Abstract
The type III secretion system (T3SS) is an important virulence factor used by several gram-negative bacteria to deliver effector proteins which subvert host cellular processes. Enterohemorrhagic Escherichia coli O157 has a well-defined T3SS involved in attachment and effacement (ETT1) and critical for virulence. A gene cluster potentially encoding an additional T3SS (ETT2), which resembles the SPI-1 system in Salmonella enterica, was found in its genome sequence. The ETT2 gene cluster has since been found in many E. coli strains, but its in vivo role is not known. Many of the ETT2 gene clusters carry mutations and deletions, raising the possibility that they are not functional. Here we show the existence in septicemic E. coli strains of an ETT2 gene cluster, ETT2sepsis, which, although degenerate, contributes to pathogenesis. ETT2sepsis has several premature stop codons and a large (5 kb) deletion, which is conserved in 11 E. coli strains from cases of septicemia and newborn meningitis. A null mutant constructed to remove genes coding for the putative inner membrane ring of the secretion complex exhibited significantly reduced virulence. These results are the first demonstration of the importance of ETT2 for pathogenesis.
Type III secretion systems (T3SSs) are utilized by gram-negative pathogenic bacteria to deliver target-specific effector proteins into eukaryotic cells (14). This energy-requiring system is composed of more than 20 largely conserved proteins, which form a secretion apparatus for direct transfer of effector proteins from bacterial cytoplasm into the host cell or the medium. These effectors interfere with specific cellular processes and facilitate bacterial survival and replication. Typical features of the T3SS are the lack of an obvious secretion signal in the delivered proteins and the requirement for binding of a chaperone to the substrate before secretion (11, 16).
Previously characterized T3SSs in members of the family Enterobacteriaceae are composed of connected inner and outer membrane rings and a needle, through which the effector proteins are most likely passed into the host cytosol. Proteins translocated by this system promote critical processes such as bacterial internalization by mammalian cells in Salmonella and Shigella spp. (34, 36), induction of macrophage apoptosis in Salmonella, Shigella, and Yersinia spp. (24, 27), and creation of attachment and effacement lesions in enteropathogenic and enterohemorrhagic Escherichia coli (18).
In enteropathogenic and enterohemorrhagic E. coli, a chromosomal pathogenicity island (PAI) called the locus of enterocyte effacement contains genes encoding the components of T3SS which are involved in the formation of attaching and effacing lesions (18).
The published sequences of the enterohemorrhagic E. coli O157:H7 genome revealed the presence of an additional putative T3SS (13, 30), which has not been previously described. This type III secretion system was termed ETT2, for E. coli type III secretion system 2 (13), to distinguish it from the locus of enterocyte effacement-encoded system now called ETT1 (E.coli type III secretion system 1). ETT2 is homologous to the Salmonella T3SS located on Salmonella pathogenicity island 1 (SPI-1). Subsequent studies have shown that, far from being peculiar to the O157:H7 genome, the ETT2 gene cluster is present (in whole or part) in many E. coli strains (12, 25), and parts of the ETT2 cluster are found in strains that have no obvious link to virulence, such as the laboratory strain K-12 (31). However, in the majority of strains, including O157:H7, the ETT2 gene cluster appears to be degenerate, suffering mutational attrition in the form of point mutations, gene deletions, and disruption by insertion sequences that are almost certain to render it incapable of producing a functional type III secretion system (31). The only apparently intact ETT2 gene cluster occurs in the genome of enteroaggregative E. coli O42, but there is no experimental evidence to suggest that even this ETT2 is expressed and functional as a secretion system (31). ETT2 would thus appear in most cases to represent a molecular example of a Darwinian “atrophied or aborted organ” (8).
Here we present evidence that not all ETT2 gene clusters can be dismissed as “baggage of history.” We describe a degenerate form of ETT2, present in invasive E. coli strains, which we refer to as ETT2sepsis. Sequence comparison to the better-characterized type III secretion systems of other pathogens indicates that ETT2sepsis has a large deletion and premature stop codons in several genes, which probably abolish the functions of the ETT2 “needle complex;” ATPase, and several other structural proteins. Nevertheless, we show that removal of the putative genes for inner membrane ring formation results in reduction in in vivo host mortality, indicating that ETT2sepsis, although degenerate, contributes to pathogenicity. This deletion does not affect growth at a broad range of conditions. Yet the ETT2sepsis mutant differs from the wild type in pellicle formation but not detectably in its secreted proteome, providing evidence that the biological role of ETT2sepsis may not involve classic secretion of effectors.
MATERIALS AND METHODS
E. coli strains and media.
E. coli serotype O78 strains from septicemic animals were isolated from spleen or bone marrow of sheep (strains 6 and 68) or chickens (strains 783, 785, 787, 788, 789, 790, 792, 793, and 794). Strains from newborn meningitis were isolated from spinal fluid (strains 285, 286, and 287). E. coli serotype O2 strains (1772, MN, Y, and OH) were isolated from bone marrow of septicemic chickens. As a nonpathogenic reference for molecular and in vivo studies we used K-12 strain MG1655. Bacteria were routinely grown in LB medium (Bactlab Diagnostics, Israel). As a minimal medium, Davis and Mingioli medium was used (10). LB medium containing bile salts no. 3 (Bactlab Diagnostics, Israel) was prepared as described (5). For the multicellular behavior assay, bacteria were grown in SOBG medium for 3 days as described (33).
Construction of a cosmid genomic library of E. coli serotype O78.
