Abstract
Pseudomonas aeruginosa poses a serious risk in individuals suffering from cystic fibrosis (CF). Strains colonizing the CF lung are generally motile but frequently convert to a nonmotile phenotype as the disease progresses. In many cases, this is coordinately regulated with the overproduction of the exopolysaccharide alginate. Both the expression of alginate (mucoidy) and the loss of flagellum synthesis may provide the bacterium with a selective advantage in the CF lung. Previously published data showed that the regulation of alginate production and flagellum biosynthesis in the CF isolate FRD1 is inversely controlled by the alternative sigma factor AlgT. In this study, we observed that in CF isolates, the mucoid and the nonmotile phenotypes occur predominantly together. Using microarrays, we compared the transcriptomes of isogenic AlgT+ and AlgT− P. aeruginosa and discovered that AlgT significantly downregulated the majority of flagellar genes. A pronounced inhibitory effect was observed in several genes essential for proper flagellum expression, including fleQ, which encodes an essential flagellar regulator. The microarray data were confirmed by reverse transcriptase PCR analysis and promoter fusion assays in isogenic AlgT+ and AlgT− strains. Transmission electron microscopy, motility assays, and Western blots showed that ectopic expression of FleQ in mucoid, nonmotile CF isolates restored flagellum biosynthesis and motility. Together, these data show that AlgT mediates the negative control of flagellum expression by inhibiting the expression of the flagellar regulator fleQ.
The gram-negative bacterium Pseudomonas aeruginosa is a ubiquitous environmental organism. As an opportunistic pathogen, it rarely causes disease in healthy individuals but poses a serious risk to those with compromised immune systems and to cystic fibrosis (CF) patients. P. aeruginosa has an arsenal of virulence factors, including elastase, hemolysin, endotoxin, type III secretion system proteins, type IV pili, the exopolysaccharide alginate, and flagella (24, 43). In acute infections, flagella are essential for P. aeruginosa pathogenesis. This has been documented in a murine thermal injury model in which nonmotile strains were dramatically attenuated in their virulence. Whereas motile strains were able to spread rapidly throughout the host, nonmotile variants were restricted to the site of inoculation and easily cleared (18). Flagella also play a critical role in the development of biofilms, where they aid in initial surface adhesion as well as biofilm dispersal (35, 39). In addition, soluble flagellin is released by motile gram-negative bacteria and acts as a potent inducer of inflammation through interaction with Toll-like receptor 5 (25, 37). Flagellin activates the NF-κB arm of the immune system and provokes the release of proinflammatory mediators such as tumor necrosis factor, interleukin 6, interleukin 8, and nitric oxide by macrophages, monocytes, and epithelial cells (14, 27, 37).
The bacterial flagellum is comprised of two substructures: the membrane-spanning hook-basal body and an external filament. The biosynthesis and assembly of a functional flagellum are subject to a highly complex and tightly controlled regulatory cascade which requires coordinate expression of approximately 50 genes encoding structural subunits, regulatory proteins, motor force generators, and chemosensory machinery (5). In P. aeruginosa, flagellar genes are clustered in three distinct regions of the chromosome, and a four-tiered transcriptional regulatory circuit controls flagellum synthesis (15). The FleQ protein, an NtrC-like transcriptional activator, has been referred to as the master regulator of this pathway, as it belongs to the top tier of the intricate flagellar hierarchy and is required for the expression of all other known flagellar genes with the exception of fliA (8, 15, 29).
In the case of chronic CF pulmonary infections, P. aeruginosa strains initially colonizing the lung are likely derived from the environment and generally have a motile phenotype (32). Flagellar components have been shown to bind to airway mucins and are necessary in establishing infection (6, 7, 42). However, as the disease progresses, the initially motile strains frequently convert to a nonmotile phenotype. Loss of the flagellar structure may reduce the release of inflammatory mediators by the host immune system and thus offer P. aeruginosa a selective advantage in the CF lung (14). Interestingly, the nonmotile phenotype of P. aeruginosa is found almost exclusively in CF isolates (32). In many cases, the loss of the flagellum is coordinately regulated with the overproduction of the exopolysaccharide alginate, thus yielding mucoid, nonmotile variants of P. aeruginosa. Previously published data revealed that in the CF isolate FRD1 (mucA22), alginate and flagellum biosynthesis are inversely regulated by the alternative sigma factor AlgT (AlgU, σ22) (23). While the role of AlgT in the biosynthesis of alginate is well understood (24, 38), not much is known about how AlgT regulates flagellum expression. It has been shown that in motile, nonmucoid P. aeruginosa strains, the activity of AlgT is minimal due to the inhibitory effect of the anti-sigma factor MucA (24, 50). However, in the majority of CF isolates, including most of those examined in this study, mucA is inactivated due to mutations that occur in vivo, which ultimately leads to a deregulation of AlgT (24).
