Abstract
The bacteriophage A2 major tail protein gene utilizes a −1 translational frameshift to generate two structural polypeptides. Frameshifting is promoted by a slippery sequence and an RNA pseudoknot located 3′ of the gene. The major head gene presents a similar recoding ability. A2 is the only phage described with two −1 frameshifts.
Programmed −1 frameshifting is generated through a backward displacement of the translating ribosome to the position −1, which results in the synthesis of two polypeptides, one that arises from translation of the gene in the 0 frame and another that is identical to it until the frameshifting position but whose sequence corresponds to the message encoded in the −1 frame from that point onwards (1). Two cis-acting elements are commonly necessary for −1 frameshifting. The first is a heptanucleotide sequence that allows tandem slippage of the tRNAs located at the functional A and P sites of the ribosome and repairing of their respective anticodon triplets with the codons that result in the −1 frame. The second is a structure that promotes ribosome pausing at the slippery sequence, such as a stem loop or a pseudoknot that starts in its 3′ vicinity (3, 5, 11) or a Shine-Dalgarno-like sequence located about 10 nucleotides 5′ of the slippery sequence (10, 15). The functional meaning of −1 frameshifting may be to produce two proteins in a defined ratio. In the case of viral genes the “frameshifted” product is usually essential for viability, both in retroviruses, where synthesis of a fixed proportion of gpGag-Pol is required for efficient packaging of reverse transcriptase (21), and in bacteriophages, for example, in lambda the fusion protein gpG-T is essential for tail assembly, even though it does not become part of the mature virion (12). However, in the case of cellular genes the frameshifted products may be dispensable, (15) although, as in the gamma subunit of DNA polymerase III of Escherichia coli (2), it may be important for the activity of the enzyme (19).
A2 is a temperate bacteriophage that belongs to the family Siphoviridae (8). The two major proteins of the capsid share their amino termini, which matched an internal sequence of orf5 in the phage genome (6). This suggested that orf5 gives rise to two polypeptides of different sizes, the smaller (gp5A) resulting from canonical translation of orf5 and the larger (gp5B) being generated by a −1 ribosomal frameshift at the penultimate codon of orf5 mRNA, resulting in a product that is 85 amino acids longer than gp5A. Frameshifting is dependent on a slippery region with the sequence CCCAAAA and on a stem loop that begins 9 nucleotides after the end of the slippery sequence. Both gp5A and gp5B appear to be essential for phage viability, because although lysogens harboring prophages that produce only one or the other protein become lysed upon induction with mitomycin C, no viable phage progeny are observed (7).
In this work, data are presented which indicate that −1 frameshifting during the phage A2 lytic cycle is not restricted to the mRNA for the major head proteins but also happens in the transcript for the major tail proteins. In addition, some requirements for this recoding event are studied.
Bacteriophage A2 was propagated and assayed on Lactobacillus casei ATCC 393 as previously described (7). Escherichia coli BL21(DE3)/pLysS was used in expression studies of the different versions of orf10 cloned into plasmid pET11a (22). Plasmid constructions, site-directed mutagenesis, and protein labeling and detection were performed as described previously (7).
Purification of gp10A and gp10B was achieved after overexpression of orf10 cloned into pET11a. Cell extracts were ultracentrifuged (100,000 × g for 90 min) and successively loaded onto Q-Sepharose, Mono Q, and Superdex 75 columns (Pharmacia). The purified proteins were digested with porcine trypsin (Promega), and the resulting peptides were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry, essentially as previously described (7).
To achieve overproduction of gp10A and gp10B in L. casei, orf10 with its 3′-adjacent region was placed under the nisin promoter (Pnis) using plasmids pEM117 and pEM110 and the procedure described previously was followed (14). The resulting plasmid, pEM110-orf10, was electroporated into an L. casei derivative that expressed the quorum-sensing system that promotes Pnis induction (9). Overexpression was obtained by addition of nisin to exponential cultures of this L. casei strain.
gp10-specific antibodies were obtained from rabbits, injected at 2-week intervals with 1 mg of pure gp10A emulsified 1:1 in incomplete Freund's adjuvant. For Western blotting of L. casei extracts, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to an Immobilon-P membrane (Millipore). The membrane was incubated with anti-gp10 rabbit immunoglobulin G (IgG) (1:1,000) and then with peroxidase-conjugated goat anti-rabbit IgG (1:5,000) (Jackson ImmunoResearch Laboratories). The Western blots were developed using the BM chemiluminescence blotting substrate (POD) (Roche Diagnostics).
