Abstract
Dihydroxyphenylalanine (DOPA) melanins formed from tyrosine by tyrosinases are found in microorganisms, plants, and animals. Most species in the soil-dwelling, gram-positive bacterial genus Streptomyces produce DOPA melanins and melanogenesis is one of the characteristics used for taxonomy. Here we report a novel melanin biosynthetic pathway involving a type III polyketide synthase (PKS), RppA, and a cytochrome P-450 enzyme, P-450mel, in Streptomyces griseus. In vitro reconstitution of the P-450mel catalyst with spinach ferredoxin-NADP+ reductase/ferredoxin revealed that it catalyzed oxidative biaryl coupling of 1,3,6,8-tetrahydroxynaphthalene (THN), which was formed from five molecules of malonyl-coenzyme A by the action of RppA to yield 1,4,6,7,9,12-hexahydroxyperylene-3,10-quinone (HPQ). HPQ readily autopolymerized to generate HPQ melanin. Disruption of either the chromosomal rppA or P-450mel gene resulted in abolishment of the HPQ melanin synthesis in S. griseus and a decrease in the resistance of spores to UV-light irradiation. These findings show that THN-derived melanins are not exclusive in eukaryotic fungal genera but an analogous pathway is conserved in prokaryotic streptomycete species as well. A vivid contrast in THN melanin biosynthesis between streptomycetes and fungi is that the THN synthesized by the action of a type III PKS is used directly for condensation in the former, while the THN synthesized by the action of type I PKSs is first reduced and the resultant 1,8-dihydroxynaphthalene is then condensed in the latter.
The genus Streptomyces comprises gram-positive, soil-dwelling, filamentous bacteria with a complex life cycle similar to that of fungi. In addition to the complex morphological differentiation, Streptomyces is also characterized by its ability to produce a wide variety of secondary metabolites, including pharmaceutically useful compounds, such as antibiotics, antitumor agents, and immunosuppressants (3). Melanins, which are high-molecular-weight dark-brown to black pigments, are secondary metabolites produced by diverse species of streptomycetes and are used for the taxonomy of this genus (24, 25). Various organisms synthesize a so-called dihydroxyphenylalanine (DOPA) melanin, which is a nonenzymatically or laccase-mediated polymerized product from DOPA. DOPA is formed from tyrosine by the oxidative action of tyrosinases. The representative of the tyrosinases responsible for the formation of DOPA melanin in Streptomyces is MelC2 (5), which is a copper-containing monooxygenase that catalyzes the o-hydroxylation of monophenols and the oxidation of o-diphenols to yield o-quinones using molecular oxygen (2). Because tyrosine occurs universally in living organisms, tyrosinases are also responsible for browning in plants and melanization in animals (2).
We previously found that a mutation in rppA, encoding a type III polyketide synthase (PKS), caused the host, Streptomyces griseus, to show an “albino” phenotype (8). Although S. griseus produces DOPA melanin, its production is apparent only when copper is added to the medium (5). RppA catalyzes condensation of five molecules of malonyl-coenzyme A (CoA) to form 1,3,6,8-tetrahydroxynaphthalene (THN) (8). These findings suggested the presence of a novel melanin biosynthetic pathway, via not DOPA but THN, in S. griseus (8), although the details of the presumptive pathway remained unknown.
For elucidation of the predicted melanin biosynthetic pathway, we focused on an open reading frame, named P-450mel, as a neighbor of rppA (Fig. 1), since functionally related genes are often organized in operons. P-450mel is a member of the cytochrome P-450 family, which are a superfamily of hemoproteins catalyzing monooxygenation of a wide range of compounds (13, 14). Because P-450mel constitutes an operon with rppA not only in S. griseus but also in several other Streptomyces species (7), we expected that P-450mel might be involved in the formation of a novel melanin by modifying THN in some way.
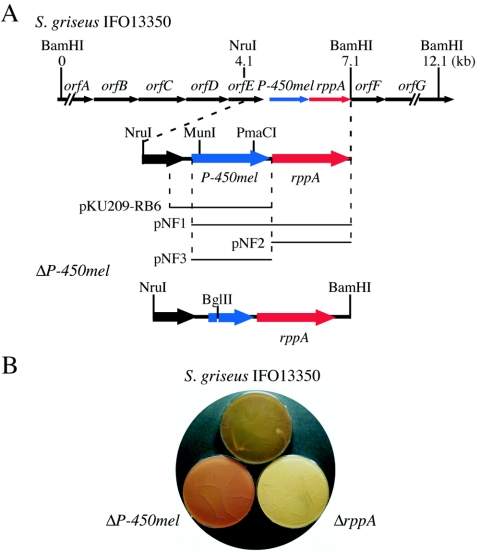
FIG. 1.