The goal of this study was to determine if the AlgT-mediated inhibition of flagellum biosynthesis is common in mucoid P. aeruginosa CF isolates and to identify the target(s) of AlgT within the flagellar regulatory hierarchy. We observed that 70% of mucoid CF isolates tested lacked flagella, and in all representative strains examined, AlgT was required for this repression. Microarray analysis, reverse transcriptase (RT)-PCR, and promoter fusion assays revealed that the expression of most genes of the flagellar regulon was significantly downregulated in the presence of AlgT, and fleQ was identified as the earliest target of AlgT within the flagellar hierarchy. To test whether the negative control of fleQ expression by AlgT is sufficient to account for the lack of flagella in mucoid CF isolates, FleQ was ectopically expressed in mucoid, nonflagellated P. aeruginosa isolates. The results obtained by Western blot analysis, motility assays, and transmission electron microscopy (TEM) indicated that increased expression of FleQ reversed the AlgT-mediated inhibition of flagellum expression in CF isolates. Together, these data suggest that the AlgT-mediated repression of P. aeruginosa flagellum biosynthesis is a common regulatory mechanism in mucoid CF isolates and occurs by inhibiting expression of the flagellar master regulator fleQ.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
P. aeruginosa PAK, PAKΔQ (kindly supplied by R. Ramphal), PAO1, WFPA50 (PAO1 fliC::xylE-aacC1), FRD1 (mucA22), FRD440 (mucA22 algT::Tn501), FRD2700 (attB::pBAD-fleQ), and PAKΔQ/pAT6 (attB::pBAD-fleQ) were used for this study. Other P. aeruginosa strains used included mucoid, nonmotile CF isolates CF1, CF2, and CF3 and their isogenic algT mutants, as well as a collection of other CF-derived mucoid strains (9, 47). Escherichia coli strain JM109 (Promega) was utilized for all cloning experiments. Strains SM10 and HB101/pRK2013 (19) were used to transfer plasmids to P. aeruginosa. Oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′ → 3′) |
|---|---|
| mucA1 | GGAATTCCAAGAGAGGTATCGCTATGAGT |
| mucA2 | GGAATTCCGGAGGTGGTGCGCATGTCTCTC |
| algT1 | CGGTCATATGCTAACCCAGGAACAGGATC |
| algT2 | CGGGATCCTCAGGCTTCTCGCAACAAAGG |
| CTX1 | CCTCGTTCCCAGTTTGTTCC |
| attB2 | GTCGCCGCCGGCGATGC |
| attB4 | CGCCCTATAGTGAGTCG |
| attB5 | CGCCCCAACCTCGCTGG |
| omlAF | CCTCGCCTGCTTCCATC |
| omlAR | GGAAGGTATCGACGATGA |
| fleQ5 | TCCCCCGGGTGAGCTGGCTGGACATCT |
| fleQ6 | CCGGAATTCATGTGGCGCGAA |
| RT-fleQ1 | GCGAAACCAAACTCTTGCT |
| RT-fleQ2 | GCTCCATCCGCGAGAT |
| RT-fliE1F | GTCAGGGTGTCGAGTTCAA |
| RT-fliE2R | AACGACACGCTGGCC |
| RT-flgC1F | AGTAACCGCCATGTCCCT |
| RT-flgC2R | TCTACTCCGCTCACTGAC |
| RT-fliC1F | GGAAATCACCATGGCCCT |
| RT-fliC2R | GTTCCCGGGCTTAGCGCA |
| RT-flgM1F | GTTTACAACCATGGTCAT |
| RT-flgM2R | TCCTTCGAGGTCAGCGCT |
| RT-omlA1F | TCGCTCACCGATGCAAAA |
| RT-omlA2R | ACGCCAGCCGTCATTGCG |
| RT-algZ21 | GAACGCTTCCTCG |
| RT-algZ27 | CTCAGGCCTGGGCCAGCTCCGC |
RT indicates primers used in RT-PCRs.
Plasmid pSW218 was generated by amplifying omlA by PCR with primers omlAF and omlAR, cutting the product with SmaI, and ligating the resulting fragment into the corresponding site of pBluescript II KS(−)1. Plasmid pAT3 (omlA::lacZ) was generated by cutting pSW218 with EcoRI and BamHI and cloning the excised omlA promoter fragment into the corresponding restriction sites of mini-CTX lacZ (10). Plasmid pAT2 (fleQ::lacZ) was generated by cutting placΩQ (8) with EcoRI and BamHI and cloning the resulting fragment into the corresponding restriction sites of mini-CTX lacZ. Plasmid pAT6 was generated by amplifying fleQ by PCR with primers fleQ5 and fleQ6 (Table 1). The PCR product was cut with EcoRI and SmaI and cloned into the corresponding sites of pSW195 (48). The three plasmids were tested for the correct insert by sequencing. Plasmid pFlp2 (26) was used to excise unwanted mini-CTX lacZ vector sequences from fusions that were integrated into the P. aeruginosa chromosome. Plasmid pJF15 (20) contains a functional copy of the algT gene and was used for complementation analysis.
Media, antibiotics, and enzyme assays.
Luria broth (LB; 10 g tryptone/liter, 5 g yeast extract/liter, 5 g NaCl/liter) and LBNS (LB without NaCl) were used throughout the study. In addition, LB and LBNS agar plates (broth and 15 g agar/liter) were used. Plasmids used in this study were maintained in E. coli by antibiotic selection with 15 μg/ml of tetracycline, 100 μg/ml of ampicillin, and 30 μg/ml of kanamycin. For P. aeruginosa, antibiotics were used at 100 μg/ml of tetracycline and 300 μg/ml of carbenicillin. For counterselection, sucrose (5%) and Irgasan (25 μg/ml) were used. For experiments that involved ectopic expression of FleQ, media were supplemented with 0.1 to 2.5% arabinose. The β-galactosidase assays were performed using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate (33). Promoter activity was calculated in amounts of ONPG hydrolyzed per minute as a function of cell density with the following equation: units = 1,000 × OD420 − (1.75 × OD550)/t × V × OD600 (where t is the time in minutes, V is the volume in ml, and ODn is the optical density at n nm).
Transcriptional profiling.
For transcriptional profiling, strain FDR1 (mucA22) and the isogenic algT mutant FRD440 (mucA22 algT::Tn501) were used. Overnight bacterial cultures were diluted to a starting OD600 of 0.01 in 25 ml of LB and grown with shaking (300 rpm) at 37°C under aerobic conditions. When cultures reached an OD600 of 0.5 (mid-exponential growth phase), 1.5 ml of culture (approximately 7.5 × 108 bacteria) was centrifuged at high speed in a microcentrifuge for 1 min. The bacterial pellet was suspended in 100 μl of lysozyme solution (1 mg/ml; Sigma) and incubated at room temperature for 5 min. RNA was purified from the lysed bacterial fraction with a commercially available RNeasy Mini RNA purification kit (bacterial protocol; QIAGEN). RNA samples were eluted with 80 μl of RNase-free distilled water and treated with 10 μl of RQ1 DNase (Promega) for 1 h at 37°C. A second purification was performed using an RNeasy Mini kit (RNA cleanup protocol) followed by elution in 60 μl of RNase-free distilled water. cDNA was synthesized, converted to cDNA targets, and hybridized to GeneChip P. aeruginosa genome arrays (Affymetrix) as described previously (46).
