Abstract
The Borrelia genome is comprised of linear and circular elements, including a group of 32-kb circular plasmids (cp32s). Earlier analyses identified a bacteriophage, ϕBB-1, that may package cp32s, suggesting that these plasmids are prophages. cp32-8, cp32-9, and cp32-1 (plasmids L, N, and P, respectively) encode virulence factors such as the factor H binding, OspE proteins (BBL39, BBN38, and BBP38). Here the expression patterns of cp32-8 open reading frames (ORFs) in in vitro-cultivated 1-methyl-3-nitroso-nitroguanidine (MNNG)-treated and untreated spirochetes and during infection were assessed. ORFs BBL42 through BBL28, which encode several bacteriophage protein homologs, were found to be cotranscribed and expression was upregulated by MNNG. Immunoblotting revealed that MNNG-induced transcription led to increased protein production. The expression of several genes that reside outside of the BBL42-BBL28 operon was not affected by MNNG. Some of these genes, including OspE (BBL39), appear to represent morons. Real-time reverse transcription-PCR of spirochetes in mouse tissue revealed that although the phage operon was not induced during infection, transcription of BBL23 (previously designated BlyA), a putative holin, was upregulated. This observation indicates that some genes within the operon can be independently transcribed from internal promoters. Additional transcriptional analyses of the operon identified multiple transcriptional start sites and provided evidence for the expression of a homologous operon from other cp32s. The data support the hypothesis put forth by C. Eggers and D. S. Samuels (J. Bacteriol. 181:7308-7313, 1999) that the cp32s are prophages, a finding with broad implications for our understanding of Borrelia pathogenesis and Borrelia genome evolution.
The genus Borrelia contains several important human pathogens, including the causative agents of Lyme disease and relapsing fever (7). Lyme disease, the most prevalent arthropod-borne infection in humans in North America and Europe, is caused by Borrelia burgdorferi, B. garinii, B. afzelii (47), and B.lonestari (50, 53). The Borrelia genome, which is comprised of linear and circular elements (6, 22), exhibits extensive genetic redundancy with over 175 paralogous gene families (24). The most notable example of genetic redundancy in the Lyme disease spirochetes are the circular plasmids of 32 kb (cp32) (14-16, 43, 49). Nine closely related cp32s are carried by some strains (14, 38, 43, 56), with additional copies or subfragments present in other circular (cp18 and cp8.3) or linear plasmids (lp56) (12, 24, 48).
A temperate B. burgdorferi bacteriophage, designated ϕBB-1, has recently been identified and partially characterized (19). The existence of bacteriophages in the Borrelia was first reported by Hayes et al. (26), who demonstrated phage-like particles in the supernatant of cultures that had undergone spontaneous lysis. Using electron microscopy, Eggers et al. demonstrated that these particles have an isometric head with a diameter of 45 to 50 nm and a contractile tail 90 nm in length and contain DNA that hybridizes with cp32 DNA (19-21). The hybridization analyses raised the possibility that the cp32s are prophages (19). It has also been demonstrated that ϕBB-1 can transduce cp32s (21). This important observation suggests that Borrelia bacteriophage can laterally transfer genetic material between strains. The present study is focused on cp32-8, a plasmid that carries 43 open reading frames (ORFs) sequentially designated BBL01 through BBL43 (Fig. 1) (15). This plasmid also encodes potential virulence factors, including BBL39, an OspE paralog that facilitates immune evasion through the binding of factor H (4, 29, 33, 40). Several of the cp32-8 ORFs exhibit various degrees of homology with known bacteriophage proteins. As annotated by The Institute for Genomic Research (TIGR), BBL01 appears to be distantly related to putative portal proteins of bacteriophages carried by Haemophilus influenzae, Xylella fastidiosa, Salmonella enterica serovar Typhi, and Enterococcus faecalis. Portal proteins are incorporated into the procapsid shell during shell assembly and are required for DNA packaging (8). BBL23 and BBL24 (also referred to as BlyB) encode a putative holin and holin accessory protein (17). Holins allow for the release of bacteriophages from bacteria (25). BBL28, designated by Yang et al. as mlp8, exhibits homology with the KVP40.0087 locus of bacteriophage KVP40 (41). BBL43 exhibits homology with the large subunit of the phage terminase from Clostridium tetani E88 (10). Phage terminases are involved in the cutting and packaging of DNA into the phage head (23). The order of the phage-related proteins on cp32-8 and other cp32s is also similar to that seen in other phages. Based on the homology of cp32 genes with phage proteins, the conserved order of the genes, and their short intergenic spacers, it has been hypothesized that BBL42 through BBL23 comprise a late phage operon (20). Late operons typically encode structural proteins and proteins involved in phage assembly and lysis.
FIG. 1.
ORF map of cp32-8. The designation assigned to each ORF by TIGR is indicated outside of the circle. Common gene name designations are indicated in parentheses. ORFs contained within the putative late phage operon of cp32-8 are indicated by black arrows, and those outside of the operon are indicated by gray arrows. The length of the DNA spacer between adjacent genes is indicated inside the circle. For genes that overlap, the length of the overlap is indicated as a negative number.
Here we investigate transcriptional expression patterns and protein production from ORFs carried by cp32-8 during normal cultivation conditions, after 1-methyl-3-nitroso-nitroguanidine (MNNG) induction, and during infection in mice. MNNG was chosen for induction since it has previously been demonstrated to stimulate bacteriophage release from some Borrelia strains and from Serpulina hyodysenteriae and S. innocens (19, 31, 32). The data demonstrate that BBL42 through BBL28 are cotranscribed at a low level during cultivation and upregulated by MNNG induction. The mammalian environment did not trigger upregulation of the cp32-8 30-gene operon, but BBL23 was selectively upregulated, indicating that one or more genes within the operon can be independently transcribed. Transcriptional start site analyses localized the operon promoter to an AT-rich region upstream of BBL42 and also provided evidence for the transcription of the analogous operon from cp32-7. Immunoblot analyses revealed that the MNNG-induced transcriptional upregulation of the late operon leads to increased protein production. Collectively, these analyses provide further support for the hypothesis that the cp32s are prophages (19, 20). These data have important implications for our understanding of Borrelia genome organization, evolution, and pathogenesis.
MATERIALS AND METHODS
Bacterial cultivation and treatment with MNNG.