The major tail protein gene of phage A2 gives rise to two polypeptides through −1 frameshifting.
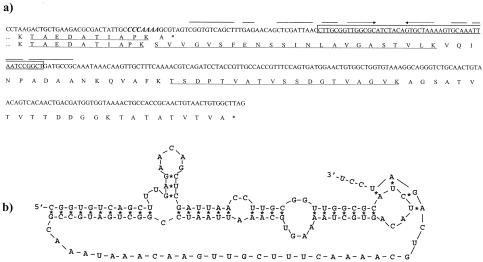
In silico analysis of the A2 genome revealed a heptanucleotide (CCCAAAA) at the end of the major tail protein gene (orf10) which was identical to the slippery sequence previously encountered in orf5 (7). This heptanucleotide was immediately followed by a putative H-type pseudoknot (ΔG = −31.9 kcal/mol), thus suggesting that a −1 frameshift might occur at the end of the orf10 transcript. This would give rise, in addition to the canonical 202 residues polypeptide, to a second one of 283 amino acids (Fig. 1a and b).
FIG. 1.
a) DNA and deduced protein sequences of the A2 genome region surrounding the 3′ end of the major tail protein gene (orf10); in the DNA, the slippery sequence is indicated in boldface and italics, and the first stem-loop is indicated with converging arrows. The 48-bp segment deleted from it is boxed. Relevant polypeptides obtained by tryptic digestion of gp10A and/or gp10B are underlined. b) Proposed secondary structure of the mRNA frameshift stimulatory element located downstream of the slippery sequence.
To determine whether these structures were functional, a DNA segment that comprises orf10 plus its 400-bp downstream stretch was cloned into pET11a and overexpressed. Two polypeptides of sizes compatible with that of the expected gp10 and with the product that would result from a −1 frameshift in the predicted position were observed (Fig. 2a, lane 1). Mass spectrometry of the tryptic peptides of these two polypeptides revealed that all those present in the smaller one (gp10A) were also present in the bigger one (gp10B), including one that would end at the lysine encoded by the second codon of the slippery sequence. In addition, gp10B generated two peptides of 2,279 and 1,792 Da, matching the expected masses of two tryptic peptides that would result from translation of the message downstream of orf10 when read in the −1 frame (Fig. 1a).
FIG. 2.
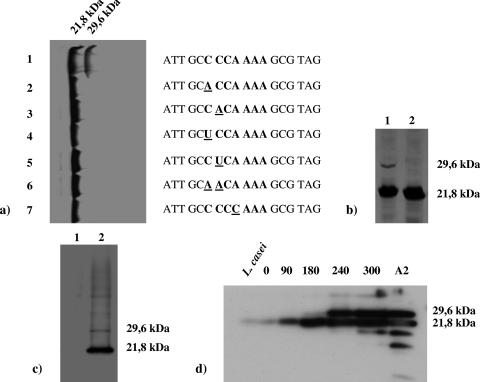
a) Effect on frameshifting of different point mutations (underlined) at the slippery sequence of orf10 (in boldface). b) Outcome of the deletion of the central part of the stem loop located downstream of the slippery sequence (lane 2); lane 1, control gene. orf10 (and its variants) were placed under the control of the P10 promoter of T7 and overexpressed in vivo in E. coli in the presence of a 35S-labeled amino acid mix and rifampin. c) Expression of orf10, cloned in pEM110 under the control of the nisin promoter, in L. casei (lane 2); lane 1, untransformed culture. d) Time course of accumulation of gp10A and gp10B in exponential cultures of an A2 lysogenic culture of L. casei induced with mitomycin C (0.5 μg/ml); the figures above the lanes indicate time postinduction in minutes, L. casei stands for an uninduced culture, and A2 corresponds to purified virions (the two bands below gp10A in this last lane are degradation products of this protein). The proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and autoradiographed (E. coli) or detected by Western blotting (L. casei).
To confirm the role of the slippery sequence, point mutations were introduced in different positions of the heptanucleotide and their effect on frameshifting was tested. Any mutations that induced a change in the dipeptide Pro-Lys encoded by the slippery sequence in both the 0 and −1 frames abolished gp10B (Fig. 2a, lanes 2 to 7), suggesting the need for compatibility of the anticodons of the tRNAs located at the A and P ribosomal sites with their complementary codons in both frames.