Microarray analysis.
Hybridization intensity data were extracted from the scanned array images, and intrachip normalizations were performed using Affymetrix Microarray Suite 5.0 software. In order to eliminate noise, we discarded those genes that yielded “absent” or “marginal” calls (based on the default setting in Microarray Suite 5.0) for both strains. The remaining transcripts that showed greater-than-threefold changes in mRNA levels were considered candidate AlgT-regulated genes (see Table S1 in the supplemental material). The summary of the entire microarray data set is provided in Table S2 in the supplemental material.
Generation of FRD2700 and PAKΔQ/pAT6.
Vector pAT6 was transferred into P. aeruginosa FRD1 or PAKΔQ by a triparental spot mating (7 μl of an overnight culture of HB101/pRK2013, 7 μl of an overnight culture of JM109/pAT6, 2 μl of an overnight culture of FRD1 or PAKΔQ), which was done by incubation at 37°C for 7 h. Colonies were subsequently streaked onto selective media containing tetracycline and Irgasan. Positive clones were verified for integration of the vector by PCR using primers CTX1 and attB2 (Table 1). Vector sequences were removed using pFlp2 (49). Final clones were screened by PCR analysis using primers attB4 and attB5 (Table 1) (49).
Genomic DNA isolation and PCR assay.
P. aeruginosa genomic DNA was purified with Wizard genomic DNA isolation reagents by following instructions from the manufacturer (Promega). PCR assays were performed in 10-μl reaction mixtures containing 100 to 150 ng of genomic DNA, the appropriate primers (0.5 μl of 50-ng/μl primer stocks) listed in Table 1, and Taq 2× Master Mix (Promega). PCRs involved 30 cycles (94°C/1 min, 55°C/1 min, and 72°C/1 min).
Total RNA isolation and RT-PCR assay.
P. aeruginosa total RNA was purified with Ambion RNA isolation reagents by following instructions from the manufacturer (Ambion). A 5-μl volume of the isolated RNA was resolved on a 1% agarose gel at 100 V for 10 min to validate the purity of RNA. RNA was converted to cDNA using SuperScript II RT (Invitrogen) as described by the manufacturer. Following this, PCRs with 10-μl portions of FRD1 and FRD440 cDNA were performed with 0.5 μl cDNA, the appropriate primers (0.5 μl of 50-ng/μl primer stocks) listed in Table 1, and Taq 2× Master Mix (Promega). The PCRs involved 25 cycles (94°C/1 min, 55°C/1 min, and 72°C/1 min) for all samples except for fliC cDNA (30 cycles under the same conditions). The reaction products were resolved on a 0.7% agarose gel. Band intensities were determined using a Kodak Image Station 2000RT system.
Static growth experiments and isolation of algT mutants.
For the static growth experiments (17, 48), an overnight shaking culture of mucoid, nonmotile P. aeruginosa was diluted 1:100 in 5 ml LBNS. The cultures were placed into test tube racks and incubated statically at 37°C for 96 h. The cultures were briefly vortexed, tenfold dilutions were performed, and 100-μl aliquots were plated onto LBNS agar plates. Nonmucoid colonies were isolated and complemented with pJF15 (algT+) (20). Clones that reverted back to the mucoid phenotype were selected and cured of pJF15 by passing on nonselective media. The algT gene was amplified by PCR using primers algT1 and algT2 and sequenced.
Flagellum-mediated motility assays.
For flagellum-mediated motility assays, bacteria were stab inoculated onto 0.3% motility agar (0.3% agar in LBNS). Arabinose (0.1 or 2.5%) was added when indicated. Motility plates were incubated overnight at 37°C. Motility was assessed by measuring the diameters of the circular zones that the colonies spread from their points of inoculation (7). Bacteria were considered motile if their zone of motility was ≥5 mm (7).
TEM.
Bacteria were grown in LBNS with or without arabinose to an OD600 of 0.5. Formvar-coated copper grids were hydrophilized by immersion in 100% ethanol. One drop of bacterial culture was added per grid. After 1 min, excess liquid was wicked off without completely drying the grid to avoid flagellum shearing. Grids were washed twice by floating them on ultrapure water. Subsequently, a drop of 2% uranyl acetate was added and wicked off after 1 min. TEM was performed on a Philips 400 transmission electron microscope operated at 80 kV.
Western blotting.
Western blotting was performed using whole-cell lysates. The lysates were prepared from P. aeruginosa grown in LBNS with or without arabinose to an OD600 of 0.5. A 1-ml volume of cells was centrifuged for 3 min at 14,000 rpm, and the pellet was resuspended in 100 μl FB (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2). A 10-μl (see Fig. 1 and 2) or 25-μl (see Fig. 6) volume of each suspension was examined by Western blotting. Rabbit anti-flagellin serotype A and anti-flagellin serotype B antiserum (23) were used at dilutions of 1:20,000. Horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin (Amersham Biosciences) was used as the secondary antibody at a dilution of 1:10,000. The blot was developed using SuperSignal West Dura extended-duration substrate (Pierce) and exposed to film for 5 to 30 s prior to development.
FIG. 1.
Flagellin expression of mucoid P. aeruginosa CF isolates. A representative Western blot of whole-cell lysates was derived from P. aeruginosa strains. Lane 1, control for flagellin A serotype; lane 2, control for flagellin B serotype; lane 3, fliC mutant WFPA50; lanes 4 to 14, mucoid CF isolates. The results indicated that 70% of the strains tested were mucoid and lacked flagellin.
FIG. 2.