Effects of rppA and P-450mel on melanin production by S. griseus. (A) Gene organization of the S. griseus chromosomal region including the rppA and P-450mel loci and the DNA fragments on the plasmids used in this study. The ΔP-450mel mutant contains an in-frame deletion from Glu-84 to Tyr-250. OrfA is similar to transcriptional factors of the AraC family. The orfBCDE operon constitutes an ABC transporter, because of the similarity of OrfB to substrate-binding extracellular domains, OrfCD to membrane integral subunits, and OrfE to ATP-binding proteins. OrfF and OrfG are similar to subtilisin-type serine proteases and chitinases, respectively. (B) Phenotypes of ΔP-450mel and ΔrppA mutants. The strains were grown at 28°C for 3 days on R5 medium.
As expected, P-450mel was found to catalyze aryl coupling of THN to yield 1,4,6,7,9,12-hexahydroxyperylene-3,10-quinone (HPQ) in an in vitro-reconstituted system with spinach ferredoxin/ferredoxin reductase. By analyzing the intermediates accumulated in rppA and/or P-450mel mutants, we also confirmed that HPQ is formed in vivo by the action of P-450mel from the THN that has been formed from malonyl-CoA by the action of RppA. HPQ polymerized nonenzymatically to yield a brownish pigment. Because the spores of the P-450mel mutant showed decreased resistance to UV irradiation, we named the brownish pigment HPQ melanin. The biosynthesis of HPQ melanin by a type III PKS and P-450 represents a novel melanin biosynthetic pathway, although THN-derived melanin is also produced by fungi. However, the THN used for melanin biosynthesis in fungi is a product of type I PKSs, which are evolutionarily distinct from type III PKSs.
MATERIALS AND METHODS
Bacterial strains, plasmids and media.
S. griseus IFO13350 was obtained from the Institute of Fermentation, Osaka (IFO). Streptomyces lividans TK21 was obtained from D. A. Hopwood (11). Escherichia coli JM109 and plasmid pUC19, used for DNA manipulation, were purchased from Takara Biochemicals. For expression of P-450mel and rppA in Streptomyces spp., pIJ6021 containing a thiostrepton-inducible tipA promoter (19) was used. pKU209 (11) with its copy number of one to two per chromosome was used as a Streptomyces vector.
For production of His-tagged proteins in E. coli BL21(DE3), pET26b (Novagen) was used. The media, growth conditions, and general recombinant DNA techniques for E. coli were described by Maniatis et al. (12). S. lividans was routinely cultured at 30°C in yeast extract-malt extract (YEME) medium (11), which was supplemented with 5 μg/ml of kanamycin or thiostrepton, when necessary. Restriction enzymes and other DNA-modifying enzymes were purchased from Takara Biochemicals. [α-32P]dCTP for DNA labeling with the Takara BcaBest DNA labeling system was purchased from Amersham Biotech. Nucleotide sequences were determined with the Amersham Thermo Sequenase fluorescently labeled primer cycle sequencing kit on an automated DNA sequencer. All PCR experiments were conducted by using the chromosomal DNA of the wild-type strain, S. griseus IFO13350, if a template is not indicated. The absence of undesired alterations during PCR was checked by nucleotide sequencing.
Disruption of P-450mel in S. griseus.
An in-frame deletion in the chromosomal P-450mel gene was introduced by replacing the region encoding from Glu-84 to Tyr-250 with a BglII recognition sequence of six nucleotides (Fig. 1A). A 0.2-kb region upstream from the Glu-84 codon was amplified by PCR with primer I, 5′-GCGAAGCTTCAATTGCTCACCGAGGAGCC-3′ (the italic letters and the underline indicate HindIII and MunI sites, respectively) and primer II: 5′-CGCGAATTCAGATCTCCGGGGTCATCCGGGG-3′ (the boldface letters correspond to the Glu-84 codon, the italic letters indicate an EcoRI site, and the underline indicates a BglII site). The amplified fragment was cloned between the EcoRI and HindIII sites of pUC19, resulting in pUC19-N-BglII. A 0.2-kb region downstream from the Tyr-250 codon was amplified with primer III, 5′-GCGAAGCTTAGATCTACCAGCTCGGCAACATCGCC-3′ (the boldface letters indicate the Tyr-250 codon, the italic letters indicate a HindIII site, and the underline indicates a BglII site), and primer IV, 5′-CGCGAATTCGTAGGACACGTGCACCACGTC-3′ (the italic letters and the underline indicate EcoRI and PmaCI sites, respectively). The amplified fragment was cloned between the EcoRI and HindIII sites of pUC19, resulting in pUC19-C-BglII.