B. burgdorferi B31MI was cultivated at 33°C in BSK-H complete medium (Sigma). To induce production of the putative phage-related proteins, the MNNG induction protocol developed by Eggers and Samuels was used (19). Briefly, cells from an actively growing culture of B. burgdorferi were recovered by centrifugation and resuspended in BSK-H complete medium containing MNNG (final concentration of 10 μg ml−1; prepared as a 50-mg ml−1 stock solution in dimethyl sulfoxide) or in BSK-H complete medium with an equivalent concentration of dimethyl sulfoxide. The cultures were maintained at 33°C for 2 h, and the cells were recovered by centrifugation, suspended in fresh media, cultivated at 33°C for 60 h, and harvested by centrifugation.
Generation of infection serum, recombinant proteins, and antisera.
To generate infection serum, C3H-HeJ mice (4 to 6 weeks old; Jackson Laboratories) were infected with 103 B. burgdorferi B31MI organisms by intradermal needle inoculation between the shoulder blades. Blood was collected by tail snip at weeks 0, 4, and 6, and sera were harvested. Infection of the mice was confirmed by using real-time PCR with a flaB primer set and SYBR Green as previously described (54).
To generate S-tagged recombinant protein for cp32-8 encoded proteins, the genes of interest were PCR amplified by using primers (Table 1) designed based on the B. burgdorferi B31MI genome sequence (www.tigr.org). The specificity of the primers was assessed through BLAST searches. The primers were designed with tail sequences to allow for ligase independent cloning (LIC) and expression using the pET-32 Ek/LIC vector (Novagen) as previously described (30, 54). To verify the sequence of all constructs, recombinant plasmids were purified from Escherichia coli NovaBlue (DE3) cells by using QiaFilter Midi Plasmid Purification kits (QIAGEN), and the inserts were sequenced (MWG Biotech). As described below, the expression of all recombinant proteins was confirmed by immunoblotting.
TABLE 1.
Primer sequences and their corresponding plasmid coordinates
| Primer | Primer sequence (5′-3′)a | Plasmid (coordinates) |
|---|---|---|
| BBL1F | CATTATCACAGGAGCTTGCTAGGC | cp32-8 (81-840) |
| BBL1R | TCCTAGTCCTTTAGCCTGCTCATT | cp32-8 (999-1022) |
| BBL4F | ACGCGGCGTTAAACTTGTTCCA | cp32-8 (2931-2952) |
| BBL4R | ACCTCCGGCCTTGATAATAACACC | cp32-8 (3133-3156) |
| BBL7F | GCTTACAAGCACAAGTTGCAGGAG | cp32-8 (4958-4981) |
| BBL7R | TGGCAGCCCATATGGTAGAAAGT | cp32-8 (5155-5177) |
| BBL11F | GGAGATGCGTCAGATGATGGACTT | cp32-8 (7079-7102) |
| BBL11R | CAGCGGCATGCCATAAGGATTTAC | cp32-8 (7310-7333) |
| BBL15F | GCTATGAGTAAAGTTGGAGGAGGC | cp32-8 (9418-9441) |
| BBL15R | GCTGCTCATAGCACCACTGGATTT | cp32-8 (9700-9723) |
| BBL16IF | AGAAAGAAATCAGGGAGATTACTCG | cp32-8 (10386-10410) |
| BBL16IR | GATGAGCGTCAAAGCGTAGCATAG | cp32-8 (10731-10754) |
| BBL17F | CAATCGCGAATTGGCAGGCAAAGA | cp32-8 (11350-11373) |
| BBL17R | GCCCATAGTCTTCTAATGCCTCGT | cp32-8 (11634-11657) |
| BBL18R | CGGTTGCTATTTTTAAGCTTGATTCG | cp32-8 (12402-12427) |
| BBL20IF | AGCACAACTTACAGTGCGGTTC | cp32-8 (13079-13100) |
| BBL20IR | TGTCTCCCTCAAGTAAAGTACCACT | cp32-8 (13274-13298) |
| BBL21IF | GCACTGACGAGTCATTTAGACTGAT | cp32-8 (13968-13992) |
| BBL21IR | GATGCAGCATATCCCGGCTTCTTT | cp32-8 (14149-14172) |
| BBL22IF | TGCCGAAGAACTTGCAGCTGAT | cp32-8 (14548-14569) |
| BBL22IR | GCTGCCATAAGCAAGATTTGATGGT | cp32-8 (14779-14803) |
| BBL24IF | TGTTGAGCTTGGACTTACGTCTT | cp32-8 (15442-15464) |
| BBL24IR | CATTTAATGGAGCGAGTGCCGCAT | cp32-8 (15621-15644) |
| BBL28F | ATGCTAAACGGCTGTAATTCTAATGAT | cp32-8 (17244-17270) |
| BBL28R | AATTCTCCAGCATCAGTTAAAGCGG | cp32-8 (17428-17452) |
| BBL1QF | GAGTGCTGCCCACAATGTTA | cp32-8 (197-216) |
| BBL1QR | CCAATCCCATGAAAACGGTA | cp32-8 (282-301) |
| BBL6QF | ATGCCCGAGCAGATAAAAGA | cp32-8 (4443-4462) |
| BBL6QR | AATTCGTCGCCCAGTTCTAA | cp32-8 (4533-4552) |
| BBL17QF | CCCTTAGACTTCACCGACGA | cp32-8 (11162-11181) |
| BBL17QR | ACCCGGATAATCAGTGCTCA | cp32-8 (11277-11296) |
| BBL23QF | TGATTTTTGTAACAGTGCTGGTTT | cp32-8 (15279-15302) |
| BBL23QR | TGTGATTTTTGCCATTACCA | cp32-8 (15382-15401) |
| L27QF | TGGAACAACTAATAGCACAAGATTT | cp32-8 (16618-16642) |
| L27QR | AGTGTTAAGTTCATTTTTGACACCA | cp32-8 (16742-16766) |
| L28QF | TGGAAAAGAAAATGGGGATG | cp32-8 (17531-17550) |
| L28QR | TAGTTGCATTGCTTGCACCA | cp32-8 (17612-17631) |
| L29QF | AAAGCACAATTTGGAGCTTGA | cp32-8 (18499-18519) |
| L29QR | GGAATTCGCGTATCAAAAGAA | cp32-8 (18385-18405) |
| L35QF | GCGTAGAAAAGAGTTTGCAAAAAG | cp32-8 (22507-22530) |
| L35QR | TGGACCTAGAATTGCTGCTG | cp32-8 (22627-22646) |
| L38QF | TGACCCCAAAAGCTATCAAAA | cp32-8 (25762-25782) |
| L38QR | CCCCTACACCCACTTATTGC | cp32-8 (25674-25693) |
| L39QF | TGATGAGCAAAGCAATGGAG | cp32-8 (26459-26478) |
| L39QR | AAATCTCCTAAGTCTGCCCAGTT | cp32-8 (26547-26569) |
| L42QF | CAAAAGTGGGAAAGTGGAGAA | cp32-8 (29090-29110) |
| L42QR | GCTCTACATCCAATTCGCTTC | cp32-8 (29211-29231) |
| L43QF | AGGGGAAGTAATTCGGCACT | cp32-8 (30010-30029) |