Unless otherwise stated, a septicemic isolate of E. coli serotype O78, strain 789 (3, 32), was used for all procedures. Chromosomal DNA from this strain was partially digested with Sau3AI, size fractionated, and treated with calf intestinal phosphatase. DNA fragments of 35 to 40 kb were ligated to SUPERCOS-1 (Stratagene, La Jolla, CA) cosmid DNA, according to the manufacturer's instructions. The ligated DNA was packaged into lambda particles by the in vitro DNA packaging system Gigapack II (Stratagene, La Jolla, CA) and used to transfect competent E. coli XL1-Blue MR cells.
PCR and sequencing.
Amplifications were carried out in a total volume of 50 μl using either cosmid or genomic DNA as the template, each deoxynucleoside triphosphate at a concentration of 0.25 mM, 10 pmol of each primer, 5 μl of 10-fold PCR ExTaq buffer (TaKaRa) and 2.5 units of ExTaq DNA polymerase (TaKaRa). Oligonucleotides used for sequencing reactions were based on the sequence of the Sakai genome (accession number NC_002695). Reaction conditions were 5 min denaturation at 94°C, 30 cycles of 40 seconds of denaturation at 94°C, 45 seconds of annealing at 56°C, and extension at 72°C for 60 to 90 seconds, and a final additional 5 min at 72°C. PCR products were purified for sequencing using the ExoSAP-IT PCR cleanup kit (US Biochemicals). Sequencing was performed on both DNA strands, using the ABI PRISM 3100 automated sequencer. The DNA sequence of the E. coli O78 strain 789 ETT2 genes was submitted to GenBank (accession number DQ077151).
Isolation of a cosmid clone containing the T3SS.
About 10,000 colonies of E.coli strain XL1-Blue MR harboring the cosmid genomic library of E. coli strain 789 were screened by colony hybridization using the digoxigenin wash-and-blot kit (Roche Diagnostics) following the manufacturer's instructions by a specific probe directed against the epaO-eivJ region. The epaO-eivJ-specific probe was amplified from E. coli strain 789 with the primers epaOF (5′ GCGCGCCATTTACACGTATCTCTA 3′) and eivJR (5′ ATGCCAATGTGCGCGAAAATGAAT 3′), and labeled by digoxigenin-11-dUTP (Roche Diagnostics). Colonies were grown on a 134-mm plate and transferred directly to a positively charged nylon membrane. The membrane was denatured in a 0.5 M NaOH, 1.5 M NaCl solution, neutralized by Tris buffer (pH 7.4), and equilibrated by washing in 2× SSC solution (0.3 M NaCl, 0.03 M sodium citrate). After UV cross-linking, the cellular debris was removed using 1.4 mg ml−1 proteinase K solution.
The membrane was prehybridized at 42°C using a 50% formamide solution, and subsequently hybridized with the denatured epaO-eivJ digoxigenin-labeled probe. Positive colonies were verified for the presence of the full T3SS region by another set of PCR amplifications using primers 3716F and 3718R (Table 1) from the eprKJI region (upstream) and primers 3736F (5′ AGGGCTTGGCGCTTCATGCA 3′) and 3737R (5′ CGGGGGCAGCAACAGATAAAAGT 3′) from the ECs3736 region (downstream).
TABLE 1.
Oligonucleotides and PCR conditions used for RT-PCRs
| Primer designation | Sequence | Annealing temp (°C) | Elongation time (s) |
|---|---|---|---|
| 3716F | 5′ GCCGCACAACAGCAGGATAAAC 3′ | 59 | 30 |
| 3718R | 5′ TCCGACCCGAAATTCCTTGC 3′ | ||
| 3722F | 5′ CGGGGCTTGCGTAAACGATAGTA 3′ | 63 | 50 |
| 3725R | 5′ TCGCGATGGACGGTTGGACAATGT 3′ | ||
| 3720F | 5′ ATGCAAGTCTTTTCCAGTGATGTC 3′ | 52 | 30 |
| 3720R | 5′ TCAACTTTCTCTTACGCAAGATTG 3′ |
Construction of eprHIJK null mutant.
The eprHIJK null mutant was constructed as described (9). Briefly, competent wild-type 789 bacteria were transformed with plasmid pKD46 (9). Transformants initially selected on LB containing 300 μg ml−1 ampicillin were grown in ampicillin-containing LB medium, induced by arabinose, made competent for electroporation, and stored at −70°C until used. Linear PCR product was made on the template of kanamycin resistance cassette flanked by FLP recombinase target (FRT) sequences from the pKD4 plasmid (9), using primers pKD4P1 (5′ TTTGGTCATGATATTTCCGTCTTGTTACACTACTCTGTGTAGGCT GGAGCTGCTTC 3′) and pKD4P2 (5′ CAAGCTTACTCGTTGAGGATGAATATAACTAATTGGACATATGAATATCCTCCTTA 3′), containing 36 nucleotides from the sequences homologous to the eprHIJK region in 789. Kanamycin-resistant recombinants were screened by means of colony PCR, using primers k2 and kt (9). The pKD46 plasmid was cured by growth on LB at 42°C. The resulting bacteria were transformed with pCP20 (9) and grown on LB at 42°C to promote both FRT-specific recombination and curing of the plasmid. The final eprHIJK-deleted strain was verified by sequencing.
Construction of an eprHIJK complementation plasmid.
Primers P1add (5′ TCATCATAATTTGGTCATGATATTTCCGTC 3′) and P2add (5′ GTGTATTATTCAAGCTTACTCGTTGAGGAT3′) were used to PCR amplify the eprHIJK region of strain 789. The resulting PCR fragment was ligated into the pGEM-T Easy vector (Promega). A single colony, sequence verified for the full eprHIJK operon, was used for preparation of the complementation plasmid pEPR using the plasmid miniprep kit (QIAGEN).