AlgT inhibits flagellum biosynthesis. Whole-cell lysates of mucoid, nonmotile P. aeruginosa CF isolates and their isogenic algT mutants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and examined for flagellin expression by Western blotting with flagellin B antiserum. Lane 1, PAO1 (flagellin B control); lanes 2 and 3, mucoid, nonmotile CF isolate 1, AlgT+ and AlgT−, respectively; lanes 4 and 5, mucoid, nonmotile CF isolate 2, AlgT+ and AlgT−, respectively; lanes 6 and 7, mucoid, nonmotile CF isolate 3, AlgT+ and AlgT−, respectively. As a loading control, a second gel was simultaneously prepared and processed by Coomassie blue staining and contained equivalent amounts of total protein.
FIG. 6.
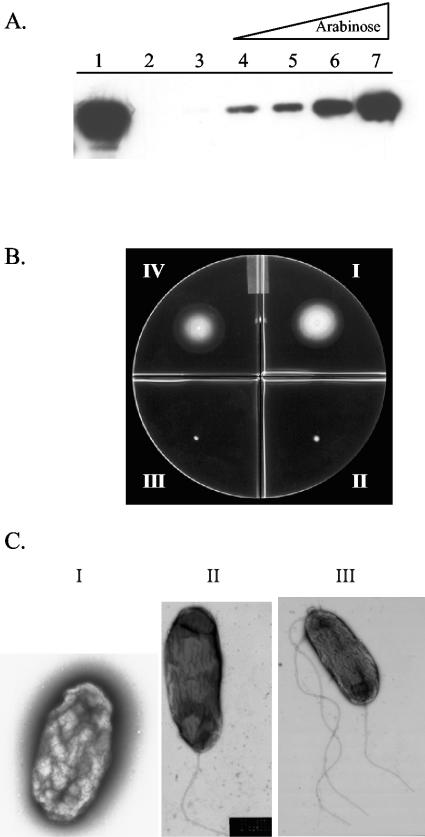
Inhibition of fleQ by AlgT is sufficient for the loss of flagellum expression. (A) Western blot of whole-cell lysates. Lane 1, type B flagellin; lane 2, FRD1; lane 3, FRD2700 (FRD1 with fleQ under the control of an arabinose-inducible promoter) with no arabinose; lanes 4 to 7, FRD2700 plus arabinose (lane 4, 0.1% arabinose; lane 5, 0.5% arabinose; lane 6, 1.0% arabinose; lane 7, 2.5% arabinose). (B) Motility assay on 0.3% agar. (I) PAO1 (motile control); (II) FRD1 (nonmotile control); (III) FRD2700 with no arabinose; (IV) FRD2700 plus 0.1% arabinose. (C) TEM of negatively stained P. aeruginosa. (I) FRD2700 with no arabinose; (II) FRD2700 plus 0.1% arabinose; (IV) FRD2700 plus 2.5% arabinose.
RESULTS
The majority of mucoid P. aeruginosa CF isolates do not express flagellin. The vast majority (98%) of P. aeruginosa isolates recovered from the environment or acute infections have a nonmucoid, motile phenotype (32). In contrast, P. aeruginosa strains obtained from chronic infections in the CF lung often display a mucoid and/or nonmotile phenotype. The mucoid phenotype has been attributed to the overexpression of the exopolysaccharide alginate upon deregulation of the alternative sigma factor AlgT (16, 24, 38). In addition, AlgT is required for the negative regulation of flagellum expression in the CF isolate FRD1 (mucA22) (23). We hypothesized that in CF isolates, the alginate phenotype and loss of flagellum expression are coordinately controlled. To address this, 20 mucoid CF isolates were examined for the presence of flagellin by Western blotting by probing with both anti-flagellin type A and B antibodies. A representative Western blot showing 11 of the 20 strains used in this study is presented in Fig. 1. The majority of mucoid P. aeruginosa strains from our collection lacked flagellin, indicating that they lacked the ability to express flagella. Of the 20 CF isolates tested, 14 (70%) were mucoid and nonflagellated, whereas 6 were mucoid and expressed flagellin. For the remainder of the study, representatives of the mucoid, nonflagellated subgroup of P. aeruginosa CF isolates were used.
Mucoid, nonmotile CF isolates contain mutations in mucA.
It has been shown that approximately 80% of mucoid P. aeruginosa CF isolates harbor a mutation in mucA, which encodes an anti-sigma factor that controls AlgT activity (24). We wanted to determine if there was a correlation between the mucA status and the inhibition of flagellin expression in mucoid, nonflagellated P. aeruginosa CF isolates. To address this, we amplified and sequenced mucA in three flagellated and five nonflagellated mucoid CF isolates. In all strains lacking flagellin, we observed mutations in mucA (data not shown). In three of these, the mutation was identical to that observed in FRD1 (mucA22) and involved the loss of a G residue, which yields a premature stop codon and thus a truncated MucA (24). In each of the mucoid, flagellated CF isolates, no mutation was detected in the mucA gene. This suggests that the AlgT-mediated inhibition of flagellum synthesis requires the inactivation of mucA and subsequent deregulation of AlgT activity.
AlgT mediates the negative regulation of flagellum biosynthesis in mucoid, nonmotile CF isolates.
Previous work has shown that in the mucoid, nonmotile CF isolate FRD1 (mucA22), AlgT is required for the inhibition of flagellum synthesis (23). Therefore, we were interested in determining whether this regulatory pathway is common among mucoid, nonflagellated P. aeruginosa isolates. We first generated algT mutants of several mucoid, nonmotile CF isolates. This was accomplished by culturing mucoid, nonmotile P. aeruginosa isolates under limiting oxygen conditions (17, 48). To identify algT mutants, the arising motile variants were complemented with algT. Subsequently, we amplified the algT gene of each mutant. Consistent with previous observations (17), the algT sequencing results revealed an algT29 mutation, which resulted in a Tyr at codon 29 being changed to Cys. The static-growth-derived algT mutants were used in subsequent experiments comparing isogenic AlgT+ and AlgT−.
