Abstract
Hexaprenyl pyrophosphate synthase (HexPPs) from Sulfolobus solfataricus catalyzes the synthesis of trans-C30-hexaprenyl pyrophosphate (HexPP) by reacting two isopentenyl pyrophosphate molecules with one geranylgeranyl pyrophosphate. The crystal structure of the homodimeric C30-HexPPs resembles those of other trans-prenyltransferases, including farnesyl pyrophosphate synthase (FPPs) and octaprenyl pyrophosphate synthase (OPPs). In both subunits, 10 core helices are arranged about a central active site cavity. Leu164 in the middle of the cavity controls the product chain length. Two protein conformers are observed in the S. solfataricus HexPPs structure, and the major difference between them occurs in the flexible region of residues 84 to 100. Several helices (αI, αJ, αK, and part of αH) and the associated loops have high-temperature factors in one monomer, which may be related to the domain motion that controls the entrance to the active site. Different side chain conformations of Trp136 in two HexPPs subunits result in weaker hydrophobic interactions at the dimer interface, in contrast to the symmetric π-π stacking interactions of aromatic side chains found in FPPs and OPPs. Finally, the three-conformer switched model may explain the catalytic process for HexPPs.
Isoprenoids are among the most diverse and widely distributed natural compounds (17, 22). Using the 5-carbon isopentenyl pyrophosphate (IPP) as building blocks, linear isoprenoids are synthesized by a group of prenyltransferases (PTases), which catalyze the multiple-IPP condensation reaction with allylic substrates, i.e., C5 dimethyl allyl pyrophosphate (DMAPP), C15 farnesyl pyrophosphate (FPP), or C20 geranylgeranyl pyrophosphate (GGPP) for chain elongation (13). These PTases have been classified as E- and Z-types, which synthesize products with trans and cis double bonds, respectively. Different enzymes synthesize varying chain length products, which are then utilized as precursors for steroids, carotenoids, quinones, dilichols, prenylated proteins, and archaeal membrane lipids.
Trans-PTases are typically classified into three groups based on the chain length of their final products, i.e., short (C10 to C25), medium (C30 to C35), and long chain (C40 to C50) (12, 16). The crystal structures of short-chain farnesyl pyrophosphate synthase (FPPs) and long-chain octaprenyl pyrophosphate synthase (OPPs) have been determined previously (6, 10, 25). These proteins are homodimers. Each subunit contains several α-helices that form a large central active site cavity with two conserved DDXXD motifs, located at the rims of the helices D and H, respectively. On the other hand, the medium-chain trans-PTases, including hexaprenyl pyrophosphate synthase (HexPPs) and heptaprenyl pyrophosphate synthase (HepPPs), are composed of either homodimers or heterodimers. The HexPPs from Sulfolobus solfataricus (23), which catalyzes the condensation of two IPP molecules with GGPP to yield all-trans- C30-HexPP, is a homodimer (8). In contrast, HexPPs from Micrococcus luteus B-P 26 and HepPPs from Bacillus subtilis are heterodimers with A and B components (5, 24, 28, 29). HexPPs synthesizes the precursor of the prenyl side chain of caldariellaqunone (3, 30). To date, no structure of medium-chain PTases has been available to explain the necessity of homodimer-heterodimer formation and the mechanism of chain length determination.
We report here the first crystal structure of a medium-chain HexPPs from S. solfataricus. On the basis of the crystal structure of HexPPs, we performed site-directed mutagenesis and identified the key amino acid for determining the product chain length. The depths of the active sites in FPPs, HexPPs, and OPPs were then compared to rationalize the mechanism of chain length determination in the short-, medium-, and long-chain enzymes. Furthermore, the dimer interfaces in these trans-PTases may provide important clues toward understanding the requirement of homo- and heterodimerization for the medium-chain prenyl-synthesizing enzymes.
MATERIALS AND METHODS
Materials.
Exsel DNA polymerase was obtained from Life Technologies, Inc. The plasmid miniprep kit, DNA gel extraction kit, and Ni-nitrilotriacetic acid resin were purchased from QIAGEN. Potato acid phosphatase (2 U/mg) was purchased from Roche Molecular Biochemicals. The protein expression kit [including the pET16b vector and competent JM109 and BL21(DE3) cells] were obtained from Novagen. The QuikChange Site-Directed Mutagenesis kit was obtained from Stratagene, Inc. Radiolabeled [14C]IPP (55 mCi/mmol) was purchased from Amersham Pharmacia Biotech, and FPP was obtained from Sigma. Reverse-phase thin-layer chromatography (TLC) plates were purchased from Merck. All commercial buffers and reagents used were of the highest purity available.
Cloning, expression, and purification.
The S. solfataricus P2 HexPPs gene was amplified by PCR from genomic DNA of Sulfolobus solfataricus P2 with primers HexPPs-NcoI (5′-CATGCCATGGCCCACCACCACCACCACCACTGAGTATTATAGAGTTCTGGTTAG-3′) and HexPPs-BamHI (5′-CGCGGATCCGCGTCATTAAATCTTATCTATGTTAGCCTCC-3′) (restriction sites are underlined). The 867-bp PCR products were digested with NcoI and BamHI, and the DNA fragments were cloned into pET16b (Novagen). This construct, with a hexa-His tag attached to the N-terminal methionine, was transferred into Escherichia coli BL21(DE3) (Novagen)-competent cells, and DNA sequencing was performed to confirm the appropriate orientation. The HexPPs protein was then overexpressed in E. coli BL21(DE3) in 6 liters of Difco Luria-Bertani broth containing 50 mg/liter ampicillin to an optical density at 600 nm of 0.5 and then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside. After additional overnight incubation, the cells were harvested and disrupted by a French press in a buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 20 mM imidazole. The homogenate was centrifuged at 27,000 × g for 30 min, and the supernatant was recovered as a crude extract which was then heated at 55°C for 30 min. The denatured proteins were removed by centrifugation at 27,000 × g for 30 min. The supernatant containing the heat-resistant HexPPs was recovered, and the enzyme was purified with an Ni-NTA column and dialyzed, each three times with 5 liters buffer (20 mM Tris-HCl, pH 8.0) for 6 h. Finally, the HexPPs enzyme was concentrated by Amicon (Millipore). About 100 mg of HexPPs was obtained, with a purity of more than 95% as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Site-directed mutagenesis of HexPPs.