Two step RT-PCR.
RNA from 400 μl of each bacterial culture (optical density at 600 nm of about 1.25) was stabilized using the RNA Protect reagent (QIAGEN) according to the manufacturer's instructions. Total RNA was isolated from the wild-type strain 789 and from the eprHIJK null mutant, using the RNeasy minikit for total RNA isolation (QIAGEN). The eluted RNA was subjected to DNase I digestion using the RQ1 RNase-free DNase (Promega) according to the manufacturer's instructions, to eliminate the possibility of DNA contamination. RNA cleanup was performed according to the RNA cleanup protocol of the RNeasy minikit (QIAGEN). As a negative control, an aliquot of the prepared RNA was digested by RNase A (Sigma) and used as a template for further reactions.
For the eprHIJK region analysis, the antisense primer 3716F (Table 1) was added to the resulting RNA to allow reverse transcription using Expand reverse transcriptase (Roche) according to the manufacturer's instructions. DNA obtained from reverse transcription was used as a template for PCR, using ExTaq DNA polymerase (TaKaRa) and primers 3716F and 3718R (Table 1). For the epaPQR region, the antisense primer was 3722F, and the resulting cDNA was subjected to a PCR with primers 3722F and 3725R (Table 1). 789 genomic DNA served as a positive control, and RNA not treated with reverse transcriptase as a negative control.
A similar reaction was performed for analysis of ECs3720 (ORF1) gene expression, using the antisense primer 3720R for cDNA formation and primers 3720F and 3720R (Table 1) for amplification. The positive and negative controls were as described above.
In vivo virulence assays.
Assessment of virulence was performed in 1-day-old chicks as previously described (32). Briefly, overnight bacterial cultures were diluted in saline, and 103 to 104 bacteria in a volume of 100 μl were injected intraperitoneally. The chicks had free access to food and water during the experiment. Infection with E. coli K-12 served as a negative control. Mortality was monitored over a period of 8 days after inoculation. Dead chicks were examined by autopsy, and the septic bacteria were isolated from the spleen and bone marrow. Groups of at least four chicks were used for each determination and the significance of the results was determined by chi-square analysis and by the Wilcoxon nonparametric test.
RESULTS
Identification of a type III secretion system in a septicemic E. coli serotype O78.
Subtractive hybridization between the highly virulent avian septicemic 789 strain of serotype O78 and a nonpathogenic K-12 strain, MG1655, revealed a 740-bp DNA fragment (O78-163) with high sequence similarity to the eivC gene (26). This gene belongs to the non-locus of enterocyte effacement T3SS, identified in E. coli serotype O157:H7 and termed ETT2 (13, 30). The eivC gene product is 63% identical to the product of Salmonella enterica serovar Typhimurium invC (accession number NP_461815), which encodes the ATPase of the secretion complex.
To obtain more of the O78 ETT2 gene cluster, a cosmid library of genomic DNA from strain 789 was screened with a digoxigenin-labeled amplicon (as described in Materials and Methods), spanning the ETT2 genes epaO and eivJ (Fig. 1A). A single clone that reacted with the probe and also gave PCR products for genes upstream and downstream (eprK-eprI and ECs3736, respectively) in the ETT2 cluster was chosen for further analysis. It should be noted that subtractive hybridization and subsequent PCRs failed to produce evidence for the existence of other T3SSs in our bacteria.
FIG.1.
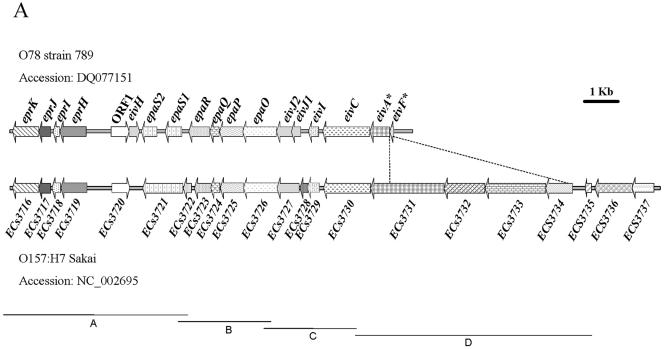
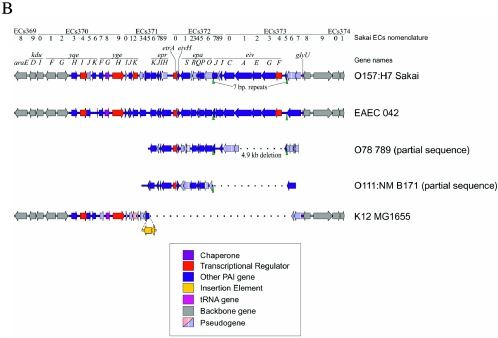
Schematic representation of the ETT2sepsis genes in a septicemic E. coli O78 strain 789 in comparison to other T3SS clusters. (A) The locations of the open reading frames (ORFs) are shown according to the nucleotide sequence prediction. Homologous reading frames have the same fill type. The large deletion (marked by a dotted line) spans the fully deleted genes eivE and eivG, resulting in the truncated genes eivA and eivF (marked with *). The gene eivH, found in O78 strain 789, is absent from the ETT2 cluster of O157:H7. The gene epaS, which is a single ORF in ETT2 cluster of O157:H7, is split into two ORFs, epaS1 and epaS2, in O78 strain 789. On the other hand, the two ORFs epaR1 and epaR2 from O157:H7 are represented by a single ORF, epaR, in O78 strain 789. In addition, premature stop codons also appear in the open reading frames of the genes eprJ, eprI, and eivC of strain 789. (B) SPI-1-like T3SSs in several enteric bacteria.