The MunI-EcoRI fragment excised from the pUC19-N-BglII and the 2.4-kb HindIII-MunI fragment, excised from pUC19-RB42 (21), were cloned into pUC19 by three-fragment ligation, resulting in pUC19-ΔP450-N. The HindIII-PmaCI fragment excised from the pUC19-C-BglII and the 2.5-kb PmaCI-EcoRI fragment excised from pUC19-RB44 (21) were similarly cloned into pUC19, resulting in pUC19-ΔP450-C. The HindIII-BglII fragment from pUC19-ΔP450-N and the BglII-EcoRI fragment from pUC19-ΔP450-C were ligated via the common BglII site and cloned between the HindIII and EcoRI sites of pUC19. The kanamycin resistance gene from Tn5 was inserted in the HindIII site of the resultant plasmid, resulting in pUC19-ΔP450. pUC19-ΔP450 was denatured with NaOH and introduced by protoplast transformation into S. griseus IFO13350.
Transformants containing pUC19-ΔP450 in the chromosome as a result of single crossover were selected among kanamycin-resistant colonies. One of the kanamycin-resistant colonies was grown for a week on YMPD agar (10) without kanamycin. Spores recovered were spread on YMPD agar without kanamycin. From these colonies, one ΔP-450mel mutant, in which the P-450mel gene on the chromosome was in-frame deleted, was isolated as a kanamycin-sensitive colony. The correct replacement was confirmed by Southern hybridization.
Construction of pKU209-RB6.
A 1.6-kb DNA fragment containing the promoter and P-450mel coding sequence was amplified with primer V, 5′-GCGAAGCTTCTCGAGGAGGTGCTGACGGAGGAGAC-3′ (the italic letters and the underline indicate HindIII and XhoI sites, respectively), and primer VI, 5′-CGCGAATTCCATGGGTTCTCCTCGTTCTCTGGA-3′ (the italic letters indicate an EcoRI site and the boldface letters represent the start codon of rppA). The HindIII-EcoRI fragment excised from the amplified DNA fragment was cloned between the HindIII and EcoRI sites of pUC19, resulting in pUC19-RB6. The XhoI-EcoRI fragment excised from pUC19-RB6 was cloned between the XhoI and EcoRI sites of pKU209, resulting in pKU209-RB6.
Construction of plasmids for expression of rppA and/or P-450mel in S. lividans.
For construction of pNF1, which contained both P-450mel and rppA under the control of the thiostrepton-inducible tipA promoter in pIJ6021 (Fig. 1A), an NdeI site was introduced at the start codon of P-450mel by PCR with primer VII, 5′-GCGGAATTCCATATGGAGAACACCTCGGTGCAGAA-3′ (the italic letters and the underline indicate EcoRI and NdeI sites, respectively), and primer VIII, 5′-GCGAAGCTTGGTTGAAGCTCTTGGCCACC-3′(the italic letters indicate a HindIII site). The amplified fragment was cloned between the EcoRI and HindIII sites of pUC19, resulting in pUC19-P-450mel-N. The NdeI-MunI fragment (an MunI site is present within the amplified fragment) excised from pUC19-P-450mel-N and the MunI-BamHI fragment excised from pUC19-RB42 were ligated via the MunI site and inserted between the NdeI and BamHI sites in pET16b, resulting in pET16b-NF1. The NdeI-BamHI fragment excised from pET16b-NF1 was cloned between the NdeI and BamHI sites of pIJ6021, resulting in pNF1.
For construction of pNF2 containing rppA under the control of the tipA promoter, the NdeI-BamHI fragment excised from pET16b-RppA (8) was cloned between the NdeI and BamHI sites of pIJ6021.
For construction of pNF3 containing P-450mel under the control of the tipA promoter, an NdeI site was introduced at the start codon of P-450mel by PCR with primer VII and primer IX, 5′-CGCGGATCCGGGTTCTCCTCGTTCTCTGGA-3′ (the italic letters indicate a BamHI site). The amplified fragment containing the region from Met-1 of P-450mel to the end of its 3′-flanking region was cloned between the EcoRI and BamHI sites of pUC19, resulting in pUC19-P-450mel. The NdeI-BamHI fragment excised from pUC19-P-450mel was cloned between the NdeI and BamHI sites of pIJ6021.