HexPPs mutants were prepared by using a QuikChange Site-Directed Mutagenesis kit in conjunction with the HexPPs gene template in the pET16b vector. The mutagenic primers for D81C, Y124A, Y174A, L164A, L164G, and W136E of HexPPs and F117E of OPPs were prepared by Biobasic Inc. (Canada). The basic procedure of mutagenesis utilizes a supercoiled double-stranded DNA (dsDNA) vector with an insert of interest and two synthetic oligonucleotide primers containing the desired mutation. The mutation was confirmed by sequencing the entire HexPPs mutant gene of the plasmid obtained from the overnight culture. The correct construct was subsequently transformed to E. coli BL21(DE3) for protein expression and purification.
Crystallization and data collection.
D81C and W136E mutant crystals were grown at room temperature by the hanging drop vapor diffusion method from a reservoir solution containing 10% (wt/vol) polyethyleneglycol 8000, 2.4 M lithium chloride, and 100 mM Tris (pH 8.0). The crystals reached the maximum size (0.2 mm) in 3 to 5 days. Crystals were frozen after cryoprotection by adding 20% (wt/vol) ethylene glycol to the mother liquor. The D81C mutant X-ray data were collected at 100 K on the Taiwan Contract BL12B2 station at SPring-8 (Hyogo, Japan). The W136E HexPPs mutant, F117E OPPs mutant, and three HexPPs multiple isomorphous replacement (MIR) data sets were collected in house using a Rigaku MicroMax007 X-ray generator equipped with an R-Axis IV++ image plate detector. The diffraction data were processed and scaled by using HKL/HKL2000 packages (20). Statistics are shown in Tables 1 and 2.
TABLE 1.
Heavy-atom derivatives and MIR statistics of HexPPs
| Data seta | Thimerosalb | C7H5ClHgO2 | C(HgOOCH3)4 |
|---|---|---|---|
| Space group | P3121 | P3121 | P3121 |
| Unit cell dimensions (Å) | a = b = 91.99, c = 121.68 | a = b = 92.28, c = 122.44 | a = b = 92.02, c = 121.28 |
| Resolution (Å) | 50-2.7 (2.8-2.7) | 50-2.8 (2.9-2.8) | 30-2.64 (2.76-2.64) |
| No. of observations | 59,726 (5,980) | 49,012 (5,014) | 93,050 (9,673) |
| Unique reflections | 16,805 (1,643) | 15,246 (1,500) | 17,777 (2,191) |
| Completeness (%) | 99.6 (99.7) | 99.3 (99.8) | 97.2 (99.5) |
| Rmerge (%) | 9.0 (48.9) | 10.4 (44.2) | 8.7 (47.8) |
| Avg I/σ(I) | 13.23 (2.69) | 10.67 (2.79) | 17.03 (2.77) |
| Phasing power | 0.28 | 0.45 | 0.26 |
| Mean overall figure of merit | 0.71 (50-2.8 Å) | ||
| No. of sites | 4 | 1 | 1 |
Numbers in parentheses are for the highest resolution shell.
Thimerosal is 2-(CH3CH2HgS)C6H4CO2Na.
TABLE 2.
Data collection and refinement for D81C, W136E HexPPs, and F117E OPPs mutants
| Statistics | D81C | W136E | F117E OPPs |
|---|---|---|---|
| Data collection statisticsa | |||
| Beam line | BL12B2, SPring-8 | Cu Kα radiation | Cu Kα radiation |
| Wavelength (Å) | 1.0000 | 1.5418 | 1.5418 |
| Space group | P3121 | P3121 | I422 |
| Unite cell dimensions (Å) | a = b = 92.49, c = 122.62 | a = b = 92.53, c = 127.19 | a = b = 151.51, c = 54.61 |
| Resolution range (Å) | 50-2.4 (2.49-2.40) | 50-2.7 (2.8-2.7) | 50-2.8 (2.9-2.8) |
| Total observations | 179,327 (17,723) | 79,446 (7,627) | 59,883 (2166) |
| Unique observations | 24,259 (2,365) | 17,750 (1,743) | 9.443 (907) |
| Completeness (%) | 99.9 (100.0) | 99.8 (99.9) | 99.0 (99.3) |
| Rmerge (%) | 6.3 (40.9) | 8.7 (49.6) | 6.4 (52.6) |
| Avg I/σ(I) | 31.8 (5.4) | 17.9 (3.1) | 27.5 (4.2) |
| Refinement statisticsb | |||
| Reflections | 23,468 (2,202) | 15,209 (1,648) | 8,732 (892) |
| R/Rfree (%) | 24.0/29.7 | 24.8/32.2 | 21.8/27.3 |
| Root mean square deviation from ideal geometry | |||
| Bonds (Å) | 0.0065 | 0.0083 | 0.0084 |
| Angles (°) | 1.08 | 1.14 | 1.25 |
| No. of nonhydrogen atoms/avg B factor (Å2) | |||
| Monomer A | 2,238/39.6 | 2,235/42.4 | 2,216/55.6 |
| Monomer B | 2,224/71.0 | 2,235/76.3 | |
| Water | 394/56.9 | 168/39.7 | 287/65.0 |
Numbers in parentheses are for the highest resolution shell.