ETT2 of septicemic strains underwent evolutionary degeneration.
PCR was used to detect the ETT2 cluster in other invasive E. coli strains from the O78 or O2 serotypes. Four primer pairs: 3715F-3722R, 3721sF-3727R, 3726F-3730R, and 3730F-3735R, were used to amplify regions A, B, C, and D, respectively (Fig. 1A and Table 2). All four regions were detected in 11 of 18 strains examined: three of three O78 strains causing human newborn meningitis (strains 285, 286, and 287), six of nine O78 strains causing avian colisepticemia (785, 787, 788, 789, 790, and 793) and two of four O2 strains causing avian colisepticemia (1772 and Y). Two O78 sheep colisepticemia strains (6 and 68) and five avian colisepticemia strains (O78 strains 783, 792, and 794, and O2 strains MN and OH) were negative. These findings are in broad agreement with our previous multilocus sequence typing studies analyses on the clonal structure of invasive strains from the O78 and O2 serotypes (1), but are in contrast to the absence of the ETT2 gene cluster from the “classic” invasive human neonatal meningitis strain RS218 (O18:K1:H7) (12).
TABLE 2.
Primers used to detect ETT2 genes in E. coli strains
| Primer designation | Sequence | Target genes in O157:H7 | Region designation | Resulting product length (kb) |
|---|---|---|---|---|
| 3715F | 5′ GCCGGGTTTGTATCGTGCCTTTTC 3′ | eprK-epaS | A | 5.4 |
| 3722R | 5′ TGCCGTTATGCTGGGTGGTGAG 3′ | |||
| 3721sF | 5′ CCGATTTCCTTTGCATATTTCC 3′ | epaR-epaO | B | 2.7 |
| 3727R | 5′ ATGCCAATGTGCGCGAAAATGAAT 3′ | |||
| 3726F | 5′ GCGCGCCATTTACACGTATCTCTA 3′ | eivJ-eivI | C | 2.7 |
| 3730R | 5′ TGGCGACAACCGTGGCAGAGTATT 3′ | |||
| 3730F | 5′ CCGCGTCATTGAGTCGATAAAAAG 3′ | eivC-eivF | D | 1.8a |
| 3735R | 5′ GGCCGCTTAGTCTCATTCCTTCC 3′ |
This is the region containing the deletion, as PCR with the same primers on a template of the O157:H7 genome is expected to result in a product of about 6.8 kb.
The ETT2 gene cluster of E. coli O78 strain 789, as represented by the cosmid library clone described above, was sequenced and found to be over 96% identical (at the DNA level) to the sequences from O157:H7 (ETT2O157). However, this septicemia-associated system, which we refer to as ETT2sepsis, has a 5-kilobase deletion relative to the ETT2O157 sequence (accession number NC_002695) (Fig. 1). Interestingly, PCR with primers from both ends of the 5-kb gap (3730F and 3735R, region D in Fig. 1A) showed that this fragment loss is conserved in all 11 invasive ETT2-positive E. coli serogroup O78 or O2 strains from our collection (data not shown). A similar deletion was reported in three related strains from the B1 subdivision of the E. coli Reference Collection (ECOR): ECOR-70 (O78:NM, isolated in the United States from a healthy gorilla), ECOR-71 (O78:NM, isolated from a case of asymptomatic bacteruria in Sweden) and ECOR-72 (O144:H8, isolated from a case of pyelonephritis in Sweden) (31).
The 5-kilobase deletion in ETT2sepsis completely eliminates two genes, eivE/ECs3732 and eivG/ECs3733 (nomenclature as in the Sakai genome sequence, accession number NC_002695), and truncates two more, eivA/ECs3731 and eivF/ECs3734. The proteins encoded by these genes are, by analogy with the well-characterized Salmonella SPI-1 T3SS, likely to play important roles in secretion: InvA is an inner membrane-associated protein essential for type III secretion in Salmonella spp., with a homologue (FlhA) in flagellar systems; InvE is required for translocation of effector proteins (20); InvG forms the outer membrane ring (16) and is similar to a type II secretion system protein; InvF is a transcriptional regulator.
Comparisons with ETT2 sequences from other E. coli strains and with the Salmonella SPI-1 revealed additional mutations leading to truncation of several other important genes (Fig. 1): the homologues of prgI, prgJ, and invJ (eprI, eprJ, and eivJ, respectively), presumably required for needle structure formation; the E. coli homologue of invC (eivC), the putative ATPase required for energizing transport; and epaS, the homologue of spaS, a putative component of the secretion apparatus. Conversely, the epaR gene, which in O157:H7 is disrupted by a premature stop codon, is intact in ETT2sepsis, implying that the O157:H7 and O78 ETT2 clusters have taken separate routes to evolutionary degeneration.
Construction of a null mutant defective in ETT2sepsis.
Although sequence analyses suggest much of the ETT2sepsis system is no longer functional, the genes encoding the proteins predicted to form the inner membrane ring of the T3SS (EprH and EprK) are intact. The homologues of these proteins in the SPI-1 system (PrgH and PrgK, respectively) have been shown to oligomerize into ring-shaped structures even in the absence of other T3SS components (17, 21). We thus speculated that such structures might form in ETT2sepsis and contribute to the highly virulent characteristics of the strain. The eprH and eprK genes flank the needle component genes eprIJ, which are inactivated by frameshifts. We therefore created a null mutant of the eprHIJK cluster to eliminate the ability to form the inner membrane ring of the complex. The eprHIJK cluster of the wild-type strain 789 was replaced by a kanamycin resistance cassette flanked with FRT sequences, which was then removed using the FLP recombinase (9). The construction of a null mutant was confirmed by Southern blotting and by sequencing the locus.