| L43QR | TAGTTTCTTGCCCGCATCTT | cp32-8 (30093-30112) |
| Fla F | TTCATGTTGGAGCAAACCAA | Flagellin (524-543) |
| Fla R | CTGAGCAGTTTGAGCTCCCT | Flagellin (602-621) |
| L42P1 | AGACCGCTCTACATCCAA | cp32-8 (29219-29236) |
| L42P2 | CCGTTAACAACACTTTCTCCACTTTC | cp32-8 (29099-29124) |
| L42P3 | GTTAACAACACTTTCTCCACTTTCCCA | cp32-8 (29096-29122) |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | Abridged anchor primer |
| AUAP | GGCCACGCGTCGACTAGTAC | Abridged universal primer |
| L1EKF | GACGACGACAAGATGTGTGATTTAAGAAAAAC AAAACTA | cp32-8 (67-92) |
| L1EKR | GAGGAGAAGCCCGGTCAAAATGAGAATAATTTCTCTTTTAA | cp32-8 (1263-1289) |
| L6EKF | GACGACGACAAGATGGAATTATTTGATGAAAATTATTATG | cp32-8 (3963-3990) |
| L6EKR | GAGGAGAAGCCCGGTTAACTTTGCTTTATGTTAACTTGCA | cp32-8 (4897-4922) |
| L11EKF | GACGACGACAAGATGCCGCAAGATACAATTAGTGTA | cp32-8 (6702-6724) |
| L11EKR | GAGGAGAAGCCCGGTTAAGCACTTAAGCTGTTTTGATAATC | cp32-8 (7787-7813) |
| L17EKF | GACGACGACAAGATGCTGTTACTACAATATGATTTTAA | cp32-8 (10952-10977) |
| L17EKR | GAGGAGAAGCCCGGTTATTTTTTACGTTTTGTATTAGAATC | cp32-8 (11876-11902) |
| L23EKF | GACGACGACAAGATGGATACTATTAAATTAACCGAACT | cp32-8 (15215-15240) |
| L23EKR | GAGGAGAAGCCCGGTTAATCTCTTTTTTTAATGTGATTTTTG | cp32-8 (15391-15418) |
Tail sequences that allow ligase-independent cloning into the pET32Ek/LIC vector are indicated by underscoring.
To generate antisera, 50 μg of each recombinant protein in Freund complete adjuvant was injected into C3H-HeJ mice (4 to 6 weeks old) with boosts at 2 and 4 weeks in incomplete Freund adjuvant. The mice were sacrificed at week 6, and the specificities of the antisera were confirmed by immunoblot analyses.
SDS-PAGE and immunoblotting.
Purified recombinant proteins, E. coli, or Borrelia cell lysates (MNNG treated or untreated) were separated in 12.5% Criterion Precast Gels (Bio-Rad) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted onto polyvinylidene difluoride membranes (Millipore) (44). Expression of recombinant proteins was confirmed by immunoblot analyses of E. coli cell lysates using horseradish peroxidase (HRP) conjugated S-Protein (Novagen; 1:40,000), which detects the N-terminal S-Tag fusion carried by recombinant proteins used in the present study. To analyze the production of Borrelia proteins in the MNNG-induced and uninduced cultures, identical immunoblots were generated and screened with α-BBL01, α-BBL06, α-BBL17, α-BBL23, and α-BBL40 antisera (1:1,000 dilution) that were generated as part of this study. Additional antisera used here, including α-OspE (BBL39), α-OspC (type A), and α-Bdr family antisera (which recognizes all Bdr paralogs including BBL27 and BBL35), were generated in separate studies from our laboratory (18, 40, 44, 45). The α-FlaB antiserum was kindly provided by Michael Theisen (Statens Serum Institute, Copenhagen, Denmark). To determine whether antibody to cp32-8 encoded proteins is elicited during infection, additional immunoblots were screened with serum (1:1,000) collected from mice infected with B. burgdorferi B31MI at 2 and 12 weeks postinfection. HRP-conjugated goat anti-mouse immunoglobulin G served as the secondary antibody (Pierce) and was used at a dilution of 1:10,000. In all cases, detection was achieved through chemiluminescence using the Super Signal West Pico chemiluminescence substrate (Pierce).
RNA and DNA isolation from cultivated bacteria and infected mouse tissue.
To isolate RNA from cultivated bacteria, the cells were harvested by centrifugation and lysed with diethyl pyrocarbonate-treated 1% SDS. RNA was recovered by using the RNeasy Midi kit as instructed by manufacturer (QIAGEN). The integrity of the RNA was assessed by electrophoresis of 5 μg of total RNA in a 1.5% agarose gel with Tris-acetate-EDTA (TAE) buffer. To isolate RNA from B. burgdorferi B31MI-infected mouse tissue, ear punch biopsies were quick-frozen on dry ice in 1.5-ml Eppendorf tubes, macerated with a mortar and pestle, and suspended in 400 μl of RLT buffer (QIAGEN), and RNA was recovered by using the RNeasy Minikit (QIAGEN). Residual DNA was removed by treatment with DNase I (37°C; 1 h; Invitrogen), followed by inactivation of the DNase with 2.5 mM EDTA (70 °C, 10 min). DNA was extracted from infected mouse tissue by using the DNeasy tissue kit as instructed by the manufacturer (QIAGEN).
PCR, reverse transcriptase PCR (RT-PCR), and real-time semiquantitative RT-PCR.
To generate template for PCR, spirochetes were harvested by centrifugation, washed with phosphate-buffered saline, and boiled for 10 min in H2O. To determine the optimal parameters for PCR with each primer set, an annealing temperature gradient of 45 to 74°C was tested. The optimal magnesium concentration was determined by testing magnesium concentrations ranging from 1.5 to 4 mM. PCR amplicons were analyzed by electrophoresis in 1.8% GTG-agarose or in 2.5% MetaPhor agarose (ISC BioExpress) gels in TAE buffer and visualized by ethidium bromide staining.