All positive reflections are used in the refinement.
The X-ray diffraction data sets for the D81C and W136E HexPPs mutants were collected to 2.4- and 2.7-Å resolution, respectively. Both D81C and W136E crystals belong to the trigonal space group P3121 with similar unit cell (Table 2). The Matthews coefficient of 2.28 Å3/Da and solvent content of 46% were consistent with one dimer per asymmetric unit.
Structure determination and refinement.
The phase angles of D81C HexPPs crystal were calculated using Thimerosal, C7H5ClHgO2, and C(HgOOCH3)4 heavy-atom derivative data by the MIR (multiple isomorphous replacement) method using SOLVE (27). The MIR map at 2.4 Å was subjected to maximum-likelihood density modification followed by autotracing using RESOLVE (26). An initial model was built using RESOLVE and XtalView (15). The model was improved by manual rebuilding using XtalView and was refined using CNS (1). Refinement statistics of D81C and W136E HexPPs mutants are summarized in Table 2. A Thermotoga maritima OPPs mutant, F117E, for probing the dimer interface was similarly refined, and its refinement statistics are also listed in Table 2.
Substrate specificity of the recombinant HexPPs.
The activity of recombinant HexPPs (1 μM) with 5 μM allylic substrate (geranyl pyrophosphate [GPP], FPP or GGPP), 50 μM [14C]IPP, 0.1% Triton X-100, 0.5 mM MgCl2, and 50 mM KCl in 100 mM HEPES buffer (pH 7.5) was measured to test the substrate specificity of the enzyme. The initial velocity of the enzyme reaction was calculated according to the first 10% of the radiolabeled substrate converted to the polyprenyl products as described previously (21).
Product analysis.
The HexPPs reaction containing 1 μM enzyme (wild-type and mutant HexPPs), 5 μM FPP, 50 μM [14C]IPP, 0.1% Triton X-100, 0.5 mM MgCl2, and 50 mM KCl in 100 mM HEPES buffer (pH 7.5) was performed for 72 h at 25°C. EDTA (10 mM) was used to terminate the reactions. Radiolabeled polyprenyl pyrophosphate products were extracted with 1-butanol. The solution was then evaporated, and a 20% propanol solution containing 4.4 U/ml acidic phosphatase, 0.1% Triton, 50 mM sodium acetate (pH 4.7) was prepared to convert polyprenyl pyrophosphate products to corresponding alcohols according to the reported procedure (4). After the pyrophosphate hydrolysis catalyzed by acidic phosphatase was completed, the polyprenols were extracted with n-hexane. The hexane volume was reduced by evaporation. The polyprenols were separated on reversed-phase TLC using acetone/water (19:1) as the mobile phase. The radiolabeled products were identified by autoradiography using a bioimaging analyzer FUJIFILM BAS-1500 (Japan) according to their reported Rf values.
Atomic coordinates and structure factors.
The atomic coordinates and structure factors (code 2AZJ for D81C mutant HexPPs, 2AZK for W136E mutant HexPPs, and 2AZL for F117E mutant OPPs) have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank.
RESULTS
Sequence comparison of trans-PTases.
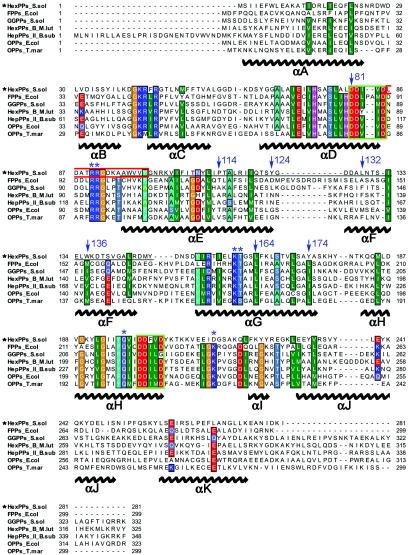
The amino acid sequences of five representative members of the trans-PTase family are compared in Fig. 1. Other trans-PTases have 22 to 26% sequence identity to S. solfataricus HexPPs. Two highly conserved catalytic DDXXD motifs, each located on helices D and H, respectively, are essential for FPP binding and IPP condensation reaction (10). The dimer interface (on helices E and F) is less conserved. Several important amino acids (Fig. 1, blue asterisk) involved in the structural-based three-metal ion mechanism for FPPs isoprenoid condensation (10) are also conserved in HexPPs, except residue 258. The amino acid located at the fifth position before the first DDXXD, which was proposed to shield product synthesis in FPPs and GGPPs (18, 19), is a small amino acid (Ala) in the long-chain T. maritima OPPs, E. coli OPPs, and S. solfataricus HexPPs. In contrast, the short-chain E. coli FPPs and S. solfataricus GGPPs have Tyr and Phe at this position, respectively. Notably, the F132 seals the bottom of OPPs's active site and determines the final product chain length (6), and the equivalent amino acid in S. solfataricus HexPPs is Leu129. The roles of residues located in the HexPPs's active site were investigated by the mutagenesis study, as elucidated below.
FIG. 1.
Multiple-sequence alignment of the trans-prenyltransferase family and secondary-structure assignment of monomer A from S. solfataricus HexPPs. Amino acid sequences of C30-HexPPs from S. solfataricus, C15-FPPs from E. coli, C20-GGPPs from S. solfataricus, component B of C30-HexPPs from M. luteus B-P 26, component II of C35-HepPPs from B. subtilis, C40-OPPs from E. coli, and C40-OPPs from T. maritima are aligned. Identical and similar amino acid residues are shaded in like colors. The two conserved catalytic DDXXD motifs are highlighted in a green box, and the amino acids associated with different structural conformations in the two HexPPs subunits are highlighted in a red box. The blue arrows indicate the amino acids mutated in this study, and the blue asterisks indicate the key amino acids that involved in a FPPs isoprenoid condensation reaction. The amino acids at the dimer interface of HexPPs are underlined. The alignment was prepared with ClustalW.