To test the functionality of the putative pseudoknot, a 48-bp deletion was generated that comprised most of its stem 1 and the intermediate loop (nomenclature used is as described in reference 16) while preserving the slippery sequence and the reading frame after the deleted DNA stretch (Fig. 1a). The clone harboring the mutant gene would be expected to generate gp10A (21.8 kDa) and a shorter form of gp10B (28.0 kDa instead of 29.6 kDa of the wild-type form), if the pseudoknot did not play a role in frameshift occurrence. However, only gp10A was visualized in the gels (Fig. 2b, lane 2), thus confirming the role of this secondary structure in frameshift occurrence.
To test whether frameshifting occurred in L. casei as well as in E. coli, orf10 with its adjacent 3′ sequence was cloned in a plasmid under the control of the nisin promoter. After induction of the system, two polypeptides were observed that reacted with gp10-specific antibodies and had masses compatible with the expected sizes of gp10A and gp10B (Fig. 2c, lane 2), indicating that the −1 frameshift signals were being recognized in L. casei.
Both gp10A and gp10B are late proteins that become incorporated into mature A2 virions.
To determine the production kinetics of both proteins, L. casei lysogenic exponential cultures were induced with mitomycin C, and samples collected during the lytic development of the phage were analyzed for their protein content by Western blotting (Fig. 2d). Expression of orf10 became evident at 90 min postinduction, but initially only gp10A was observed. Detection of gp10B was delayed until 240 min postinduction (under those conditions the cultures start to lyse at about 300 min postinduction). This might reflect a very different rate of biosynthesis for these proteins, the canonical form being very favored over the frameshifted product, which would reach a concentration detectable by the antibodies only after a long period of accumulation. Both gp10A and gp10B were present in the virions (Fig. 2d, lane A2), indicating that they contribute to the final structure of the mature particle.
Ribosomal −1 frameshifting appears to be quite common among genes involved in virion morphogenesis of tailed bacteriophages. It was first described for the gene that encodes the major head proteins of T3 and T7 (4) and later for generation of the G-T fusion proteins of lambda (12) and many other siphoviruses and myoviruses infecting both Eubacteria and Archaea (23). What makes A2 peculiar is that −1 frameshifting occurs upon translation of both the major head and tail genes and that the four resulting polypeptides are part of the virion particle. In addition, A2 does not have a system similar to the one that promotes formation of the G-T fusion protein in other phages: the orf located in a position similar to that of the lambda gene g is immediately followed by several stop codons in all three possible frames, thus precluding formation of any fusion product.
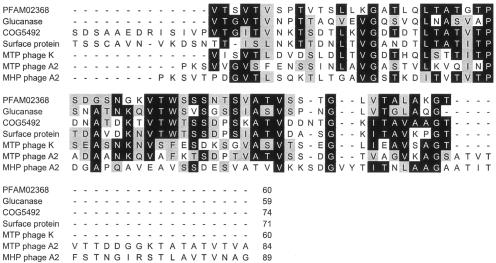
No amino acid sequence similarities were found between the canonical products of the genes that encode the major head and tail proteins of A2, i.e., gp5A and gp10A; however, their frameshifted counterparts, gp5B and gp10B, showed extensive homology in their extra carboxy-terminal ends. These stretches, of 85 and 81 amino acids, respectively, are 42% identical (63% similar) (Fig. 3). The predicted structures of these two protein segments are related to bacterial immunoglobulin-like folding domains defined by pfam02368 (Big 2) and COG5492 (13). Similar domains are found in tail components of phages such as N15 and K (18, 20), bacterial surface proteins involved in adherence (17), and enzymes that recognize polysaccharides as substrates, such as an agarase from Microscilla spp. or a β-1-3-endoglucanase from Bacillus circulans (24, 25) (Fig. 3). The occurrence of Ig-like domains in structural phage proteins and in bacterial products mainly involved in carbohydrate recognition might indicate that the phage proteins function in the stabilization of the phage particle on the polysaccharide-based cell envelope once the specific binding by the tail tip has occurred (A. Davidson, personal communication). The phage A2 has an extraordinarily long tail of about 280 nm, which would justify the need of these “molecular hooks” at the level of both the head and tail of the infecting virion to retain its infectivity, thus explaining why gp5B is essential for phage propagation (7).
FIG. 3.