For RT-PCR analyses, the absence of DNA in the RNA preparation was verified by PCR using a FlaB primer set and Taq DNA polymerase. cDNA was generated by using Superscript II RT (Invitrogen) with 1 μg of RNA, 1.5 pmol of each specific primer (Table 1), and reagents supplied by the manufacturer (42°C, 50 min). The RT was inactivated by incubation at 70 °C for 15 min. As a negative control, reactions were also run without RT. The cDNA was used as the template for amplification in PCR with Taq polymerase with the following cycle parameters: 40 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 1.5 min. The resulting amplicons were analyzed in 1.8% agarose gels.
Real-time RT-PCR was performed in triplicate using the DNA Engine Opticon 2 System (MJ Research) and the SYBR Green PCR Master Mix (Applied Biosystems) as instructed by the supplier. Primer pairs (final concentration, 0.5 μM) were designed to amplify a 100- to 150-bp fragment of each target. A standard curve was generated by using serial dilutions of a known amount of genomic DNA as a template. PCR was performed by using the following cycling parameters: 1 cycle of 10 min at 95°C, followed by 40 cycles of 10 s at 94°C, 20 s at 60°C, and 20 s at 72°C. Melting curves were generated over a temperature range of 55 to 95°C. PCR product was quantified, and the data were analyzed by using software provided by the thermocycler manufacturer (MJ Research).
Localization of promoter elements by using a 5′RACE (5′ rapid amplification of cDNA ends) approach.
To localize the promoter element of the late operon of cp32-8, the 5′RACE was used as previously described (54). In brief, primers (2.5 pmol) were annealed with 3 μg of total cellular RNA, and cDNA was generated by using SuperScript II RT (42°C, 50 min; Gibco-BRL). The RNA was degraded using RNase, and the cDNA was purified by using the GlassMAX spin cartridge (Invitrogen). A poly(C) tail was added to the 3′ end of the cDNA using terminal nucleotidyltransferase. The manufacturer supplied abridged anchor primer (AAP), which hybridizes to the 3′ poly(C) tail of the cDNA, was used in conjunction with a nested gene specific primer to reamplify the cDNA. A final PCR was performed using the abridged universal amplification primer (AUAP) with a nested, gene-specific primer. The amplicons were analyzed by agarose gel electrophoresis and cloned into the TOPO pCR2.1 vector as instructed by the manufacturer (Invitrogen). DNA sequence analysis was performed by using automated methods.
RESULTS
Demonstration of the cotranscription of a large cp32-8 gene cluster.
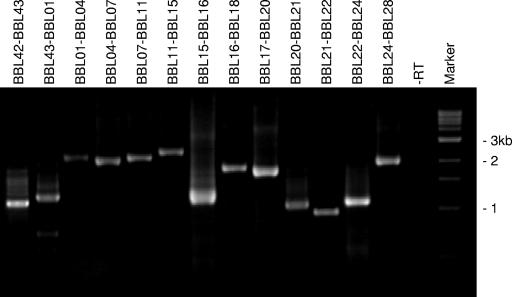
Cp32-8 encodes 43 genes, many of which partially overlap or are separated by short intergenic spacers (Fig. 1). Table 2 provides a summary of the properties of the cp32-8 carried genes and their predicted gene products as inferred from annotation of the genome sequence (24). The possible cotranscription of a series of genes that form a putative cp32-8 phage operon was assessed by using an RT-PCR approach. Since most cp32-8 genes are members of paralogous gene families, the first step was to design primers that are as paralog specific as possible. The potential specificity of primers was assessed through BLAST searches. For many ORFs, paralog-specific primers could not be made since identical copies of some cp32-8 genes are present on other plasmids. However, specific primers could be made for some genes, and their specificity was confirmed by PCR (data not shown). Using combinations of primers designed to amplify across adjacent genes, a 30-gene operon that initiates with BBL42 and extends through BBL28 was demonstrated in bacteria grown under standard cultivation conditions (Fig. 2). Attempts to detect this transcript by Northern hybridization were unsuccessful, presumably due to its size and low abundance. Based on the properties of the genes present within this operon, it appears to constitute a typical late phage operon.
TABLE 2.
Summary of the properties of cp32-8 ORFs and the proteins they encode
| TIGR designation | Common name and protein information | 5′ End | 3′ End | Pfam | No. of paralogs | Protein mass (kDa) |
|---|---|---|---|---|---|---|
| BBL01 | Putative bacteriophage portal protein | 66 | 1289 | 146 | 9 | 47.037 |
| BBL02 | Hypothetical protein | 1306 | 1998 | 147 | 9 | 26.666 |
| BBL03 | Hypothetical protein | 2011 | 2568 | 148 | 26 | 20.003 |
| BBL04 | Hypothetical protein | 2575 | 3339 | 148 | 26 | 28.041 |
| BBL05 | Hypothetical protein | 3369 | 3941 | 148 | 26 | 21.244 |
| BBL06 | Conserved hypothetical protein | 3948 | 4922 | 149 | 9 | 36.721 |
| BBL07 | Conserved hypothetical protein | 4936 | 5397 | 150 | 9 | 17.204 |
| BBL08 | Conserved hypothetical protein | 5379 | 5780 | 107 | 9 | 15.554 |
| BBL09 | Conserved hypothetical protein | 5768 | 6157 | 108 | 8 | 14.502 |
| BBL10 | Conserved hypothetical protein | 6154 | 6720 | 151 | 9 | 21.639 |
| BBL11 | Hypothetical protein | 6701 | 7813 | 152 | 9 | 42.102 |
| BBL12 | Hypothetical protein | 7828 | 8256 | 153 | 9 | 16.247 |
| BBL13 | Hypothetical protein | 8272 | 8727 | 154 | 9 | 17.888 |
| BBL14 | Hypothetical protein | 8724 | 8960 | 155 | 9 | 9.406 |
| BBL15 | Hypothetical protein | 8968 | 10242 | 156 | 8 | 47.288 |
| BBL16 | Hypothetical protein | 10265 | 10948 | 157 | 9 | 26.1 |
| BBL17 | Hypothetical protein | 10952 | 11902 | 159 | 9 | 36.023 |
| BBL18 | Hypothetical protein | 11920 | 12465 | 160 | 9 | 20.609 |
| BBL19 | Conserved hypothetical protein | 12495 | 12827 | 139 | 9 | 12.849 |
| BBL20 | Conserved hypothetical protein | 12824 | 13699 | 140 | 9 | 33.176 |
| BBL21 | Conserved hypothetical protein | 13709 | 14314 | 141 | 9 | 22.855 |