Overall structure of HexPPs.
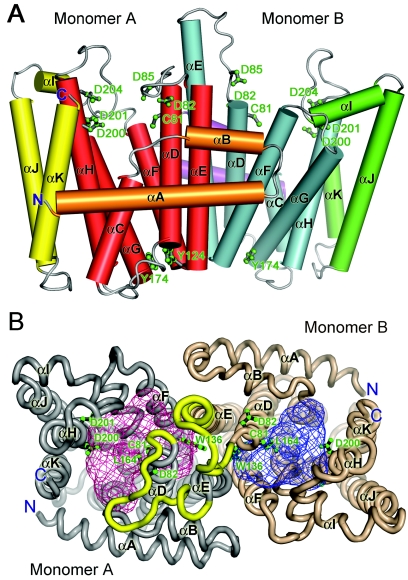
The crystal structure of S. solfataricus HexPPs was determined by a multiple isomorphous replacement (MIR) method, and the refinement was carried out using the 2.4-Å resolution data of the D81C mutant (Fig. 2A and Table 2). The first amino acid, D81, of the first DDXXD motif was mutated to cysteine in order to bind heavy atoms (mercuric ion) for phase determination. The final refined D81C HexPPs mutant structure consists of residues 1 to 276 in chain A and residues 3 to 276 in chain B. Each asymmetric unit of the crystal contains one dimer, which is the active form of the enzyme. Its secondary structures (Fig. 1) were assigned according to the three-dimensional structure of monomer A.
FIG. 2.
(A) Overall structure of S. solfataricus D81C HexPPs is shown using a cylinder diagram. The residues of two active-site DDXXD motifs are shown in a ball-and-stick model. (B) The top-view model of S. solfataricus D81C HexPPs is shown as “worm” tracing. The two subunits of HexPPs are shown in gray and light brown, respectively. Their active-site cavities are shown in red and blue, and the cavity volumes are 1,331 and 804 Å3, respectively. The residues 84 to 100 (in yellow) have different conformation in two subunits. These figures were produced using PyMOL.
The structure is composed entirely of all α-helices joined by connecting loops. The 10 α-helices (all α-helices, except helix I) form a large cavity as the active site at the center of each monomer. The substrate-binding site is formed by two conserved DDXXD motifs, each located on the respective helices D and H. The bottom of the central cavity is occupied by two bulky side chains of Y174 and Y124. A complementary hydrophobic interface between helices E and F mainly mediates the HexPPs dimerization. The two subunits of HexPPs adopt different conformations in residues 84 to 100 comprising two α-helices, αD and αE (Fig. S1 in the supplemental material). The N terminus of helix E has two additional turns and the connecting loop to helix D is stretched out in monomer B, whereas monomer A exhibits a similar conformation to the corresponding regions in FPPs and OPPs. In the wild-type HexPPs structure (data not shown), similar conformation of monomer B is observed, suggesting that it is not a result of the D81C mutation. Helix E in monomer B is involved in lattice contact interactions, which should be responsible for capturing the HexPPs molecules in such an asymmetric conformation (Fig. S2 in the supplemental material). The volume of the active-site cavity is 1331 Å3 and 804 Å3 in monomers A and B, respectively (Fig. 2B). In monomer A, these helices are partly unraveled to create a different active-site cavity from that of monomer B. Owing to the steric hindrance effect in the dimer interface, the bulky residue W136 of monomer B protrudes inward to the active-site cavity. This side chain conformational change significantly reduces the cavity volume of monomer B.
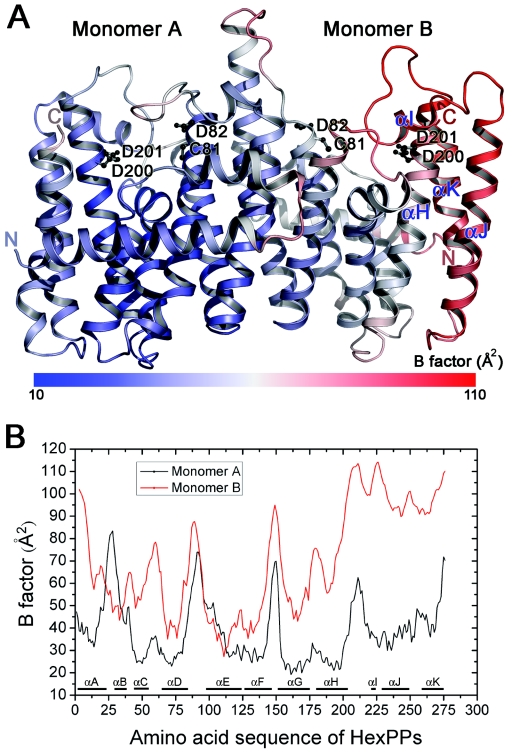
Flexibility analysis of HexPPs by temperature factors.
The B-factor analysis was performed over the entire sequences and for different protein subunits (Fig. 3). The results show that the residues with low B factors occur predominantly in the central portions of the helix bundles, and the residues with high B factors are associated with the interconnecting loops between the helices. This observation suggests that the active-site residues, in general, are less flexible than the non-active-site residues. The protein can thus provide a hydrophobic pocket within the helices along with the stabilization of specific charged groups at the active site. On the other hand, the C-terminal region of monomer B (helices I, J, and K) has the highest B factors (Fig. 3), a result of loose crystal packing interaction (Fig. S2). Although the overall dispositions of these helices are not different in the two monomers, since the conformational flexibility is likely correlated with B factors, this region may also adopt alternative conformation during substrate binding or catalysis.
