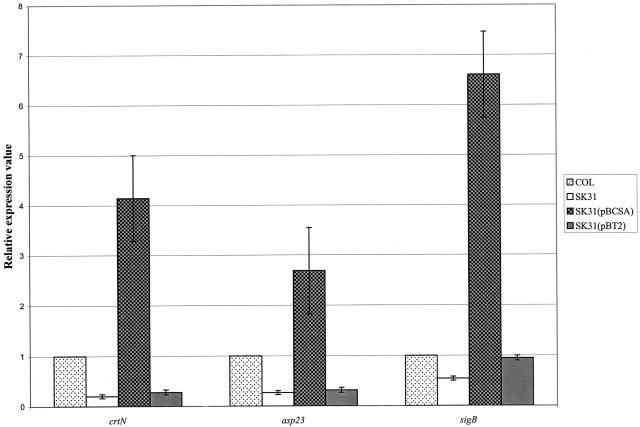
We asked whether CspA and SigB act independently or together to modulate the level of pigment. Compared to that in parental strain COL, the level of the crtN transcript was reduced (Fig. 2) by five- and fourfold in strains SKC31 and SKC31(pBT2), respectively. In contrast, expression of cspA in trans (SKC31/pBCSA) resulted in a fourfold increase in the level of the crtN transcript compared to parental strain COL and a nearly 16-fold increase relative to strains SKC31 and SKC31(pBT2), which was most likely due to the copy number of cspA expressed from pBCSA. However, the level of the asp23 transcript mirrored that of the crtN transcript in being decreased in SKC31 and SKC31(pBT2) and elevated in SKC31(pBCSA). This result prompted us to test whether loss of cspA expression would impact expression of sigB, which regulates expression of crtN (18). As is also shown in Fig. 2, the sigB transcript level was decreased by twofold in strain SKC31 compared to strain COL. Although the presence of vector plasmid pBT2 increased expression of sigB in strain SKC31, the capacity of CspA to regulate sigB expression was more evident when its gene was expressed in trans. Thus, as is shown in Fig. 2, sigB expression in strain SKC31pBCSA was 6.6- and 12.7-fold greater than that observed for strains COL and SK31, respectively.
FIG. 2.
qRT-PCR comparison of transcription of selected S. aureus genes. Shown are the relative levels of transcripts of the asp23, crtN, and sigB genes from S. aureus strains COL, SKC31, SKC31(pBT2), and SKC31(pBCSA). Expression of each gene was normalized to the expression of the reference gene for 16S rRNA from each strain. Relative values of expression in the test strains were calculated and compared to that of parental strain COL. All values are averages (±standard deviation) from triplicate analyses that used three independent preparations of RNA.
In order to determine whether CspA regulates pigment production by stabilizing transcripts involved in pigment biosynthesis or its regulation, we compared the decay rate for the sigB, asp23, and crtN transcripts. An analysis of the decay rate of each transcript revealed the loss of CspA did not significantly impact mRNA stability. In this respect, the half-life for the asp23 mRNA in strain COL (mean ± standard deviation) was 38 ± 10.9 versus 36.3 ± 4.7 seconds in strain SKC31; sigB mRNA in strain COL was 33.4 ± 3.8 versus 33.8 ± 1.2 seconds in strain SKC31; and crtN mRNA in strain COL was 42.9 ± 8.5 versus 55.2 ± 9.3 seconds in strain SKC31; statistical analyses showed that none of these differences were significant (all Pvalues were >0.05).
Based on the above results and similar findings (data not presented) made with pigmented strains Newman and SH1000 (12), we propose that CspA modulates pigment production in S. aureus. This regulatory scheme likely involves control of expression of the crtMN operon and possibly other genes (e.g., orf1-orf4) (28). It is not yet clear whether CspA controls crtMN expression directly or indirectly because loss of CspA production was shown to modulate expression of sigB and a SigB-regulated gene (asp23) by two- and fourfold, respectively. In our hands, loss of the sigB operon in strain COL (strain IK183) has a more dramatic impact on levels of pigment than loss of cspA alone; a strain (SKC32) with mutations in both the rsbUVWsigB operon and cspA produces a level of carotenoid similar to that of the single mutant lacking SigB (Table 3). Importantly, neither SigB nor pigment production is phenotypically linked to antimicrobial peptide susceptibility (16).
Although the mechanism by which CspA regulates pigment production in a SigB-dependent manner is not yet clear, our data indicate that it does not involve stabilization of transcripts. Rather, it appears that CspA acts as an enhancer of transcription. In support of this hypothesis, the CspB protein of Bacillus subtilis (21) has been reported (10) to bind to ATTGG box elements and the complementary pentanucleotide sequence CCAAT in promoter regions of certain genes. Such binding in vivo of CspB to partial single-stranded DNA (32) could function to stabilize an open complex of RNA polymerase. The pentanucleotide sequence has been termed the cold shock domain (Y-box) that is recognized by a family of eukaryotic transcription factors (30). In E. coli, CspA binds to the promoter of gyrA, facilitating its transcription during cold shock (15), and it can also bind to the hns promoter region (3). It has also been proposed to bind RNA (14). Our analysis of the genes involved in pigment production in strain COL (www.tigr.org) revealed a potential CCAAT DNA-binding site for CspA upstream of the promoter for crtMN transcription, a site within crtM, six sites within or adjacent to orf3 and orf4, and one site within sigB (data not presented). The importance of these sites in CspA-mediated regulation of pigment production will require additional investigation.