High-performance liquid chromatography analysis of in vivo products.
S. lividans TK21 harboring pNF1, pNF2, pNF3, or pIJ6021 was grown at 30°C for 24 h in YEME medium. Thiostrepton was added at a final concentration of 5 μg/ml to induce the tipA promoter and the culture was continued for 36 h. The material in the culture broth was extracted with ethyl acetate, dried, and applied to high-performance liquid chromatography (HPLC). The HPLC conditions were an ODS-80Ts column (Tosoh) eluted with a linear gradient from 5 to 40% CH3CN in water (each containing 2% acetic acid) over 30 min and then 100% CH3CN within 10 min at a flow rate of 0.8 ml/min. UV absorbance was detected at 254 nm and 440 nm. In vivo products of wild-type, ΔrppA, and ΔP-450mel S. griseus IFO13350 strains were similarly analyzed, except that these strains were grown at 28°C for 4 days on R5 agar medium (11) and the material produced was extracted by ethyl acetate after homogenization of the agar.
Isolation and identification of polyketides produced by S. lividans harboring pNF1.
S. lividans TK21 harboring pNF1 was grown at 30°C in 1 liter of YEME medium containing 5 μg/ml of kanamycin. After 24 h, thiostrepton was added to give a final concentration of 5 μg/ml for induction of the tipA promoter and the culture was continued for a further 24 h. The culture broth was adjusted to pH 1.0 with 6 M HCl and 500 ml of ethyl acetate was added and vigorously mixed. The mixture was then passed through a pad of Celite to remove the emulsion and the organic layer was collected. The Celite was washed five times with 100 ml of ethyl acetate and twice with 50 ml of acetone. The organic layers were combined, washed with brine, and dried with Na2SO4. After evaporation to dryness, the residue was methylated by trimethylsilyl-diazomethane in methanol. The resultant mixture was separated by silica gel flash chromatography using ethyl acetate containing 1% acetic acid as an eluant, and the major compound was further purified by reversed-phase preparative HPLC (Docosil B [C22], 20 by 250 mm) by a linear gradient of 80 to 100% CH3CN in water (each containing 1% acetic acid) at a flow rate of 6 ml/min to provide 2 mg of 4,9-dihydroxy-1,6,7,12-tetramethoxyperylene-3,10-quinone as a red solid. 1H nuclear magnetic resonance (NMR) (500 MHz, CDCl3): δ 6.53 (s, 4H, C-2, C-5, C-8, C-11), 4.01 (s, 12H, OCH3 × 4); 13C NMR (125 MHz, CDCl3) δ 178.3 (C-3, C-4, C-9, C-10), 165.9 (C-1, C-6, C-7, C-12), 127.8 (C-9b, C-12c), 119.5 (C-6a, C-6b, C-12a, C-12b), 102.7 (C-2, C-5, C-8, C-11), 102.2 (C-3a, C-9a), 56.3 (OCH3); high-resolution electrospray ionization (positive)/time-of-flight mass spectrum (HRESI+/TOF-MS) m/z 435.10733 (calculated for C24H19O8, 0.66 mmu error).
Production and purification of P-450mel.
The nucleotide sequence (TGACGA) covering the TGA stop codon of P-450mel was changed to CTCGAG to create an XhoI site by PCR with primer X, 5′-GCGAAGCTTCATATGGAGAACACCTCG-3′ (the italic letters indicate a HindIII site, the underline indicates an NdeI site, and the boldface letters indicate the start codon of P-450mel), and primer XI, 5′-GCGGAATTCCTCGAGCCATGTGACGGGC-3′ (the italic letters indicate an EcoRI site and the underline indicates an XhoI site). The amplified 1.2-kb fragment was cloned between the HindIII and EcoRI sites of pUC19, resulting in pUC19-P-450mel-Xho. The NdeI-XhoI fragment excised from pUC19-P-450mel-Xho was cloned between the NdeI and XhoI sites of pET26b, resulting in pET26b-P-450mel.