FIG. 3.
(A) S. solfataricus HexPPs is colored from blue to red according to B factor from 10 to 110 Å2. The figure was produced using PyMOL. (B) Temperature factor plot for monomers A and B of HexPPs, independently. The average temperature factor of monomer B (red line) is apparently higher than monomer A (black line). Notably, the B factor is higher than 100 Å2 in the C-terminal region (residues 200 to 276) of monomer B.
Substrate specificity of the recombinant HexPPs.
Prenyltransferases utilized IPP for chain elongation of allylic substrate FPP or GGPP. Since in S. solfataricus there is a GGPP but not FPP synthase enzyme, it was suggested that HexPPs should utilize GGPP rather than FPP as the substrate. We tested the reaction products and measured the activity by using C15-FPP, C20-GGPP, and shorter C10-GPP as a substrate. We found that all of the above substrates led to the same C30 final product by HexPPs. In term of enzyme activity, GGPP (specific activity, 0.016 s−1) displayed threefold better activity than that using FPP (specific activity, 0.005 s−1) as a substrate under the experimental conditions. The activity was threefold further reduced by using shorter substrate GPP (specific activity, 0.002 s−1). Apparently, GGPP is a native substrate for HexPPs. However, in the test of the products for the mutant enzymes, we used FPP, which is available in the laboratory for the reaction, since both FPP and GGPP gave the same products.
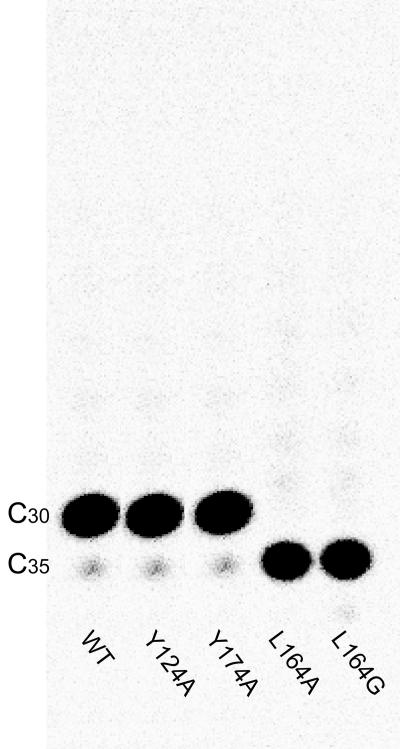
Essential amino acid for product chain length determinant.
On the basis of the crystal structure, we aimed to identify the key residues for determining the ultimate chain length of HexPPs. Two large amino acid residues (Y124 and Y174) at the bottom of the internal cavity, analogous to F132 in OPPs (6), were initially predicted to be the chain length determinant of the C30 product. We replaced these large residues with Ala and examined the chain lengths of the products synthesized by the mutant enzymes using TLC analysis. Surprisingly, both Y124A and Y174A mutants generated the same C30 product (Fig. 4, lanes 2 and 3). Another residue, L164, located in the middle of the cavity was then predicted to be a suitable candidate. Conversion of L164 to alanine, making the L164A mutant, produced only one C5-prenyl unit longer C35 product (Fig. 4, lane 4).
FIG. 4.
The products synthesized by wild-type, Y124A, Y174A, L164A, and L164G HexPPs. The reaction mixture containing 1 μM of each mutant enzyme, 5 μM FPP, and 50 μM [14C]IPP was incubated at 25°C for 72 h to complete the reaction. The final products were extracted and analyzed using TLC and a phosphorimager. Among the mutants, L164A and L164G synthesize a slightly longer product (major product, C35) than that synthesized by the wild type (C30).
Comparison of active sites among trans-PTases.
The Protein Data Bank (PDB) (2) was used to search for other structural homologues of HexPPs using the DALI server (9). The best match for the A monomer of S. solfataricus HexPPs was obtained from the A chain of the E. coli FPPs ternary complex (PDB ID code 1RQI), with a Z-score of 23.5, which corresponds to the root-mean-square deviation (RMSD) of 2.4 Å for 233 equivalent Cα atoms. The second highest score goes to the A chain of the T. maritima OPPs (PDB ID code 1V4E), with a Z-score of 23.3, which corresponds to the root-mean-square deviation (RMSD) of 2.9 Å for 242 equivalent Cα atoms. The structures of these three enzymes share the similar overall fold and topology (Fig. S3 in the supplemental material), despite relatively low sequence identity (about 25%). Most of the backbones are superimposed very well, except for the two regions in αD-αE and αH-αI loops. Notably, conserved Arg116 and Lys258 emanating from the conformationally variable αD-αE and αH-αI loops in FPPs shield the reaction from bulk solvent and stabilize the catalytic base of pyrophosphate oxygen by their positive charges (10). FPPs undergoes the substrate-induced active-site rearrangements that protect the centrally located cavity from bulk solvent and position active-site residues in a catalytically competent conformation. Similar conformation in the αD-αE loop can be found in monomer A of HexPPs.