To determine whether the AlgT-mediated inhibition of flagellum expression is conserved among mucoid, nonmotile CF isolates, we compared the flagellin expression of mucoid, nonflagellated CF isolates and that of their isogenic algT mutants (Fig. 2). The results showed that while the mucoid, nonmotile parental strains clearly lacked flagellin (lanes 2, 4, and 6), flagellin expression was restored in the algT mutants (lanes 3, 5, and 7). All algT mutants analyzed expressed type B flagellin. In addition, the algT mutants tested exhibited flagellum-mediated swimming motility. When these mutants were complemented with algT, alginate production was restored, and flagellar motility and flagellin expression were abolished (data not shown). These results designate AlgT as a regulator of flagellum biosynthesis in mucoid, nonmotile CF isolates of P. aeruginosa.
Microarray analysis of isogenic AlgT+ and AlgT− P. aeruginosa strains.
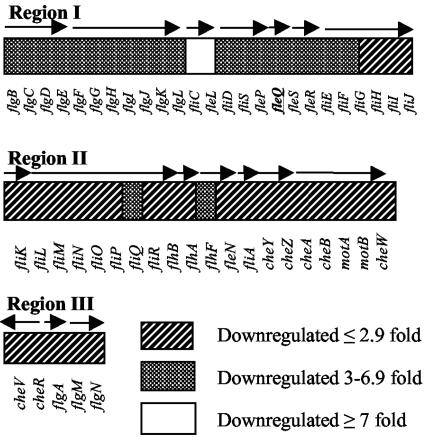
Upon identification of AlgT as a regulator of flagellum expression, we wanted to determine its target(s) within the complex flagellar hierarchy. Previous work in our laboratory showed that the AlgT-mediated control of flagellum biosynthesis within the flagellar hierarchy occurs downstream of rpoN, which codes for an alternative sigma factor involved in flagellum expression, and upstream of fliC, which encodes flagellin, the major component of the flagellar filament (23). In order to identify candidate AlgT-regulated genes, we performed single replicate microarray analysis of the mucoid, nonflagellated strain FRD1 (mucA22) and its isogenic nonmucoid, flagellated algT mutant, FRD440 (mucA22 algT::Tn501), by using a P. aeruginosa whole-genome microarray (Affymetrix). The summary of the entire microarray data set is provided in Table S2 in the supplemental material. Based on our analysis, we identified 552 candidate AlgT-controlled genes out of the 5,666 predicted open reading frames included on the microarray (see Table S1 in the supplemental material). As expected, the microarray data showed significantly higher mRNA levels for alginate biosynthetic and regulatory genes in the presence of AlgT. In contrast, the mRNA levels of many flagellar genes were significantly reduced in the AlgT+ strain FRD1. P. aeruginosa contains approximately 50 genes encoding structural and assembly components of the flagellum or proteins of the chemotaxis machinery, which are clustered in three regions of the bacterial chromosome (15). The most pronounced inhibitory effect of AlgT was seen in flagellar genes clustered in region I, which contains regulatory and structural genes involved in flagellum expression. These include fleQ and fleSR, coding for essential regulators of flagellum biosynthesis (15), and fliC, which encodes the main structural component of the flagellar filament. In region I, mRNA levels of fliC were 16-fold lower in the AlgT+ strain than in the AlgT− strain. The fliI and fliH genes, which code for an ATPase and a regulator of the ATPase protein, respectively (15), were least affected by AlgT. The genes clustered in region II and III were moderately affected by AlgT and were downregulated between two- and threefold. The effect of AlgT on mRNA levels of known flagellar genes is summarized in Fig. 3.
FIG. 3.
Organization of P. aeruginosa flagellar genes and the effect of AlgT on their expression. Microarray analysis comparing transcriptomes of isogenic AlgT+ and AlgT− P. aeruginosa strains was performed. Significantly lower levels of mRNA were observed for the majority of flagellar genes. fleQ, which encodes the flagellar master switch, was identified as the highest target of AlgT within the flagellar hierarchy. Arrows indicate the direction of transcription of the flagellar operons.
In addition to alginate and flagellar genes, a number of other genes encoding unidentified transcriptional regulators and two-component systems as well as toxins and antibiotic resistance proteins were also affected by AlgT (see Table S1 in the supplemental material). The data obtained by microarray analysis indicate that AlgT exerts broad-range regulatory effects on approximately 6% of the P. aeruginosa genome, with alginate and flagellar genes accounting for one-fifth of AlgT-regulated genes.
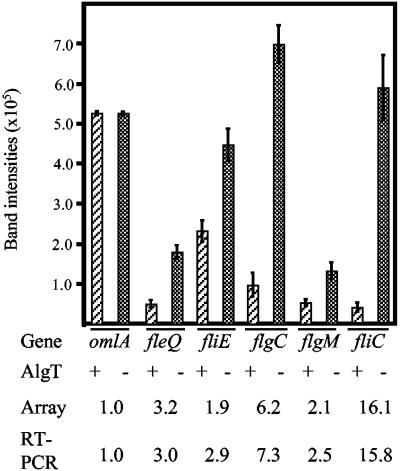
While the single replicate microarray analysis provided an overview of the AlgT effect on flagellar genes, we wanted to validate the obtained results and performed RT-PCR as an independent assay. We chose a representative for each class of the flagellar genes and analyzed mRNA levels in isogenic AlgT+ and AlgT− P. aeruginosa strains (Fig. 4). The class I gene fleQ was downregulated threefold in the presence of AlgT, which was comparable to the array data (3.2-fold). The class II gene fliE, which encodes a flagellar hook-basal body complex protein (15), was repressed 1.9-fold (array data, 2.9-fold) in the AlgT+ strain. Expression of flgC, which codes for a basal body rod protein (15) and belongs to the third class of flagellar genes, was reduced 7.3-fold in the presence of AlgT (array data, 6.2-fold). Class IV genes flgM, which codes for an anti-sigma factor (22), and fliC were downregulated 2.5- and 15.8-fold, respectively, which also corresponded to the array data (2.1- and 16.1-fold, respectively). As an internal control, we evaluated expression of the omlA gene, which encodes a constitutively expressed outer membrane lipoprotein (34). This was validated by the array data and the RT-PCR assay. Thus, the RT-PCR data provided strong support for the microarray data and independently confirmed the role of AlgT in the inhibition of flagellar gene expression.