| BBL22 | Conserved hypothetical protein | 14324 | 15139 | 142 | 9 | 30.646 |
| BBL23 | BlyA (putative holin) | 15215 | 15418 | 109 | 8 | 7.478 |
| BBL24 | BlyB (putative holin accessory protein) | 15422 | 15769 | 111 | 8 | 13.191 |
| BBL25 | Conserved hypothetical protein | 15759 | 16094 | 112 | 8 | 12.772 |
| BBL26 | Conserved hypothetical protein | 16081 | 16440 | 143 | 10 | 13.876 |
| BBL27 | (BdrE1) conserved hypothetical protein | 16515 | 17099 | 80 | 18 | 22.421 |
| BBL28 | (mlpH) lipoprotein | 17205 | 17651 | 113 | 8 | 16.626 |
| BBL29 | Putative phage ss binding protein | 18761 | 17688 | 161 | 10 | 42.221 |
| BBL30 | Conserved hypothetical protein, downregulated by blood | 19091 | 20188 | 57 | 42 | 43.923 |
| BBL31 | Conserved hypothetical protein, downregulated by blood | 20198 | 20764 | 50 | 23 | 22.58 |
| BBL32 | Putative plasmid partition protein, par A homolog of Ecoli P1 phage, downregulated by blood | 20740 | 21480 | 32 | 29 | 28.594 |
| BBL33 | Hypothetical protein | 21467 | 21559 | 3.909 | ||
| BBL34 | Conserved hypothetical protein, downregulated by blood | 21540 | 22100 | 49 | 26 | 22.112 |
| BBL35 | (BdrD1) conserved hypothetical protein, Bdr proteins exhibit genuswide distribution | 22113 | 22691 | 80 | 18 | 22.05 |
| BBL36 | (bppA) conserved hypothetical protein | 23306 | 24691 | 165 | 9 | 53.496 |
| BBL37 | (bppB) conserved hypothetical protein | 24744 | 25169 | 144 | 9 | 16.829 |
| BBL38 | (bppC) putative phage integrase | 25951 | 25175 | 96 | 10 | 30.267 |
| BBL39 | ospE (factor H-binding protein) | 26370 | 26903 | 162 | 5 | 20.106 |
| BBL40 | E1p protein | 26931 | 28067 | 163 | 6 | 43.636 |
| BBL41 | Hypothetical protein | 28209 | 28790 | 114 | 6 | 23.412 |
| BBL42 | Hypothetical protein | 28970 | 29536 | 115 | 8 | 21.682 |
| BBL43 | Conserved hypothetical protein, homology with the phage terminase large subunit from Clostridium tetani E88 | 29533 | 30885 | 145 | 9 | 52.008 |
FIG. 2.
Identification of a large polycistronic cp32-8 encoded operon by using RT-PCR. RNA was isolated and RT-PCR was performed as described in the text. Primer pairs spanning across two or more of the cp32-8 genes (indicated above each lane) were used to determine whether adjacent or nearby genes are cotranscribed. A negative control, in which RT was omitted, was performed for every primer set (only one negative control is presented in the figure). The resulting amplicons were analyzed by electrophoresis in 1.8% GTG-agarose gels. Molecular sizes markers in kilobases are indicated.
Treatment of B. burgdorferi with MNNG induces transcription of the phage operon of cp32-8.
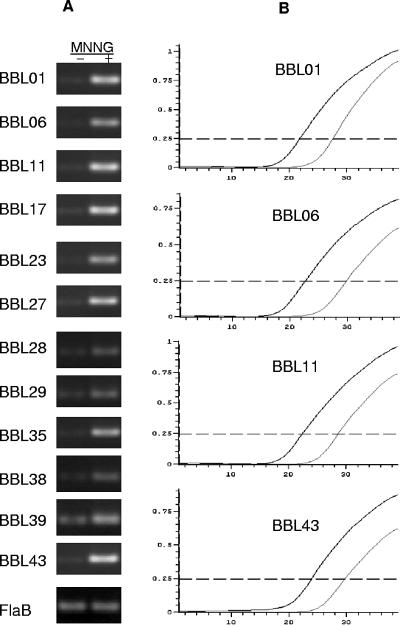
The DNA alkylating agent, MNNG, which has been widely used to study bacteriophage gene expression in several bacteria, upregulates the expression of some phage genes and can induce phage production (19, 20). To determine whether the expression of cp32-8 genes is influenced by MNNG treatment, RNA was isolated from treated and untreated cells, and real-time RT-PCR was performed. MNNG treatment led to a dramatic induction of all genes within the BBL42-BBL28 operon. Most cp32-8 genes outside of this cluster, as well as other genes tested that are carried on other plasmids (ospC) or the chromosome (flaB), were not induced by MNNG (Fig. 3). Typically, increases in transcription as determined by real-time RT-PCR are presented in terms of fold increase. Calculation of such values revealed that most genes in the operon were upregulated 2,000- to 3,000-fold. However, due to several technical considerations, such as the multicopy nature of most of the ORFs, the percentage of total transcript derived from a specific locus cannot be accurately determined. Hence, we do not present fold induction values as part of this report. Even with this caveat, the data presented in Fig. 3 clearly demonstrate significant upregulation of ORFs within the BBL42-BBL28 cluster upon MNNG induction.
FIG. 3.
Demonstration of the transcriptional upregulation of the cp32-8 phage operon with MNNG treatment. Real-time RT-PCR was performed with RNA isolated from cells treated with or without MNNG as described in the text. (A) Ethidium bromide-stained gels of the resulting amplicons obtained from MNNG-induced (+) or uninduced cultures (−). (B) Representative real-time RT-PCR amplification curves. The thick and thin lines indicate the curves obtained with induced and uninduced cultures, respectively.
Localization of the promoter elements for the cp32-8 late operon by using 5′ RACE analysis.
To localize the promoter element of the cp32-8 late operon, 5′RACE analyses were performed. Several amplicons, presumably derived from expression of the late operon from multiple cp32s as well as from multiple promoters on each cp32, were obtained. The amplicons were TA cloned (in batch), and several E. coli clones were picked, plasmids were harvested, and the sequences of the inserts were determined. BLAST searches were performed to identify the origin of the sequences. Amplicons derived from two different plasmids, cp32-8 and cp32-7, were identified. The BBL42 homolog on cp32-7 is designated BBO43 (cp32-7 has an extra ORF that is not carried by cp32-8). The 5′ end of the amplicon (i.e., the transcriptional start site) derived from cp32-8 terminated at a T residue located 54 bases upstream from the putative translational start codon of BBL42 (Fig. 4), whereas the amplicon from cp32-7 terminated at an A located 38 bases upstream from the putative translational start site of BBO43. These sites are at the 3′ border of an AT-rich region (∼85% over a 100-bp stretch).
FIG. 4.