For production of P-450mel with a His tag at its C terminus, E. coli BL21(DE3) harboring pET26b-P-450mel was grown at 37°C for 2 h in M9 medium containing 100 μM FeSO4 and 100 μg/ml of ampicillin, followed by addition of 5-aminolevulinic acid hydrochloride (final, 80 μg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 100 μM). After further incubation at 22°C for 1 day, the cells were collected by centrifugation. A crude cell lysate was prepared by sonication and removal of cell debris by centrifugation at 10,000 × g for 30 min. P-450mel was purified by using a Ni-nitrilotriacetic acid column (QIAGEN), according to the manual from the manufacturer. Protein concentrations were measured with a Bio-Rad protein assay kit using bovine serum albumin as a standard. The sodium dithionite-reduced carbon monoxide difference spectrum of purified P-450mel was measured by the method described by Omura and Sato (16).
In vitro reconstitution of P-450mel activity.
The reactions, containing 100 mM sodium phosphate (pH 7.3), 1 mM dithiothreitol, 1 mM EDTA, 10% glycerol, 1 mM NADPH, 0.5 U of spinach ferredoxin-NADP+ reductase, 40 μg of spinach ferredoxin, 23.4 μg of P-450mel, and 400 μM of THN, were performed in a total volume of 500 μl. Ferredoxin and ferredoxin-NADP+ reductase from spinach were purchased from Sigma. THN was synthesized according to the method of Ichinose et al. (9). The reactions were carried out at 30°C for 30 min and terminated by adding 50 μl of 6 M HCl, and extracted with 200 μl of ethyl acetate. The organic layer was collected and evaporated and the residual material was dissolved in 20 μl of methanol for HPLC analysis. Conditions of HPLC were as follows: ODS-80Ts column (4.6 by 150 mm, Tosoh), maintained at 40°C, eluted with 25 mM KH2PO4 (pH 4.7) containing 13% CH3CN at a flow rate of 1 ml/min.
Spore survival after UV irradiation.
Approximately 9 × 107 spores of wild-type IFO13350, ΔP-450mel, and ΔrppA S. griseus strains were suspended in 20 ml of 10% glycerol, stirred at room temperature, and then irradiated with 254-nm UV light at a distance of 30 cm. A portion (0.1 ml) of the UV-irradiated spores of appropriate dilutions at different time intervals was spread on R5 medium. Surviving spores formed visible colonies after incubation at 30°C for 1 day, and colonies were counted the next day. Spore survival was the ratio of the number of colonies that appeared after administration of an appropriate dose of UV irradiation to that of colonies that appeared without UV irradiation.
Nucleotide sequence accession number.
The nucleotide sequence of the 12.1-kb BamHI fragment including P-450mel has been deposited in the DDBJ database with accession number AB218878.
RESULTS
Involvement of P-450mel and rppA in melanogenesis of S. griseus.
The mycelium and spores of S. griseus IFO13350 are brownish and greenish, respectively, when grown on routine YMPD and R5 media. We previously found that disruption of the chromosomal rppA gene in S. griseus caused the host strain to show an albino phenotype (8); the mycelium and spores of the ΔrppA mutant remained colorless. The DNA databases revealed that in some actinobacteria, such as Streptomyces coelicolor A3(2), Streptomyces avermitilis, and Saccharopolyspora erythraea, rppA appeared to form an operon with a gene encoding a P-450 enzyme (7). These observations led us to hypothesize that the THN formed from malonyl-CoA by the action of RppA would be further modified by the P-450s to yield a precursor for melanin.
We inactivated the chromosomal P-450mel gene by means of in-frame deletion to examine possible involvement of this gene, as a neighbor of rppA, in pigmentation (Fig. 1A). Correct deletion was checked by Southern hybridization with the 6.5-kb BamHI fragment as the 32P-labeled probe against the chromosomal DNA digested with BamHI (data not shown). The ΔP-450mel mutant constructed in this way had a deletion of the region encoding Glu-84 to Tyr-250. Because THN is readily converted into flaviolin by auto-oxidation, we expected that the ΔP-450mel mutant would be red due to accumulation of flaviolin. In fact, the mycelium and spores of mutant ΔP-450mel were light red, which were apparently different from those of the wild-type strain IFO13350 and mutant ΔrppA (Fig. 1B). Introduction of pKU209-RB6 containing P-450mel alone into mutant ΔP-450mel caused the host to produce the same brownish pigment, as did strain IFO13350 (data not shown). The red-brown pigment accumulated in mutant ΔP-450mel was identified to be flaviolin, which was nonenzymatically derived from THN, a product from malonyl-CoA by the action of RppA. These data suggested that the brownish pigment, which we later named HPQ melanin, was formed as a result of polymerization of an unstable compound derived from malonyl-CoA via THN.