FIG. 4.
RT-PCR comparing flagellar gene expression in isogenic AlgT+ and AlgT− P. aeruginosa strains. RT-PCR was performed on representatives of the four classes of flagellar genes to compare their mRNA levels in isogenic AlgT+ and AlgT− P. aeruginosa strains. The RT-PCRs were separated on an agarose gel, and band intensities were compared using a Kodak Image Station 2000RT system. omlA was included as constitutive control (34). Shown are the averages of four independent experiments and standard deviations. Differences (n-fold) in mRNA levels for both microarray and RT-PCRs are indicated.
AlgT inhibits the expression of the flagellar key regulator fleQ.
The results obtained from the microarray and the RT-PCR analysis showed that the majority of flagellar genes were downregulated in the presence of AlgT and that fleQ represented the highest target within the flagellar hierarchy. Since FleQ ultimately drives the expression of all other known flagellar genes (8, 15) with the exception of fliA, it is likely that the observed repression of other flagellar genes occurs as a result of the inhibition of fleQ. To further evaluate the AlgT effect on fleQ, we fused the fleQ promoter with a promoterless lacZ and integrated the fusion into the chromosomes of the mucoid, nonflagellated strain FRD1 and the isogenic algT mutant FRD440. As an internal control, we generated lacZ fusions to the constitutive omlA promoter (34). The activities of the fleQ promoter in the AlgT+ and AlgT− strains were determined with β-galactosidase assays (33). In FRD1, fleQ promoter activity is significantly reduced in the presence of AlgT (Fig. 5). To determine whether this AlgT-mediated inhibition of fleQ is conserved among mucoid, nonmotile CF isolates, we introduced fleQ-lacZ fusions into two mucoid, nonflagellated CF isolates and their isogenic algT mutants. The results of this experiment revealed that, as in FRD1, the fleQ promoter activity in these mucoid, nonmotile CF isolates was reduced in the presence of AlgT (Fig. 5).
FIG. 5.
AlgT inhibits fleQ expression. fleQ::lacZ fusions were integrated into the chromosomes of mucoid, nonmotile P. aeruginosa CF isolates and their isogenic algT mutants at the neutral attB site. Promoter activity was measured with β-galactosidase assays using ONPG as a substrate and determined as amounts of ONPG hydrolyzed min−1 as a function of cell density. omlA::lacZ fusions were included as a constitutive control (34). Shown are the averages of four independent experiments and standard deviations.
Negative control of fleQ by AlgT is sufficient to inhibit P. aeruginosa flagellum biosynthesis.
While we demonstrated that high levels of active AlgT result in the negative transcriptional regulation of fleQ, we also needed to determine whether this inhibition of fleQ is sufficient for the loss of flagellum expression. We hypothesized that if the repression of fleQ by AlgT is sufficient to abolish flagellum biosynthesis, increased expression of FleQ in mucoid, nonmotile cells should overcome the inhibitory AlgT effect. We constructed pAT6 by placing fleQ under the control of an arabinose-inducible promoter. Plasmid pAT6 was introduced into the chromosomes of FRD1 and the fleQ deletion strain PAKΔQ. The resulting strains, FRD2700 and PAKΔQ/pAT6, were grown in the absence and presence of arabinose and examined for flagellin expression by Western blot (Fig. 6). In the absence of the inducer, FRD2700 did not express flagellin (Fig. 6A, lane 3). However, as the concentration of inducer was increased, a dose response increase in expression of flagellin was observed (6A, lanes 4 to 7). The results obtained for PAKΔQ/pAT6 mirrored those observed for FRD2700 (data not shown).
To determine if elevated levels of FleQ also promote motility, we performed motility assays using soft agar with and without arabinose. In the absence of the inducer, FRD2700 was nonmotile, as indicated by the lack of a motility zone observed on the motility plates (Fig. 6B, section III). In contrast, overexpression of FleQ in FRD2700 restored flagellum-mediated motility, resulting in a zone of motility (32 mm) comparable to that observed for the motile control, PAO1 (33 mm) (Fig. 6B, sections IV and I, respectively). Similar results were obtained when the motility of strain PAKΔQ/pAT6 was examined on soft agar plates with or without arabinose (data not shown). In addition to performing these experiments, we examined FRD2700 by TEM to determine whether the bacteria were expressing a normal polar flagellum. Cells grown in the absence of the inducer did not express flagella (Fig. 6C, panel I). In the presence of the 0.1% arabinose, FRD2700 expressed a single polar flagellum characteristic for P. aeruginosa (Fig. 6C, panel II). Upon addition of higher concentrations of arabinose, FRD2700 became hyperflagellated. Flagella were observed on both poles of the cells, and flagellar numbers varied between three and eight (Fig. 6C, panel III is a representative). The mucoid phenotype of FRD2700 was unaltered upon induction of flagellar motility, which implies that the mucoid and the motile phenotypes are not mutually exclusive.