Identification of transcriptional start sites associated with the late phage operon of the cp32s. To identify transcriptional start sites for the late operon, the 5′RACE approach was used as described in the text, and the amplicons were cloned and sequenced. The origin of each sequence was determined through a BLAST search. The alignment presents the sequence determined for the upstream and 5′ domain of the family 115 paralogs, BBL42 and BBO43, the first genes of the late operon of cp32-8 and cp32-7, respectively. The transcriptional start sites (TSS), ribosome-binding sites (RBS; underlining), and translational start codons (TLS) are indicated. Identical residues are indicated by an asterisk, and the coding sequences are capitalized. The transcriptional start sites are indicated by boldface capital letters.
Demonstration of the MNNG-induced production of cp32-8-encoded proteins.
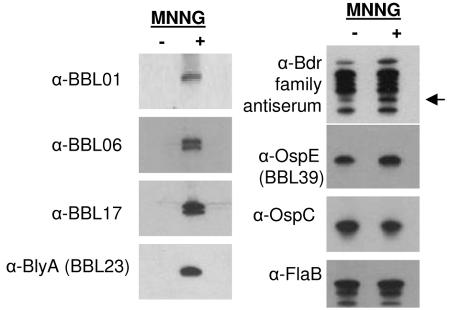
To determine whether the MNNG-induced transcription of cp32-8 genes resulted in increased protein production, several cp32-8 genes were cloned, and recombinant protein was generated for each. The recombinant proteins were purified and used to generate antisera in mice. After preabsorption of the antisera with an irrelevant S-Tagged protein to remove S-Tag-directed antibody, the specificity of the antisera was confirmed by immunoblot analyses using the recombinant proteins as the test antigens. With the caveat that some of the antisera cannot differentiate among identical paralogs encoded by different plasmids, the antisera were determined to be specific (data not shown). The antisera were then used to screen immunoblots of B. burgdorferi B31MI cultures that had been treated or untreated with MNNG. The production of BBL01, BBL06, BBL17, and BBL23, all of which are encoded within the 30-gene operon, was dramatically upregulated by induction (Fig. 5). In untreated cultures these proteins were not detectable, indicating that their transcription and production are tightly linked to MNNG induction. The upregulation of BBL23 upon MNNG treatment is consistent with an earlier study (17). Two proteins were detected with both the α-BBL06 and α-BBL17 antisera, indicating that other paralogs of these proteins are also induced by MNNG. Using α-Bdr subfamily-specific antisera, we also demonstrate that production of at least one Bdr paralog is upregulated by MNNG treatment. The mass of the upregulated protein is consistent with BdrE1 (BBL27), the gene for which is present within the late operon of cp32-8. It is also evident that not all 18 paralogs of the Bdr family are upregulated by MNNG. This is consistent with earlier studies that demonstrated differential expression of bdr paralogs (44). The production of the OspE paralog, BBL39, which is located outside of the late operon and appears to be a protein of bacterial origin, was not influenced by MNNG induction. As a control, recombinant protein was generated for proteins encoded by genes on other plasmids or on the chromosome, and the influence of MNNG on their production was assessed by immunoblotting. The production of OspC and FlaB, which are encoded by cp26 (36, 46) and the chromosome (24), respectively, was not affected by MNNG induction. It can be concluded from these analyses that increased production of late operon-encoded proteins upon treatment with MNNG is not a genomewide, nonspecific phenomenon.
FIG. 5.
Analysis of protein production in B. burgdorferi cultures treated with or without MNNG. Bacteria were recovered from cultures that were either MNNG treated (+) or untreated (−) as described in the text. Cell lysates were generated, fractionated by SDS-PAGE, and immunoblotted. Identical immunoblots were screened with antisera specific for cp32-8-encoded proteins or other Borrelia proteins as indicated to the left of each panel. The migration position of the Bdr paralogs, BBL27 (BdrE1) and BBL35 (BdrD1), which comigrate are indicated by an arrow.
The transcriptional expression and production of most late operon encoded proteins is not induced by the mammalian environment.
To determine whether the host environment stimulates the expression of the late operon, real-time RT-PCR was performed with RNA extracted from infected mouse tissue and primers targeting several genes within the operon. Although flaB transcript (positive control) was readily detected, transcript from BBL01 (Fig. 6), BBL06, BBL11, BBL17, BBL27, BBL28, BBL29, BBL35, BBL38, BBL42, and BBL43 (data not shown) was not. However, BBL23 transcript was detected (Fig. 6), indicating that this putative holin-encoding gene, possibly in conjunction with its holin accessory protein gene (BBL24), can be independently transcribed from the late operon by an internal promoter. This observation is consistent with an earlier report by Damman et al. (17). Transcript for ospE (BBL39) was also detected in mouse tissue (Fig. 6). However, transcripts for BBL29, BBL35 (BdrD1), and BBL38, which also reside outside of the late operon, were not detected (data not shown). Hence, whereas transcription of the cp32-8 late operon as a whole is not stimulated by the host environment, at least one gene within the operon (BBL23) can be independently expressed during infection. In addition, the data also demonstrate that most cp32-8 genes outside of the late operon, with the exception of BBL39 (OspE) and BBL40 (see immunoblot data below), are not expressed in mouse tissue.
FIG. 6.
Analysis of expression of cp32-8 genes in the tissue of infected mice by real-time RT PCR. RNA was extracted from ear punch biopsies from mice infected with B. burgdorferi B31MI and used as a template for real-time RT-PCR analyses as described in the text. The expression level of two genes within the late phage operon (BBL01 and BBL23) and OspE (BBL39), a cp32-8 carried moron and flaB (a chromosomal gene), was assessed. The left panel presents the ethidium bromide-stained gels of the amplification products after 40 cycles. The DNA containing positive amplification control and the with or without RT reactions (+RT or −RT lanes) are indicated. The right hand panel depicts the amplification curves for each, with the thick lines indicating the +RT reaction and the thin lines indicating the DNA positive control. The curve for the −RT negative control is not discernible since it was essentially at baseline.