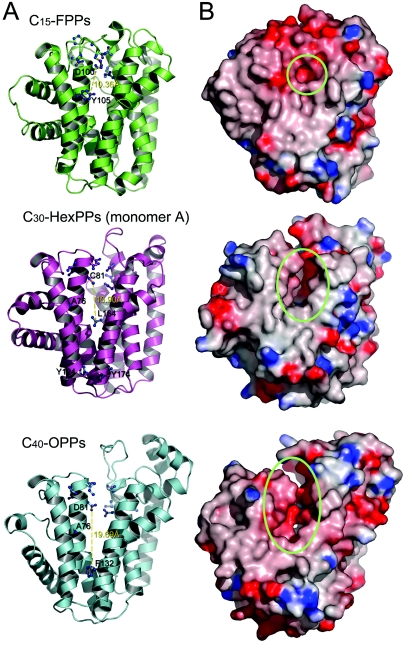
It has been proposed that the size of an elongated hydrophobic tunnel in the active site determines the product size of PTases (6, 7), because a bulky side chain at the distal end of the cavity blocking the product from penetrating the tunnel. To further understand the product specificity of FPPs, HexPPs, and OPPs, we calculated the depth of the active-site cavities for these three trans-PTases (Fig. 5A). The direct distances between the first DDXXD motif and the residue responsible for chain length determination are 10.36 Å, 10.90 Å, and 18.69 Å for FPPs, HexPPs, and OPPs, respectively. Although there is little difference between FPPs and HexPPs in the distance, the smaller size of A76 in HexPPs compared to the corresponding residue Y79 in FPPs apparently provides more space for HexPPs to accommodate longer products. In contrast, OPPs with deeper active sites provides sufficient space for the C40 isoprenoid. Figure 5B illustrates that the larger space within the active-site cavities gives rise to longer chain lengths of the final products.
FIG.5.
Ribbon diagram (A) and electrostatic surface potential diagram (B) are shown from red to blue according to the charge potential from −15 to 15 kBT. Structural comparison of E. coli FPPs, S. solfataricus HexPPs, and T. maritima OPPs are shown in lime, magenta, and cyan, respectively. (A) The first Asp of the first DDXXD motif and the key residues for chain length determination are shown in a ball-and-stick model. The distances between the first DDXXD motif and the residue for chain length determination are 10.36 Å, 10.90 Å, and 18.69 Å for FPPs, HexPPs, and OPPs, respectively. The active-site cavities of the enzymes become deeper and wider as the lengths of the ultimate products increase. The diagrams were produced with PyMOL and GRASP.
Comparison of the dimer interfaces in trans-PTases.
The dimer interface parameters for three trans-PTases were calculated using the Protein-Protein Interaction Server (11) (Table 3). The accessible solvent areas at the dimer interface of FPPs, HexPPs, and OPPs are 1,800, 1,820, and 1,712 Å2, respectively, above the mean 1,685 Å2 of many other homodimers. We also measured the “complementarity” between the interacting surfaces using a gap index, defined as the enclosed volume between any two molecules divided by the surface area. For FPPs, HexPPs, and OPPs dimers, the gap index values are 1.09, 1.63, and 2.01, respectively (compared with the mean value of 2.2 of other homodimers), indicative of a highly complementary dimer interface. Van der Waals interactions between hydrophobic residues contribute most to the stability of these dimers; about 70% of the atoms involved in the dimer interface are nonpolar. Among the three trans-PTases, the HexPPs dimer has fewer hydrogen bonds and no salt bridge at the interface. Most of the dimer surface interactions in the three trans-PTases are between helices E and F and part of helix B.
TABLE 3.
Parameters of protein-protein interaction for E. coli FPPs, S. solfataricus HexPPs, and T. maritima OPPsa
| Interface parameter | FPPs | HexPPs | OPPs | Homodimera |
|---|---|---|---|---|
| ΔASAb (Å2) | 1,800 | 1,820 | 1,712 | 1,685 |
| % ΔASA | 14.12 | 12.68 | 11.35 | |
| % Polar atoms in interface | 28.25 | 27.05 | 31.76 | |
| % Nonpolar atoms in interface | 71.70 | 72.90 | 68.20 | |
| Hydrogen bonds | 9 | 5 | 6 | |
| Salt bridges | 2 | 0 | 4 | |
| Gap volumec (Å2) | 3,814 | 5,809 | 6,829 | |
| Gap volume index | 1.09 | 1.63 | 2.01 | 2.2 |
A mean value from the data set of 32 nonhomologous homodimers (11).
The protein-protein interface has been defined based on the change in their solvent accessible surface (ΔASA).
The gap volume between the two components of the complexes was calculated using SURFNET.
Interactions at the dimer interface were analyzed using the Protein-Protein Interaction Server (11).
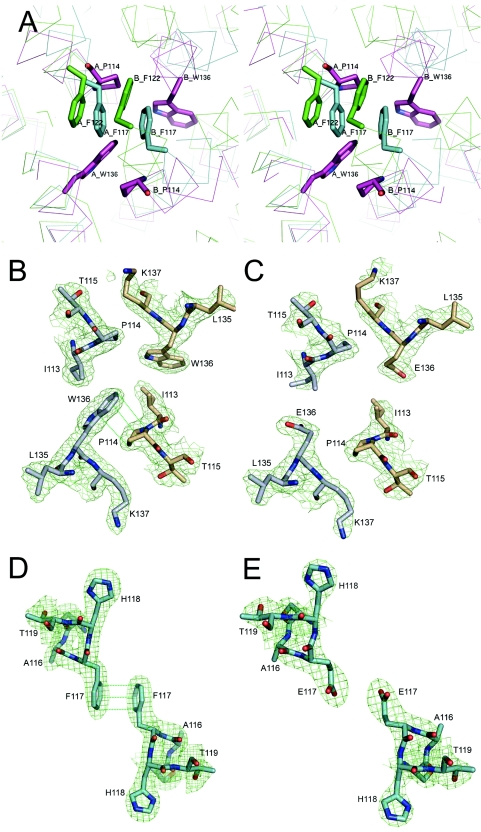
Several planar residues, such as F122 of FPPs, P114 and W136 of HexPPs, and F117 of OPPs, are found at the dimer interface through strong stacking interactions (Fig. 6A). In FPPs and OPPs, the side chains of F122 and F117, respectively, are involved in the π-π interaction with the same residues from the other subunit. In contrast, HexPPs forms the crossing stacking interaction between W136 on helix F from monomer A and P114 on helix E from monomer B (Fig. 6A). In an attempt to disrupt the dimer, the hydrophobic residues in the dimer interface of OPPs (F117) and HexPPs (W136) (Fig. 6B and D) were replaced by a negatively charged Glu residue. The mutants still formed dimers (Fig. 6C and E). Apparently, the dimer interface is held together by a wide range of hydrophobic interactions. A single-site mutation is not sufficient to disrupt the dimer formation, although some side chain conformational changes occur in the mutational region.