Genes in ETT2sepsis are expressed.
Given the mutational attrition suffered by the ETT2sepsis cluster, it was necessary to determine if the genes in the cluster are expressed. Reverse transcription (RT)-PCR was used to detect transcription of the epr and epa operons (Fig. 2). Both operons were transcribed, consistent with speculation that the system might be physiologically functional. As expected, there were no RT-PCR products of the eprKIJ genes in the null mutant (Fig. 2A), confirming the deletion of the eprHIJK cluster.
FIG. 2.
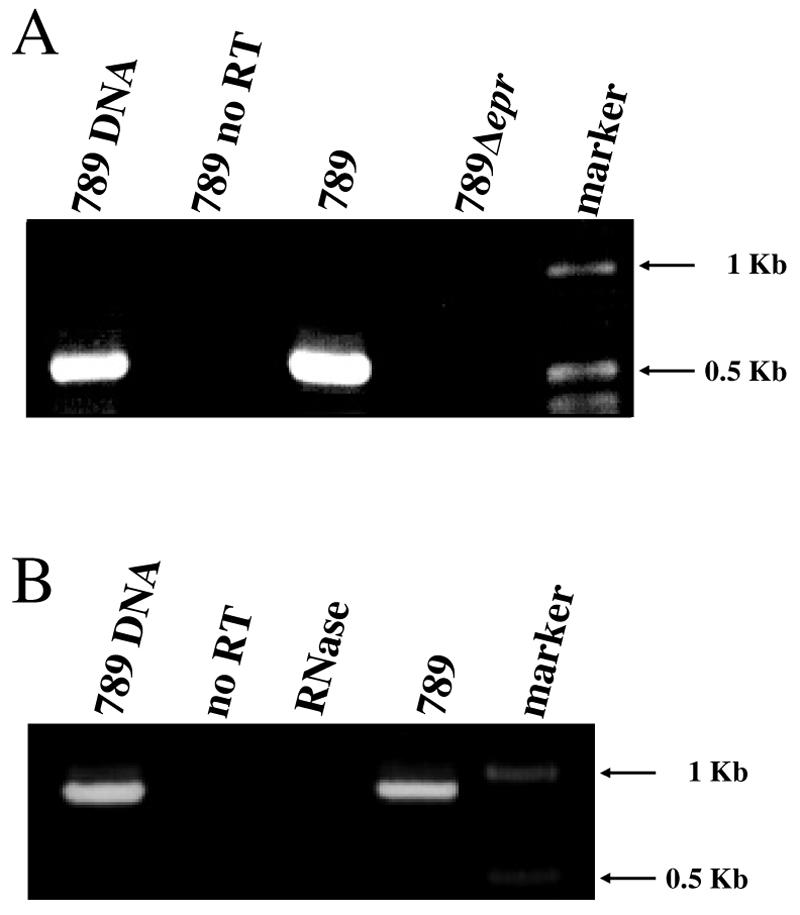
Transcription of the ETT2sepsis epr and epa operons. Agarose gel electrophoresis of RT-PCR products amplifying the epr region of E. coli 789 was carried out as described in the text, using primers 3716F and 3718R (A). Primers 3722F and 3725R were used to amplify the epa region (B). As a negative control, the RT reaction was performed without the addition of the reverse transcriptase enzyme (no RT). Marker, 1-kb DNA ladder (Epicenter); RNase, RT reaction performed on a template of RNase A-treated RNA; 789 DNA, product obtained from amplification of genomic DNA.
An epr null mutation of ETT2sepsis does not seem to affect the secreted proteome.
Secretion by T3SSs is generally induced by contact with a host cell, but several laboratory conditions can trigger secretion of the effectors into the extracellular medium. Such conditions are growth of Yersinia spp. at 37°C in a medium containing low Ca2+ (15, 23), or Congo red for Shigella spp. (4, 29). Secretion of the SPI-1-encoded T3SS in Salmonella is induced upon a shift to a higher pH (7).
Because ETT2sepsis is similar to SPI-1, we determined the effect of a shift from pH 6 to pH 8 on the pattern of protein secretion by strain 789. When comparing the wild-type 789 strain to the epr null mutant following the pH shift, no unique secreted proteins could be reproducibly detected by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Furthermore, no secreted proteins unique to the wild-type 789 strain could be detected following growth in calcium-depleted medium or addition of Congo red dye (results not shown). Although it is theoretically possible that appropriate conditions for induction are still to be discovered for ETT2sepsis, these data, in conjunction with sequence analysis, do not support the assumption that the degenerate ETT2sepsis is involved in secretion of effector proteins.
epr null mutation of ETT2sepsis does not affect growth.
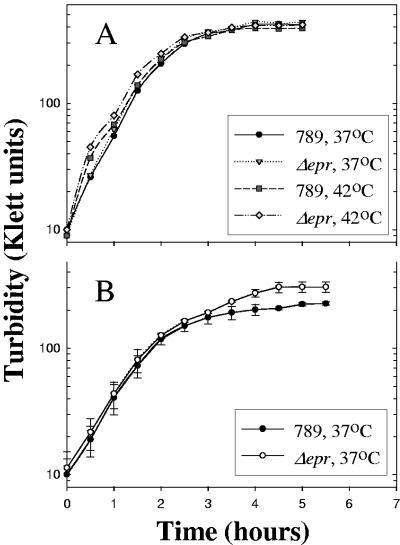
The growth of the epr-deleted mutant at 37°C or 42°C was similar to that of the wild type in LB medium (Fig. 3A) in Davis minimal medium and in serum (6), even when higher concentrations of serum were used (data not shown). It should be noted that strain 789 is very resistant to serum, as addition of 5 ml serum and even 9 ml serum to 1 ml bacterial culture (mid-logarithmic phase) did not inhibit growth. Several experiments were performed to test the effect of various detergents, and the only difference between the mutant and the wild type was detected in the presence of 2% bile salts. Under these conditions, growth of the epr deletion mutant (Fig. 3B) was somewhat better than that of the wild-type strain 789.