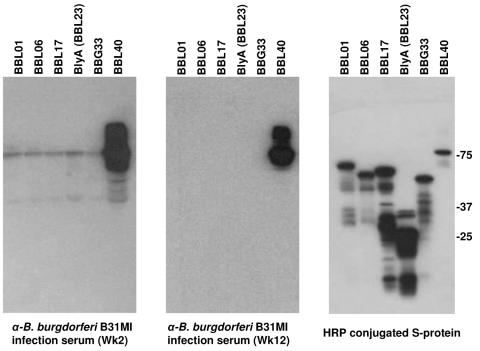
The lack of detection of cp32-8 late operon transcript in spirochetes in the host environment could have resulted from sampling of the tissue at a time point during which late operon mRNA levels were not abundant. As a second approach to investigate phage protein production, immunoblot analyses were performed. An antibody response to BBL01, BBL06, BBL17, and BBL23 was not detected in spite of the relatively high antigenic index of several domains within each of these proteins (Fig. 7). This coupled with the mRNA analyses suggests that these proteins are poorly expressed during infection. The lack of an antibody response to BBL23, which yielded detectable transcript in the real-time RT-PCR analyses, suggests that this protein as presented during infection is not antigenic. Positive controls for the immunoblot analyses included BBL40 and OspE (27, 40, 51), both of which elicited an antibody response. BdrF2 (BBG33), an inner-membrane-anchored, nonantigenic protein (45) that is upregulated during infection (54), served as a negative control and, as expected, did not elicit an antibody response. Collectively, the transcriptional and immunoblot analyses suggest that host environment does not stimulate expression of the late phage operon.
FIG. 7.
Demonstration that proteins encoded by the BBL01-BBL28 operon do not elicit an antibody response during infection in mice. To determine whether representative proteins encoded by the late operon are antigenic during infection, recombinant proteins were generated as S-Tag fusions, immunoblotted, and screened with sera collected from a mouse infected with B. burgdorferi B31MI at weeks 2 and 12 postinfection. The expression of all recombinant proteins in E. coli was confirmed by screening an immunoblot with HRP-conjugated S protein. Note that the S-Tag adds approximately 17 kDa to the mass of each recombinant protein. BBL40 and BBG33 served as positive (+) and negative (−) controls, respectively. Molecular mass standards are indicated.
DISCUSSION
Temperate bacteriophages can influence the virulence, genomic evolution, and overall fitness of bacteria in several ways. They may serve as hotspots for genomic rearrangement, disrupt bacterial genes, introduce new fitness or virulence factors, or lyse competing strains through prophage induction (11, 13). Earlier studies demonstrated bacteriophage in B. burgdorferi, and it was suggested that the cp32s, which carry several genes with homology to bacteriophage proteins, may be prophage (17, 19-21, 26). The goals of the present study were to determine whether the cp32-8 genes exhibit transcriptional expression properties consistent with that of bacteriophage late operons and, if so, to determine the boundaries of the operon. Although B. burgdorferi strains may carry as many as 9 cp32s, we focused on cp32-8 because it carries BBL39, an ospE paralog gene (35, 38). The OspE paralogs are important virulence factors that facilitate immune evasion through the binding of the complement regulatory protein, factor H (3, 29, 34, 39). Binding of factor H facilitates the local downregulation of complement by increasing the efficiency of the disruption of the C3Bb convertase complex and of the factor I-mediated cleavage of C3b (55). C3b is an important opsonin that stimulates the phagocytosis of C3b-coated cells (55). OspE and other nonphage genes on the cp32s may represent phage morons, which are defined by Brussow et al. as “all extra genes present in prophage genomes which do not have phage function but may act as fitness factors for the lysogen” (11). OspE would appear to fit this criteria and its presence on a potentially transferrable genetic element may have important implications for Borrelia pathogenesis.
Based on the spacing or overlapping nature of the cp32-8 genes, BBL43 through BBL22, and the homology of several with phage late proteins, it has been speculated that this gene cluster forms a late operon (20, 42). However, the cotranscription of these genes and the boundaries of the putative late operon have not been assessed. Using RT-PCR and real-time RT-PCR, we demonstrate low-level coexpression of ORFs BBL42 through BBL28 of cp32-8 of B. burgdorferi B31MI grown under standard cultivation conditions as a single transcriptional unit. Transcription of this operon was dramatically upregulated in response to treatment with the stress-inducing, DNA-alkylating agent, MNNG. Consistent with this, immunoblot analyses revealed that BBL01, BBL06, BBL17, and BBL23 are detectable in MNNG-induced cultures but not in uninduced cultures. Damman et al. previously assessed the influence of MNNG on BBL23 and BBL24 expression specifically and reported upregulation as well (17). BBL23 and BBL24, which appear to constitute a holin system, have also been reported to be upregulated upon exposure of B. burgdorferi to the OspB targeting monoclonal antibody (MAb), CB2 (5). The molecular mechanism by which CB2 triggers upregulation has not yet been defined, but it was hypothesized that its apparent bactericidal activity may result from high-level expression of the BBL23-BBL24 holin system. However, while we observed significant BBL23 upregulation upon MNNG treatment, we did not observe cell death, suggesting that additional mechanisms and factors may be responsible for killing by CB2 MAb. CB2 MAb treatment was also found to stimulate transcription of BBL25 and BBL26 (5), which are contained within the late operon defined here. It is evident that stress induced by a variety of agents (MNNG or CB2 MAb) triggers elevated transcription of at least a subset of cp32 genes. It is not clear at this time if MNNG treatment induces the expression of other genes located elsewhere in the genome that are typically involved in stress responses.
The natural environmental conditions that trigger phage induction in B. burgdorferi are yet to be identified. In general, it has been hypothesized that bacteriophage production and release during infection in humans could contribute to the pathogenesis of bacterial infections (1). Transduction in the mammalian environment might allow for lateral transfer of fitness-enhancing morons such as the factor H-binding OspE (BBL39) protein. Lateral transfer and expression of this gene in the mammalian environment could facilitate the successful completion of the enzootic cycle and thereby ensure population maintenance in nature. Here we used real-time RT-PCR and immunoblotting to assess expression of the late operon in mammals. Although flaB transcription was detected in mouse skin, transcripts derived from BBL01, BBL06, BBL11, BBL17, BBL27, BBL28, BBL42, and BBL43 and several cp32-8 genes outside of the operon, including BBL29, BBL35, and BBL38, were not. This is consistent with a study by Brooks et al. (9) that demonstrated downregulation of BBL15, BBL28, and BBL29 in spirochetes propagated using the dialysis membrane chamber implant model; a model that approximates the host environment (2). In another important study, Tokarz et al. demonstrated by using microarrays that when (52) blood was added to spirochetes cultivated at 37°C in BSK-H complete medium, the putative phage portal protein, BBL01, and the putative holin, BBL23, were downregulated. Consistent with the transcriptional data, we did not detect an antibody response during infection to BBL01, BBL06, BBL17, and BBL23. It is evident from the data presented here and in earlier studies that late operon expression in tissue or under “host-simulated” conditions either does not occur or occurs at a low level. However, BBL23, which can be expressed independently from the late phage operon in tissue, is an exception and, as demonstrated by real-time RT-PCR, appears to be expressed at a level similar to that of OspE. In contrast to OspE, BBL23 does not elicit an antibody response, indicating that it is not antigenic or that there may be some form of posttranscriptional regulation. Posttranscriptional control and accumulation of mRNA has been demonstrated for other Borrelia genes (28, 37). The biological role for BBL23 in host tissues is unclear.