FIG. 6.
(A) Stereo diagram of the Cα backbone in the dimer interface area. E. coli FPPs (in lime), S. solfataricus HexPPs (in magenta), and T. maritima OPPs (in cyan) are superimposed. The 2Fo-Fc maps are contoured at the 1.0-σ level about the residues at the dimer interface. The model is shown as sticks in gray and gold for the two subunits of HexPPs and in cyan for OPPs. (B) Wild-type HexPPs. (C) W136E HexPPs mutant. (D) Wild-type OPPs. (E) F117E OPPs mutant. These diagrams were produced with PyMOL.
DISCUSSION
Several enzymes, including the crenarchaeal medium-chain S. solfataricus HexPPs (code XXXX), short-chain E. coli (code 1RQI) FPPs, and long-chain T. maritima OPPs (code 1V4E), are shown here to illustrate their similar structural features. However, a large amino acid occurring at varying locations along the active site cavity can block the chain elongation of a defined product. Y105 (the fifth amino acid prior to the first DDXXD motif), L164, and F132 are the predominant key amino acids in FPPs, HexPPs, and OPPs, respectively. The space of the active-site area constrained by the large amino acid is proportional to the chain length of the product. This molecular ruler mechanism has been proposed for several trans-PTases (7, 14, 25).
The size of a C30 product, if arranged linearly, is estimated to be 32 Å, but the distance from the first DDXXD to L164 is only about 11 Å. This can be explained by the fact that the C30-HexPP is flexible enough and can fold to adapt to the cavity of the active site. In addition, the fifth amino acid before the first DDXXD motif in HexPPs is A76, smaller than the corresponding residue Y79 in E. coli FPPs and F79 in S. solfataricus GGPPs. This provides a wider cavity for HexPPs than those in E. coli FPPs and S. solfataricus GGPPs. In conclusion, chain length determination cannot be accurately predicted simply through the active-site cavity depth alone. The size of the amino acid near the active site is also significant in determining the product chain length.
It is interesting that only medium-chain (C30 and C35) trans-PTases contain either homodimers or heterodimers in different species. From the above description, the homodimer of HexPPs from S. solfataricus is not held as tightly as short- and long-chain trans-PTases. The sequence of each monomer is homologous to component B of the heterodimer for other medium-chain trans-PTases, e.g., HexPPs of Micrococcus luteus B-P 26 and HepPPs of Bacillus subtilis (Fig. 1). However, component A of the heterodimer does not show sequence homology to any other protein. From the structure presented here, we identified the characteristic interactions at the dimer interface in the homodimeric HexPPs. This part of the sequence, especially in helix F, is least conserved in component B of the heterodimeric synthases, possibly rendering them unable to form a homodimer.
The conformations of the two subunits of S. solfataricus HexPPs are nearly identical, except in the αD-αE loop, which is relevant to the catalytic function of HexPPs (Fig. S1). The loop may mediate the interaction of IPP with GGPP, necessary to initiate the condensation reaction and to serve as a hinge to control the substrate binding and product release. In addition, the B-factor distribution provides information on the protein dynamics, flexibility, and stability. Therefore, it is tempting to propose that monomer A with an inward αD-αE loop represents the substrate-free conformation because of its similar conformation to the corresponding regions in substrate-free OPPs (Fig. S3) (10). Monomer B may correspond to the product-released conformation with a flexible region (helices H to K) and two outward loops. It is unlikely that a rigid conformation of HexPPs releases the final product, since there is no exit hole through the bottom for the product. The outward loop in monomer B provides a putative conformation for product release.
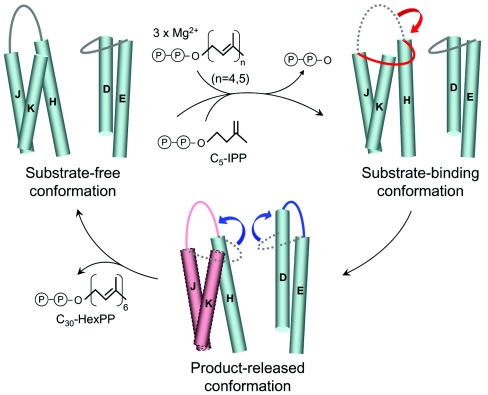
We conclude here with a three-conformer switch model (Fig. 7) based on the two conformers observed in HexPPs and FPPs-substrate complex (Fig. S1 and S3). The monomer A with the inward αD-αE loop represents a substrate-free conformation. As the substrates bind, the αH-αJ loop moves inward toward the active site to facilitate catalysis involving with some positive-charged amino acids (R90, R91), analogous to the FPPs-substrate complex. When the product chain length reaches C30, the αD-αE and αH-αJ loops at the top of the active site moves away from the closed position to the open position and the flexible region (αI, αJ, αK) moves outward for product release. The open conformation will also facilitate binding of the next substrate molecule. According to this catalysis model, HexPPs can switch among the three conformations with its flexibility in certain regions of the enzyme.
FIG. 7.
A three-conformer switch model of HexPPs catalysis. As described in the text, three conformations of HexPPs are represented by the substrate-free conformation (left), the substrate-binding conformation (right), and the product-released conformation (middle) in schematic cylindrical diagram. Substrate binding and product release might trigger the movement of the αD-αE and αH-αJ loops inward and outward of the active site. The flexible region (helices H to K) with high B factor observed in monomer B of HexPPs is highlighted in red.