Acknowledgments
We thank J. Iandolo, C. Lee, M. Bischoff, A. Peschel, and K. Bayles for providing bacterial strains, phages, and plasmids, P. Dunman for the RNA extraction procedure, L. Pucko for manuscript preparation, C. P. Moran and T. Romeo for critical review of the manuscript, and P. Johnson for help in preparing the figures.
This work was supported by NIH grant AI43316 (W.M.S.). W.M.S. is the recipient of a Senior Research Career Scientist Award from the VA Medical Service.
REFERENCES
- 1.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigma B regulons. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandi, A., C. L. Pond, and C. O. Gualerzi. 1994. Interaction of the main cold shock protein CS7.4 of Escherichia coli with the promoter region of hns. Biochimie 76:1090-1098. [DOI] [PubMed] [Google Scholar]
- 4.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain, N. R., B. G. Mehrtens, Z. Xiong, F. A. Kapral, J. L. Boardman, and J. I. Rearick. 1991. Correlation of carotenoid production, decreased membrane fluidity, and resistance to oleic acid killing in Staphylococcus aureus 18Z. Infect. Immun. 59:4332-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcus coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 7.Dye, E. S., and F. A. Kapral. 1981. Partial characterization of a bactericidal system in staphylococcal abscesses. Infect. Immun. 30:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 9.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graumann, P., and M. A. Marahiel. 1994. The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides. FEBS Lett. 338:157-160. [DOI] [PubMed] [Google Scholar]
- 11.Grinstead, J., and R. W. Lacey. 1973. Ecological and genetic implications of pigmentation in Staphylococcus aureus. J. Gen. Microbiol. 75:256-267. [DOI] [PubMed] [Google Scholar]
- 12.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. Sigma B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janzon, L., and S. Arvidson. 1990. The role of the δ-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus 8325-4. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 15.Jones, P. G., R. Krah, S. R. Tafaci, and A. P. Wolffe. 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174:5798-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzif, S., D. Danavall, S. Bowers, J. T. Balthazar, and W. Shafer. 2003. The major cold shock gene cspA is involved in the susceptibility of Staphylococcus aureus to an antimicrobial peptide of human cathepsin G. Infect. Immun. 71:4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreisworth, B., S. Lofdahl, M. J. Betley, M. O'Reilly, M. P. Shlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 35:709-7121. [DOI] [PubMed] [Google Scholar]
- 18.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, A. M., and. M. S. Bergdoll. 1985. Spontaneous occurrence of Staphylococcus aureus mutants with different pigmentation and ability to produce toxic shock syndrome toxin 1. J. Clin. Microbiol. 22:308-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, G. Y., A. Essex, J. T. Buchanan, V. Datta, H. M. Hoffman, J. F. Bastian, J. Fierer, and V. Nizet. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez, M. M., K. Yutani, and G. I. Makhatadze. 1999. Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 274:33601-33608. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, J. H., and G. J. Wilmoth. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, J. H., and G. J. Wilmoth. 1981. Proposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutants. J. Bacteriol. 147:914-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morikawa, K., A. Maruyama, Y. Inose, M. Higashide, H. Hayashi, and T. Ohta. 2001. Overexpression of sigma factor, σB, urges Staphylococcus aureus to thicken the cell wall and to resist β-lactams. Biochem. Biophys. Res. Commun. 288:385-389. [DOI] [PubMed] [Google Scholar]
- 25.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raisig, A., and G. Sandmann. 1999. 4,4′-Diapophytoene desaturase: catalytic properties of an enzyme from the C30 carotenoid pathway of Staphylococcus aureus. J. Bacteriol. 181:6184-61877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wieland, K. P., and F. Goetz. 1996. Regulation of the staphyloxanthin biosynthesis in Staphylococcus aureus Newman. http://www.ncbi.nlm.nih.gov/entrez/viewer. fcgi?db=nucleotide&val=1340127. (Online.)
- 29.Wieland, B., C. Feil, E. Gloria-Maercker, E. G. Thumm, M. Lechner, J-M. Bravo, K. Poralla, and F. Götz. 1994. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176:7719-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolffe, A. P., S. Tafari, M. Ranjan, and M. Familari. 1992. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 4:290-298. [PubMed] [Google Scholar]
- 31.Wu, S., H. De Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeeb, M., and J. Balbach. 2003. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 12:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]