In summary, we demonstrate here the existence of a 30-gene late phage operon on cp32-8 that is induced by MNNG treatment. These analyses provide further evidence for the hypothesis that the cp32s are prophages (19-21). Although the environmental conditions encountered in mice during infection did not lead to phage induction, it is possible other environmental conditions such as that encountered in the tick may elicit phage production and transduction. In fact, the tick environment might represent the most likely environment for phage-mediated genetic exchange due to the high density of spirochetes in fed ticks. Future analyses will seek to define the influence of the tick environment on the transcription of bacteriophage genes. The demonstration that cp32-8 is a prophage has important implications for our understanding of Borrelia pathogenesis, genome organization, and evolution.
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. R. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alitalo, A., M. T., H. Lankinen, I. Seppälä, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 5.Anderton, J., R. Tokarz, C. Thill, C. Kuhlow, C. Brooks, D. Akins, L. Katona, and J. Benach. 2004. Whole-genome DNA array analysis of the response of Borrelia burgdorferi to a bactericidal monoclonal antibody. Infect. Immun. 72:2035-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour, A. G., and C. F. Garon. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409-411. [DOI] [PubMed] [Google Scholar]
- 7.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50:381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazinet, C., J. Benbasat, J. King, J. M. Carazo, and J. L. Carrascosa. 1988. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27:1849-1856. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, C. S., P. S. Hefty, S. E. Joliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brussow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caimano, M. J., X. Yang, T. G. Popova, M. L. Clawson, D. R. Akins, M. V. Norgard, and J. D. Radolf. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canchaya, C., G. Fournous, and H. Brussow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 14.Carlyon, J. A., C. LaVoie, S. Y. Sung, and R. T. Marconi. 1998. Analysis of the organization of multi-copy linear and circular plasmid carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect. Immun. 66:1149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 16.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids in Lyme Disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damman, C. J., C. H. Eggers, D. S. Samuels, and D. B. Oliver. 2000. Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J. Bacteriol. 182:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnhart, C. G., E. L. Buckles, J. S. Dumler, and R. T. Marconi. 2005. Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC antibody response. Infect. Immun. 73:7869-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eggers, C., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 4:365-373. [PubMed] [Google Scholar]
- 21.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferdows, M. S., and A. G. Barbour. 1989. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc. Natl. Acad. Sci. USA 86:5969-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frackman, S., D. A. Siegele, and M. Feiss. 1985. A functional domain of bacteriophage lambda terminase for prohead binding. J. Mol. Biol. 180:283-300. [DOI] [PubMed] [Google Scholar]
- 24.Fraser, C., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischman, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 25.Grundling, A., M. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes, S. F., W. Burgdorfer, and A. G. Barbour. 1983. Bacteriophage in the Ixodes dammini spirochete, etiological agent of Lyme disease. J. Bacteriol. 154:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hefty, P. S., C. S. Brooks, A. M. Jett, G. L. White, S. K. Wikel, R. C. Kennedy, and D. R. Akins. 2002. OspE-related, OspF-related and ELp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of the OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 30.Hovis, K., J. V. McDowell, L. Griffin, and R. T. Marconi. 2004. Identification and characterization of a linear plasmid encoded factor H-binding protein (FhbA) of the relapsing fever spirochete, Borrelia hermsii. J. Bacteriol. 186:2612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey, S. B., T. B. Stanton, and N. S. Jense. 1995. Mitomycin C induction of bacteriophages from Serpulina hyodysenteriae and Serpulina innocens. FEMS Microbiol. Lett. 134:97-101. [DOI] [PubMed] [Google Scholar]
- 32.Humphrey, S. B., T. B. Stanton, N. S. Jensen, and R. L. Zuerner. 1997. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J. Bacteriol. 179:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. European J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 34.Kraiczy, P., C. Skerka, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface protein E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J.Bacteriol. 175:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marconi, R. T., D. S. Samuels, T. G. Schwan, and C. F. Garon. 1993. Identification of a protein in several Borrelia species which is related to OspC of the Lyme disease spirochetes. J. Clin. Microbiol. 31:2577-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marconi, R. T., S. Y. Sung, C. N. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metts, S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in the binding of factor H and OspE targeting antibodies elicited during infection in mice. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. G. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. R. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, D., M. Caimano, J. McDowell, M. Theisen, A. Holm, E. Orff, D. Nelson, S. Wikel, J. Radolf, and R. Marconi. 2002. Environmental regulation and differential expression of members of the Bdr protein family of Borrelia burgdorferi. Infect. Immun. 70:7033-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, D. M., M. Theisen, and R. T. Marconi. 2000. Analysis of the cellular localization of Bdr paralogs in Borrelia burgdorferi, a causative agent of Lyme disease: evidence for functional diversity. J. Bacteriol. 182:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadziene, A., B. Wilske, M. S. Ferdows, and A. G. Barbour. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson, B., S. Casjens, R. van Vugt, S. F. Porcella, K. Tilly, J. L. Bono, and P. Rosa. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stromdahl, E. Y., P. C. Williamson, T. M. Kollars, Jr., S. R. Evans, R. K. Barry, M. A. Vince, and N. A. Dobbs. 2003. Evidence of Borrelia lonestari DNA in Amblyomma americanum (Acari: Ixodidae) removed from humans. J. Clin. Microbiol. 41:5557-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung, S. Y., J. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box flanked ospE related genes of the Lyme disease spirochetes results in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokarz, R., J. Anderton, L. Katona, and J. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varela, A. S., M. P. Luttrell, E. W. Howerth, V. A. Moore, W. R. Davidson, D. E. Stallknecht, and S. E. Little. 2004. First culture isolation of Borrelia lonestari, putative agent of southern tick-associated rash illness. J. Clin. Microbiol. 42:1163-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, H., A. Raji, M. Theisen, P. R. Hansen, and R. T. Marconi. 2005. bdrF2 of the Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J. Bacteriol. 187:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zipfel, P. F., C. Skerka, J. Hellwage, S. T. Jokiranta, S. Meri, V. Brade, P. Kraiczy, M. Noris, and G. Remuzzi. 2002. Structure-function studies of the complement system. Biochem. Soc. Trans. 30:971-978. [DOI] [PubMed] [Google Scholar]
- 56.Zuckert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme Disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]