FIG. 3.
Growth of E. coli O78 strain 789 and its eprHIJK deletion mutant. Overnight cultures of strain 789 and its ΔeprHIJK mutant were diluted 1:100 and grown with aeration by shaking. A, LB medium; B, LB containing 2% bile salts no. 3. Shown is a summary of three experiments.
ETT2sepsis deletion affects multicellular behavior in vitro.
Although ETT2sepsis is degenerate, we demonstrated that it is transcribed and, therefore, tested the deletion mutant to determine if it had a phenotype. One possibility was related to multicellular behavior, as it is altered in Erwinia chrysanthemi type III secretion mutants, which do not form stable pellicles in SOBG medium at 36°C (33). We compared the wild type and the epr null mutant of 789 in SOBG medium at several growth temperatures. While the two cultures were similar at 25°C (Fig. 4A), a visible difference was found between the wild-type 789 and the ETT2sepsis deletion mutant cultures at 37°C (Fig. 4B) and at 42°C. Strong mixing of the mutant culture restored the homogenous turbidity, indicating that the phenotype was due to pellicle formation and not to cell lysis. Complementation of the mutant by pEPR, a plasmid encoding the full eprHIJK region, restored the phenotype of the wild-type 789 culture (Fig. 4B).
FIG. 4.
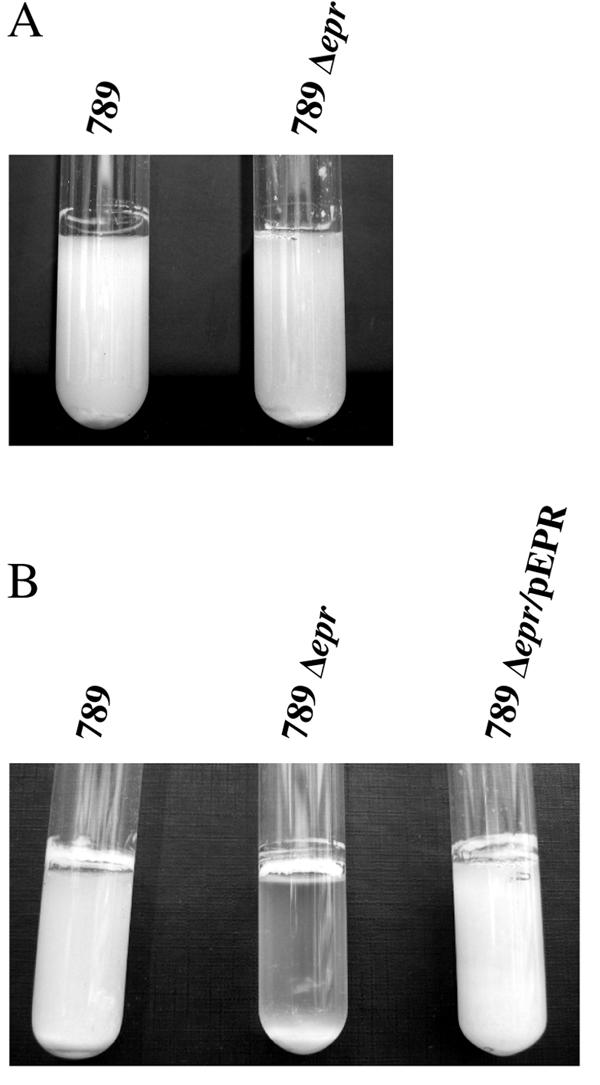
Comparison between the wild-type strain 789 and the eprHIJK null mutant phenotypes in vitro. SOBG medium was inoculated with about 106 bacteria. The cultures were grown for 3 days without aeration at 25°C (A) or at 37°C (B). 789Δepr/pEPR, complemented mutant.
ETT2sepsis is required for virulence.
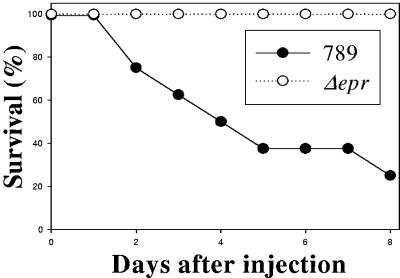
As strain 789 is from a case of septicemia, it was important to determine if the deletion of ETT2sepsis affects virulence. The wild-type strain and two independently isolated eprHIJK null mutants were injected intraperitoneally into 1-day-old chicks, which are a natural host for colisepticemia. Three concentrations of bacteria were tested, and the chicks were monitored for 8 days after infection. The results, summarized in Fig. 5, indicated that the mutant bacteria are highly attenuated compared to the wild type (P < 0.05). There was 75% mortality (six of eight) in the chicks injected with 103 CFU of the wild-type 789, but no killing was observed with the mutant at this concentration (0 of 16 chicks died). Furthermore, only one chick died following injection of a higher dose, 104 CFU, of the mutant bacteria (1 of 16), a concentration resulting in 100% killing by the wild type. Similar results were obtained with an independently isolated mutant, with no mortality following injection of either 103 or 104 CFU (zero of eight chicks dead). The reduced virulence phenotype of the null mutation could be partially complemented (four of nine chicks dead) by plasmid pEPR. These results demonstrate that although the eprHIJK genes are part of a degenerate system, they are important to virulence.