Supplementary Material
Acknowledgments
We thank S.-S. Chern and Y.-S. Cheng for technical assistance.
This work was supported by grants from Academia Sinica and the National Science Council (NSC91-3112-P-001-019-Y) to A.H.J.W. The synchrotron data collections were conducted using the Biological Crystallography Facilities (Beamline BL17B2 at NSSRC in HsinChu and Taiwan Beamline BL12B2 at SPring-8), supported by the National Science Council (NSC).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bahadur, R. P., P. Chakrabarti, F. Rodier, and J. Janin. 2003. Dissecting subunit interfaces in homodimeric proteins. Proteins 53:708-719. [DOI] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rosa, M., S. De Rosa, A. Gambacorta, and L. Minale. 1977. Caldariellaquinone, a unique benzo(b)thiophen-4,7-quinone from Caldariella acidophila, an extremely thermophilic and acidophilic bacterium. J. Chem. Soc. Perkin Trans. I 1977:653-657. [DOI] [PubMed] [Google Scholar]
- 4.Fujii, H., T. Koyama, and K. Ogura. 1982. Efficient enzymatic hydrolysis of polyprenyl pyrophosphates. Biochim. Biophys. Acta 712:716-718. [PubMed] [Google Scholar]
- 5.Fujii, H., T. Koyama, and K. Ogura. 1983. Essential protein factors for polyprenyl pyrophosphate synthetases. Separation of heptaprenyl pyrophosphate synthetase into two components. FEBS Lett. 161:257-260. [DOI] [PubMed] [Google Scholar]
- 6.Guo, R. T., C. J. Kuo, C. C. Chou, T. P. Ko, H. L. Shr, P. H. Liang, and A. H. Wang. 2004. Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J. Biol. Chem. 279:4903-4912. [DOI] [PubMed] [Google Scholar]
- 7.Guo, R. T., C. J. Kuo, T. P. Ko, C. C. Chou, P. H. Liang, and A. H. Wang. 2004. A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products. Biochemistry 43:7678-7686. [DOI] [PubMed] [Google Scholar]
- 8.Hemmi, H., S. Ikejiri, S. Yamashita, and T. Nishino. 2002. Novel medium-chain prenyl diphosphate synthase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 184:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 10.Hosfield, D. J., Y. Zhang, D. R. Dougan, A. Broun, L. W. Tari, R. V. Swanson, and J. Finn. 2004. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J. Biol. Chem. 279:8526-8529. [DOI] [PubMed] [Google Scholar]
- 11.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koyama, T. 1999. Molecular analysis of prenyl chain elongating enzymes. Biosci. Biotechnol. Biochem. 63:1671-1676. [DOI] [PubMed] [Google Scholar]
- 13.Liang, P. H., T. P. Ko, and A. H. Wang. 2002. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269:3339-3354. [DOI] [PubMed] [Google Scholar]
- 14.Long, S. B., P. J. Casey, and L. S. Beese. 1998. Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Biochemistry 37:9612-9618. [DOI] [PubMed] [Google Scholar]
- 15.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 16.Ogura, K., and T. Koyama. 1998. Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98:1263-1276. [DOI] [PubMed] [Google Scholar]
- 17.Ogura, K., T. Koyama, and H. Sagami. 1997. Polyprenyl diphosphate synthases. Subcell. Biochem. 28:57-87. [DOI] [PubMed] [Google Scholar]
- 18.Ohnuma, S., K. Hirooka, H. Hemmi, C. Ishida, C. Ohto, and T. Nishino. 1996. Conversion of product specificity of archaebacterial geranylgeranyl-diphosphate synthase. Identification of essential amino acid residues for chain length determination of prenyltransferase reaction. J. Biol. Chem. 271:18831-18837. [DOI] [PubMed] [Google Scholar]
- 19.Ohnuma, S., K. Narita, T. Nakazawa, C. Ishida, Y. Takeuchi, C. Ohto, and T. Nishino. 1996. A role of the amino acid residue located on the fifth position before the first aspartate-rich motif of farnesyl diphosphate synthase on determination of the final product. J. Biol. Chem. 271:30748-30754. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, vol. 276. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 21.Pan, J. J., T. H. Kuo, Y. K. Chen, L. W. Yang, and P. H. Liang. 2002. Insight into the activation mechanism of Escherichia coli octaprenyl pyrophosphate synthase derived from pre-steady-state kinetic analysis. Biochim. Biophysica. Acta 1594:64-73. [DOI] [PubMed] [Google Scholar]
- 22.Poulter, C. D. 1974. Model studies in terpene biosynthesis. J. Agric. Food Chem. 22:167-173. [DOI] [PubMed] [Google Scholar]
- 23.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi, I., K. Ogura, and S. Seto. 1980. Heptaprenyl pyrophosphate synthetase from Bacillus subtilis. J. Biol. Chem. 255:4539-4543. [PubMed] [Google Scholar]
- 25.Tarshis, L. C., P. J. Proteau, B. A. Kellogg, J. C. Sacchettini, and C. D. Poulter. 1996. Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. USA 93:15018-15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D. Biol. Crystallogr. 56(Part 8):965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 55(Part 4):849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida, I., T. Koyama, and K. Ogura. 1987. Dynamic interaction between components of hexaprenyl diphosphate synthase from Micrococcus luteus BP-26. Biochemistry 26:6840-6845. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, I., T. Koyama, and K. Ogura. 1989. Formation of a stable and catalytically active complex of the two essential components of hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26. Biochem. Biophys. Res. Commun. 160:448-452. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, D., and R. H. White. 1989. Biosynthesis of caldariellaquinone in Sulfolobus spp. J. Bacteriol. 171:6610-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.