DISCUSSION
In the United States, approximately 30,000 individuals suffer from the recessive genetic disorder cystic fibrosis. Pulmonary complications as a result of this disease remain the primary concern for these patients, as they are exceedingly difficult to treat, especially upon colonization with P. aeruginosa (13). Two critical factors allowing P. aeruginosa to persist in the airways of CF patients include its ability to form biofilms and its conversion to a mucoid phenotype, which is frequently accompanied by the loss of flagellum expression. Previous work performed in our laboratory as well as this study shows a link between alginate biosynthesis and flagellum expression, which are inversely regulated by the alternative sigma factor AlgT (23). While the role of AlgT in alginate expression is well defined (24, 38), its function in the negative control of flagellum biosynthesis remains to be elucidated. In this report, we show that expression of flagellar genes is significantly reduced in the presence of AlgT. The fleQ gene, a crucial regulator in the flagellar regulon, was identified as the highest target of AlgT within the flagellar hierarchy. In addition, we show that increased expression of FleQ in mucoid, nonflagellated P. aeruginosa CF isolates reverses the AlgT-mediated inhibition of flagellum expression. Taken together, our data indicate that AlgT represses P. aeruginosa flagellum biosynthesis by inhibiting expression of the flagellar regulator fleQ.
While it has been suggested that there is no correlation between mucoidy and loss of flagellar motility in CF isolates (32), we observed that 70% of mucoid P. aeruginosa strains from CF patients did not express flagellin. These data indicated that in a subgroup of mucoid CF isolates, flagellum expression and alginate expression are inversely regulated. In most cases (80%), the conversion of P. aeruginosa to a mucoid phenotype in the CF lung is due to an inactivating mutation in mucA which results in deregulation of the alternative sigma factor AlgT (24). We observed that each of the mucoid CF isolates that lacked flagellin harbored a mutation in mucA. Sequencing of mucA in the mucoid, flagellated CF isolates of P. aeruginosa revealed that this gene was intact. In the latter case, overexpression of alginate is MucA independent and may involve the secondary, RpoN-dependent pathway (12).
It has been suggested that the mucoid phenotype and the lack of flagella provide P. aeruginosa with a selective advantage in the CF lung (32, 38). The copious quantities of exopolysaccharide form a barrier that shields the bacteria from inflammatory mediators as well as antibiotics. Moreover, infection of airway epithelial cells with mucoid P. aeruginosa results in increased expression of epithelial cellular genes with antiapoptotic effects. The presence of alginate not only attenuates host responses but also aids in the bacterial circumvention of host defenses (14). Flagella, on the other hand, are potent inducers of inflammatory mediators (14, 27, 28, 37). While flagella are essential in the colonization of the CF lung (6, 7, 42), they may become detrimental as the disease progresses. Thus, both loss of the flagellar structure and overproduction of alginate are advantageous for P. aeruginosa, as those allow it to evade the immune system and facilitate its persistence in the CF lung.
The inhibition/regulation of flagellum biosynthesis in response to external conditions is commonly observed among gram-negative bacteria. Usually, if the environmental conditions impose stress on the cells, flagellum synthesis is reduced or eliminated (2, 30). These conditions include nutrient availability, temperature, ionic composition, pH, and surface interactions (3, 30, 31, 40). Similar effects of environmental conditions have also been observed for Salmonella enterica serovar Typhimurium (3, 31). In both E. coli and Salmonella serovar Typhimurium, environmental conditions have been shown to regulate flagellum expression at the level of transcription. H-NS, a nucleoid protein, and a wide variety of transcription factors and two-component response regulators often modulate motility (1, 11, 21). In Bordetella bronchiseptica, the two-component system BvgAS is involved in the downregulation of flagella in response to external stimuli, such as MgSO4 or nicotinic acid (4). Therefore, it is possible that the FleQ-dependent repression of flagellum expression we uncovered in P. aeruginosa also occurs in response to environmental stress signals. In this context, Wolfgang et al. demonstrated that P. aeruginosa reduces flagellum synthesis when grown in the presence of mucopurulent respiratory liquids derived from CF patients. Upon exposure to CF airway fluid, an 80% reduction of fliC mRNA was observed, indicating the presence of a repressive signal in this environment (45).
We have shown that AlgT mediates alginate and flagellum biosynthesis in P. aeruginosa in a reciprocal fashion. There is evidence in other bacterial species, including E. coli (36) and Vibrio cholerae (44), for a similar inverse control of flagellum and exopolysaccharide biosynthesis. In E. coli, downregulation of flagellum synthesis is often accompanied by an increase of colanic acid exopolysaccharide expression (36). In V. cholerae, mutations in flrA and flrC, which code for regulatory genes of flagellum biosynthesis, resulted in the induction of exopolysaccharide synthesis (44). These examples suggest that the coordinate regulation of exopolysaccharide production and flagellum expression may be a common feature of gram-negative bacterial species.
The assembly of a functional flagellum requires the coordinate expression of approximately 50 genes and is energetically costly. In E. coli, as much as 2% of the total biosynthetic energy expenditure is required for the synthesis and function of flagella (31). Flagellum genes are organized in an ordered regulatory cascade in which the expression of later genes requires the presence of earlier gene products. In peritrichously flagellated bacterial species, such as E. coli or Salmonella serovar Typhimurium, flagellum biosynthesis follows a three-tiered hierarchy, with flagellar genes being grouped into classes I to III. In contrast, monoflagellates, such as P. aeruginosa and Vibrio parahaemolyticus, regulate flagellum biosynthesis with a four-tiered cascade (classes I to IV) (5). In this report, we observed that the repression of flagellum synthesis in P. aeruginosa occurs by transcriptional inhibition of fleQ, a class I gene. The fleQ gene is the earliest in the transcriptional regulatory hierarchy and controls expression of all remaining genes of the flagellar regulon with the exception of fliA. As fleQ forms the master switch of the P. aeruginosa flagellar regulatory network, its reduced expression would ultimately have an inhibitory effect on downstream genes within the cascade. This is similar to the shutoff of flagellum synthesis in E. coli, where the class I master operon flhDC is transcriptionally repressed (40, 41).