FIG. 5.
Virulence of eprHIJK null mutant of E. coli strain 789. Overnight cultures of the wild-type 789 strain and its eprHIJK null mutant were serially diluted in sterile saline, and 100 μl of the suspension containing 103 bacteria was injected intraperitoneally into 1-day-old chicks. The survival of the chicks was recorded for 8 days following the injection.
epr null mutation does not impair the expression of the adjacent ORF1, homologous to ECs3720.
One of the genes adjacent to the eprHIJK operon in strain 789 is designated ECs3720 in O157:H7 (accession number NC_002695). This gene encodes a regulator that has a profound effect on expression of several virulence-related genes outside of the ETT2 gene cluster in O157:H7 (35). To ensure that the attenuation of virulence in the eprHIJK null mutant of strain 789 is not a result of altered expression of the adjacent ECs3720-like gene, RT-PCR analysis of this gene was performed (Fig. 6). The results demonstrate that the gene is transcribed both in the wild-type 789 and in the eprHIJK null mutant, indicating that the decreased virulence of the mutant is due to the deletion of the eprHIJK operon rather than polar effects on the ECs3720 regulon.
FIG. 6.
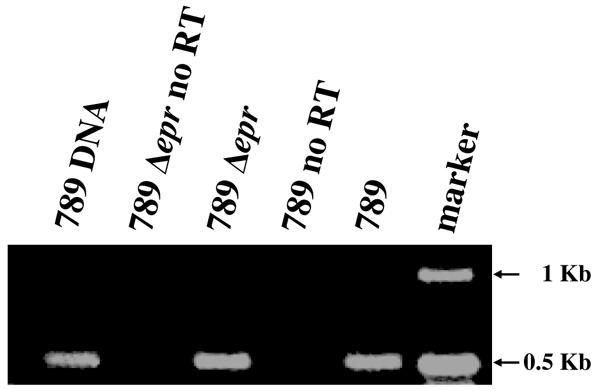
Transcription of ORF1, the homologue of the ECs3720 gene of E. coli O157:H7 Sakai. Agarose gel electrophoresis of RT-PCR products amplifying ORF1 of E. coli 789 and the eprHIJK null mutant was carried out as described in the text, using primers 3720F and 3720R. As a negative control, the RT reaction was performed without the addition of the reverse transcriptase enzyme (no RT). Marker, 1-kb DNA ladder (Epicenter); 789 DNA, product obtained from amplification of genomic DNA.
DISCUSSION
Type III secretion systems are important virulence mechanisms in bacteria (14). One of the most prominent findings from the elucidation of two E. coli O157:H7 genome sequences was the discovery of a second gene cluster potentially associated with type III secretion in addition to the well-characterized locus for enterocyte effacement (13, 30). Genes for this type III secretion system (ETT2) are present, in whole or part, in the majority of E. coli strains, including many commensal strains (12, 31). Here we extend the spectrum of E. coli strains containing the ETT2 gene cluster to include O78 and O2 strains causing sepsis and neonatal meningitis. However, even though the ETT2 gene cluster is widespread, its role has not been determined in any E. coli strain. In fact, so far as we are aware, this is the first report that ETT2 genes are even expressed.
The physical structure of the type III secretion apparatus, referred to as the needle complex, can be visualized by electron microscopy. The conserved morphology of the structures, revealed by analysis of Salmonella and Shigella T3SSs, includes a basal complex (a membrane-bound structure composed of inner membrane and outer membrane rings joined by a rod) and a hollow needle. Kimbrough and Miller (17) have dissected the SPI-1 needle complex, showing that the needle consists of PrgI and PrgJ, and that PrgK and PrgH form the inner membrane ring of the basal complex. A needleless prgI null mutant of the Salmonella T3SS was completely defective in its ability to invade epithelial cells (22).
Sequence analysis of the ETT2sepsis genes reveals premature stop codons in both genes predicted to encode the needle (eprI and eprJ) and deletion of the invG gene, which encodes a highly conserved component of the outer membrane ring. ETT2sepsis also lacks the gene for the cytoplasmic ATPase that energizes secretion (eivI) and some other conserved components of T3SSs (e.g., epaS). In addition, no activation of ETT2sepsis-specific secretion was observed when using inducers of T3SS known for other enterobacteria. Thus, it was surprising to find that, although degenerate, ETT2sepsis has a role in virulence. This role is demonstrated by the results of in vivo studies with 1-day-old chicks, where mortality was unpredictably greatly reduced by the deletion of the epr cluster.
Confirmation for a direct role of the cluster in virulence came from the complementation of the deletion in vivo by the addition of a plasmid containing the eprHIJK genes. The complementation restored virulence and lethality for chicks, even at the low bacterial concentration, although not to the same level as the wild type (45% versus 75% mortality, respectively, and no mortality with the mutant). The difference between the complemented mutant and the wild type, which was statistically insignificant, could be attributed to loss of the plasmid or its insert during the infection. This possibility is very likely, as it is difficult to maintain transformed plasmids in wild-type strain 789.
A possible explanation for the differences in virulence of the wild type and the eprHIJK mutant is that the deletion of membrane proteins causes a structural alteration. Such alterations occur, as phenotypic differences (pellicle formation) were found between the wild type and the mutant grown in SOBG. It should be noted that the visible differences between the wild type and the epr null mutant grown in SOBG medium were not paralleled by differences in the pattern of proteins secreted to the culture medium, suggesting that the effect of ETT2sepsis may originate from physical changes in the membrane properties rather than in secretion. The fact that these changes occurred at elevated temperatures but not at 25°C implies that the structural alteration in the membrane of the mutant could lead to a disadvantage in the host. Thus, the membrane structure itself could have a role in fitness within the host, in a way unrelated to secretion, either by modifying bacterial surface properties and affecting interaction with host cells and immune system, or by modulating bacterium-bacterium interactions, as recently observed for Erwinia chrysanthemi (33).