Alignment of the amino acid sequences encoded by the stretches located in the 3′ vicinity of the major head (MHP) and tail (MTP) A2 genes after −1 frameshifting with the COG5492 and PFAM02368 (bacterial and phage surface proteins containing immunoglobulin-like domains) consensus motifs. The sequences of the portions of three individual proteins with different functions (a β-1-3-endoglucanase from Bacillus circulans, an adhesion factor from Clostridium acetobutylicum, and the major tail protein of phage K; see the text for references) that contain these motifs are also included for comparison.
Acknowledgments
This work was supported by CICYT grants BMC2002-0638 and SAF2004-0033 from the Ministry of Science and Technology (Spain) and the FEDER Plan. P.G. and I.R. are holders, respectively, of a fellowship and a scholarship associated with these grants.
We thank K. F. Chater for critical reading of the manuscript. The proteomics service of the National Biotechnology Centre (CSIC) is acknowledged.
REFERENCES
- 1.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2002. Recoding: translational bifurcations in gene expression. Gene 286:187-201. [DOI] [PubMed] [Google Scholar]
- 2.Blinkova, A., C. Hervas, P. T. Stukenberg, R. Onrust, M. E. O'Donnell, and J. R. Walker. 1993. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J. Bacteriol. 175:6018-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 76:1885-1892. [DOI] [PubMed] [Google Scholar]
- 4.Condron, B. G., J. F. Atkins, and R. F. Gesteland. 1991. Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol. 173:6998-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condron, B. G., R. F. Gesteland, and J. F. Atkins. 1991. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 19:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia, P., V. Ladero, and J. E. Suarez. 2003. Analysis of the morphogenetic cluster and genome of the temperate Lactobacillus casei bacteriophage A2. Arch. Virol. 148:1051-1070. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, P., I. Rodriguez, and J. E. Suarez. 2004. A −1 ribosomal frameshift in the transcript that encodes the major head protein of bacteriophage A2 mediates biosynthesis of a second essential component of the capsid. J. Bacteriol. 186:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrero, M., C. G. de los Reyes-Gavilán, J. L. Caso, and J. E. Suárez. 1994. Characterization of φ393-A2, a bacteriophage that infects Lactobacillus casei. Microbiology 140:2585-2590. [Google Scholar]
- 9.Kuipers, O. P., P. G. G. A. De Ruyter, M. Kleerebezem, and W. M. De Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 10.Larsen, B., N. M. Wills, R. F. Gesteland, and J. F. Atkins. 1994. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176:6842-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen, B., R. F. Gesteland, and J. F. Atkins. 1997. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol. 271:47-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 13.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [Online.] [DOI] [PMC free article] [PubMed]
- 14.Martin, M. C., M. Fernandez, J. M. Martin-Alonso, F. Parra, J. A. Boga, and M. A. Alvarez. 2004. Nisin-controlled expression of Norwalk virus VP60 protein in Lactobacillus casei. FEMS Microbiol. Lett. 237:385-391. [DOI] [PubMed] [Google Scholar]
- 15.Mejlhede, N., J. F. Atkins, and J. Neuhard. 1999. Ribosomal −1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol. 181:2930-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon, S., Y. Byun, H. J. Kim, S. Jeong, and K. Han. 2004. Predicting genes expressed via −1 and +1 frameshifts. Nucleic Acids Res. 32:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Markarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard, A. E., H. G. Dallmann, B. P. Glover, and C. S. McHenry. 2000. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of deltadelta′ with DnaX(4) forms DnaX(3)deltadelta′. EMBO J. 19:6536-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 21.Ribas, J. C., and R. B. Wickner. 1998. The Gag domain of the Gag-Pol fusion protein directs incorporation into the L-A double-stranded RNA viral particles in Saccharomyces cerevisiae. J. Biol. Chem. 273:9306-9311. [DOI] [PubMed] [Google Scholar]
- 22.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 23.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto, M., R. Aono, and K. Horikoshi. 1993. Structure of the 87-kDa beta-1,3-glucanase gene of Bacillus circulans IAM1165 and properties of the enzyme accumulated in the periplasm of Escherichia coli carrying the gene. Biosci. Biotechnol. Biochem. 57:1518-1525. [DOI] [PubMed] [Google Scholar]
- 25.Zhong, Z., A. Toukdarian, D. Helinski, V. Knauf, S. Sykes, J. E. Wilkinson, C. O'Bryne, T. Shea, C. DeLoughery, and R. Caspi. 2001. Sequence analysis of a 101-kilobase plasmid required for agar degradation by a Microscilla isolate. Appl. Environ. Microbiol. 67:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]