Flagella are common among gram-negative bacteria and allow the microorganisms to reach favorable envirommental conditions and avoid detrimental environmental conditions. Thus, bacteria can successfully compete with other microbial species and colonize ecological niches optimal for their growth. Interestingly, flagella are also involved in virulence, adhesion, colonization, and biofilm formation. Stressful external conditions affect flagellar motility in similar fashions and often result in the inhibition of flagellum biosynthesis. The ability to tightly regulate the expression of flagella provides bacteria with the capacity to adapt to their changing environments. In opportunistic pathogens such as P. aeruginosa, for instance, the requirements for flagellum expression vary depending on their mode of growth within their host or within the environment (free swimming versus biofilm), and in each case, such expression or loss thereof provides the bacterium with a selective advantage. To persist in the CF lung, P. aeruginosa must be able to circumvent the immune defenses of the host, and repression of flagellum biosynthesis aids in accomplishing this goal. In mucoid, nonflagellated CF isolates of P. aeruginosa, flagellum expression is shut off as the result of the transcriptional inhibition of fleQ by the alternative sigma factor AlgT. A detailed knowledge of the mechanism of this inhibitory process in P. aeruginosa not only may be useful in understanding the adaptability of this opportunistic pathogen to its various environments but may also elucidate similar pathways in other flagellated gram-negative microorganisms.
Supplementary Material
Acknowledgments
We thank Rajendar Deora and Yolanda Sanchez for helpful comments during the preparation of this work. We also thank Ken Grant for assistance in the electron microscopy assays. We thank Stephen Lory for providing RNA controls and access to GeneChip hybridization and scanning instrumentation. Finally, we acknowledge Reuben Ramphal for kindly providing us with requested strains and plasmids and Sam Woolwine for generating pSW218.
The work was supported by Public Health Service grants AI-35177 and HL-58334 (D.J.W.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, P., R. Fowler, N. Kinsella, G. Howell, M. Farris, P. Coote, and C. D. O'Connor. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J., and B. Templeton. 1967. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46:175-184. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa, S.-I. 2004. The desk encyclopedia of microbiology. Academic Press, San Diego, Calif.
- 4.Akerley, B. J., and J. F. Miller. 1993. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J. Bacteriol. 175:3468-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 6.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1996. Cloning and characterization of Pseudomonas aeruginosa fliF, necessary for flagellar assembly and bacterial adherence to mucin. Infect. Immun. 64:2130-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. [DOI] [PubMed] [Google Scholar]
- 10.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-952. [DOI] [PubMed] [Google Scholar]
- 11.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucher, J. C., M. J. Schurr, and V. Deretic. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 36:341-351. [DOI] [PubMed] [Google Scholar]
- 13.Boucher, R. C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146-158. [DOI] [PubMed] [Google Scholar]
- 14.Cobb, L. M., J. C. Mychalecky, D. J. Wozniak, and Y. S. Lopez-Boado. 2004. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: implications for cystic fibrosis disease. J. Immunol. 173:5659-5670. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 16.Deretic, V., M. J. Schurr, and H. Yu. 1995. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 3:351-356. [DOI] [PubMed] [Google Scholar]
- 17.Devries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake, D., and T. C. Montie. 1988. Flagella, motility and invasive virulence of Pseudomonas aeruginosa. J. Gen. Microbiol. 134:43-52. [DOI] [PubMed] [Google Scholar]
- 19.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn, J. L., and D. E. Ohman. 1988. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J. Bacteriol. 170:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 22.Frisk, A., J. Jyot, S. K. Arora, and R. Ramphal. 2002. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J. Bacteriol. 184:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett, E., and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi, F., K. D. Smith, A. Oznisky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 26.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: applications for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 27.Honko, A. N., and S. B. Mizel. 2004. Mucosal administration of flagellin induces innate immunity in the mouse lung. Infect. Immun. 72:6676-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hybiske, K., J. K. Ichikawa, V. Huang, S. J. Lory, and T. E. Machen. 2004. Cystic fibrosis airway epithelial cell polarity and bacterial flagellin determine host response to Pseudomonas aeruginosa. Cell. Microbiol. 6:49-63. [DOI] [PubMed] [Google Scholar]
- 29.Jyot, J., N. Dasgupta, and R. Ramphal. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 184:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, C., C. J. Louise, W. Shi, and J. Adler. 1993. Adverse conditions which cause lack of flagella in Escherichia coli. Mol. Microbiol. 175:2229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 32.Mahenthiralingam, E., M. E. Campbell, and D. P. Speert. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62:596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 34.Ochsner, U. A., A. I. Vasil, Z. Johnson, and M. L. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181:1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 36.Prigent-Combaret, C., C. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos, H. C., M. Rumbo, and J. C. Sirard. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509-517. [DOI] [PubMed] [Google Scholar]
- 38.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 39.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, W., C. Li, C. J. Louise, and J. Adler. 1993. Mechanism of adverse conditions causing lack of flagella in Escherichia coli. J. Bacteriol. 175:2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson, D. A., R. Ramphal, and S. Lory. 1995. Characterization of fliO, a gene involved in flagellar biosynthesis and adherence. Infect. Immun. 63:2950-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasil, M. L. 1986. Pseudomonas aeruginosa: biology, mechanisms of virulence, epidemiology. J. Pediatr. 108:800-805. [DOI] [PubMed] [Google Scholar]
- 44.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfgang, M. C., J. Jyot, A. L. Goodman, R. Ramphal, and S. Lory. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 101:6664-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 47.Wozniak, D. J., A. B. Sprinkle, and P. J. Baynham. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 185:7297-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyckoff, T. J. O., B. Thomas, D. J. Hassett, and D. J. Wozniak. 2002. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 148:3423-3430. [DOI] [PubMed] [Google Scholar]
- 49.Wyckoff, T. J. O., and D. J. Wozniak. 2001. Transcriptional analysis of genes involved in Pseudomonas aeruginosa biofilms. Methods Enzymol. 336:144-151. [DOI] [PubMed] [Google Scholar]
- 50.Xie, Z. D., C. D. Hershberger, S. Shankar, R. W. Ye, and A. M. Chakrabarty. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J. Bacteriol. 178:4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.