Alternatively, the structural change in the membrane could trigger a signal that activates expression of genes and/or secretion of proteins via another secretion system. In support of this idea, a TMHMM search (19) with the ETT2sepsis EprH sequence predicted that the protein possessed a cytoplasmic N-terminal domain (residues 1 to 144) related by using PSI-BLAST (2) to members of the YscD/EscD family of type III secretion proteins. The homology links the N-terminal cytoplasmic domain of EprH to the variant forkhead-associated domains commonly involved in reversible protein-protein interactions modulated by phosphorylation of threonine residues (28) and in signal transduction. The finding that the complex degenerate ETT2 mechanism improves survival in the host may explain the fact that it was conserved through evolution even after losing its secretion functions.
Although it was tempting to dismiss ETT2sepsis as nonfunctional baggage of history, the results presented here provide an important lesson against ignoring apparently degenerate gene clusters. We show that the degenerate and probably not secretion-competent ETT2sepsis cluster contributes to virulence in chicks. In this regard, the ETT2sepsis cluster might have powerful phenotypic effects that outlive extensive degeneration through mutational attrition. Indeed, as a second example of this phenomenon, it has been discovered that regulators encoded within the degenerate ETT2 gene cluster from E. coli O157:H7 can affect the expression of virulence-associated genes outside the cluster (35). However, from a broad evolutionary perspective, the role of the structural genes of ETT2sepsis is more striking, because it provides a rare example where a part of a complex can have alternative roles and therefore persists while the complex as a whole is no longer functional.
Acknowledgments
We thank Tslil Ophir for help with automated sequencing and Ran Rosen and Ayelet Shachar of the Miman Institute of Proteomics at Tel-Aviv University for their help. D.I. and U.G. thank the Constantiner Institute for Molecular Genetics.
This work was supported by the Manja and Morris Leigh Chair for Biophysics and Biotechnology, the Israel Center for Emerging Diseases, and European Community project COLIRISK.
REFERENCES
- 1.Adiri, R. S., U. Gophna, and E. Z. Ron. 2003. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 222:199-203. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babai, R., G. Blum-Oehler, B. E. Stern, J. Hacker, and E. Z. Ron. 1997. Virulence patterns from septicemic Escherichia coli O78 strains. FEMS Microbiol. Lett. 149:99-105. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidlack, J. E., and P. M. Silverman. 2004. An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J. Bacteriol. 186:5202-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daefler, S. 1999. Type III secretion by Salmonella typhimurium does not require contact with a eukaryotic host. Mol. Microbiol. 31:45-51. [DOI] [PubMed] [Google Scholar]
- 8.Darwin, C. 1859. The origin of species by means of natural selection. J.Murray, London, United Kingdom.
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 12.Hartleib, S., R. Prager, I. Hedenstrom, S. Lofdahl, and H. Tschape. 2003. Prevalence of the new, SPI1-like, pathogenicity island ETT2 among Escherichia coli. Int. J. Med. Microbiol. 292:487-493. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobi, C. A., A. Roggenkamp, A. Rakin, R. Zumbihl, L. Leitritz, and J. Heesemann. 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol. 30:865-882. [DOI] [PubMed] [Google Scholar]
- 16.Kimbrough, T. G., and S. I. Miller. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75-82. [DOI] [PubMed] [Google Scholar]
- 17.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 20.Kubori, T., and J. E. Galan. 2002. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J. Bacteriol. 184:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 22.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michiels, T., P. Wattiau, R. Brasseur, J. M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills, S. D., A. Boland, M. P. Sory, P. van der Smissen, C. Kerbourch, B. B. Finlay, and G. R. Cornelis. 1997. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc. Natl. Acad. Sci. USA 94:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki, J., W. Ba-Thein, T. Kumao, H. Akaza, and H. Hayashi. 2002. Identification of a type III secretion system in uropathogenic Escherichia coli. FEMS Microbiol. Lett. 212:221-228. [DOI] [PubMed] [Google Scholar]
- 26.Mokady, D., U. Gophna, and E. Z. Ron. 2005. Extensive gene diversity in septicemic Escherichia coli strains. J. Clin. Microbiol. 43:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monack, D. M., J. Mecsas, N. Ghori, and S. Falkow. 1997. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. USA 94:10385-10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallen, M., R. Chaudhuri, and A. Khan. 2002. Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol. 10:556-563. [DOI] [PubMed] [Google Scholar]
- 29.Parsot, C., R. Menard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 30.Perna, N. T., G. Plunkett 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 31.Ren, C. P., R. R. Chaudhuri, A. Fivian, C. M. Bailey, M. Antonio, W. M. Barnes, and M. J. Pallen. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186:3547-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ron, E. Z., Z. Yerushalmi, and M. W. Naveh. 1991. Adherence pili of E. coli O78 determine host and tissue specificity, p. 61-68. In E. Z. Ron and S. Rottem (ed.), Microbial surface components and toxins in relation to pathogenesis. Plenum Press, New York, N.Y.
- 33.Yap, M. N., C. H. Yang, J. D. Barak, C. E. Jahn, and A. O. Charkowski. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, S., E. Katayama, A. Kuwae, H. Mimuro, T. Suzuki, and C. Sasakawa. 2002. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 21:2923-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. USA 96:10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]