Identification of HPQ as a precursor of melanin in S. griseus.
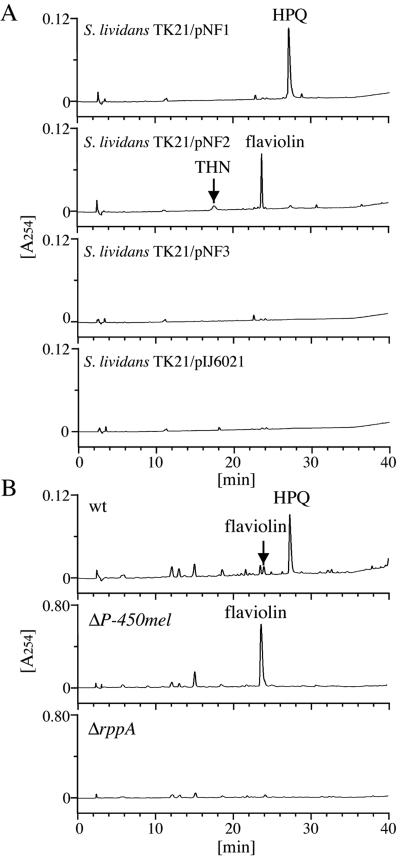
We constructed three plasmids, pNF1 carrying both P-450mel and rppA, pNF2 carrying rppA alone, and pNF3 carrying P-450mel alone (Fig. 1A), by using pIJ6021, and introduced them by protoplast transformation into S. lividans TK21. These genes were all under the control of a strong, thiostrepton-inducible tipA promoter. After the S. lividans cells had been grown in the presence of thiostrepton, cell extracts were prepared and analyzed by HPLC (Fig. 2A). S. lividans harboring pNF2 accumulated THN and flaviolin (Fig. 3A), as we detected both compounds as products from the in vitro reaction of RppA on malonyl-CoA (8). Flaviolin was probably produced mainly as a result of auto-oxidation of THN and as a result of oxidation of THN by MomA (7). S. lividans harboring pNF1 produced a dark green pigment, which was later identified to be HPQ (see below), whereas no HPQ was produced by S. lividans harboring pNF2 or the vector pIJ6021 (Fig. 2A).
FIG. 2.
HPLC analysis of in vivo products. (A) HPLC patterns of compounds produced by S. lividans TK21 expressing P-450mel alone (pNF3), rppA alone (pNF2), and P-450mel and rppA (pNF1). As a negative control, the culture broth of S. lividans harboring the vector pIJ6021 was also analyzed. Both THN and flaviolin were identified by their comigration with authentic samples. (B) HPLC patterns of compounds produced by S. griseus wild-type (IFO13350), ΔP-450mel, and ΔrppA strains.
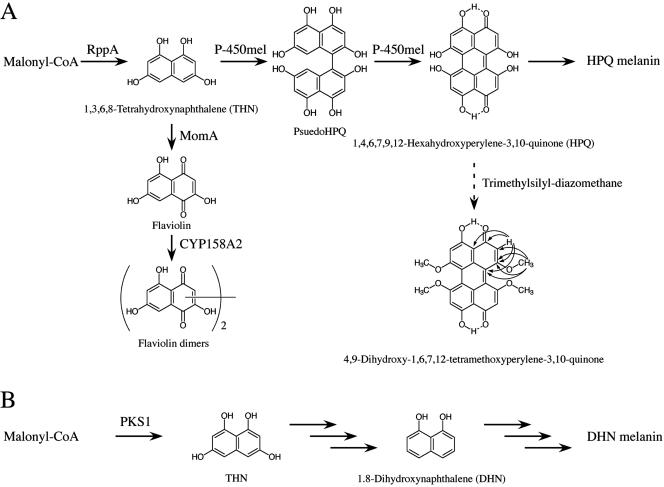
FIG. 3.
Biosynthesis of melanin from THN in streptomycetes and fungi. (A) Proposed HPQ melanin biosynthetic pathway in S. griseus. The structure and HMBC correlation of methylated HPQ are also shown. In S. coelicolor A3(2), instead of producing HPQ melanin, CYP158A2 dimerizes flaviolin, which is formed from THN by MomA (7) to yield flaviolin dimers. (B) The DHN melanin biosynthetic pathway in fungi.
Structural elucidation of the dark green pigment accumulated in S. lividans harboring pNF1 was difficult because the pigment was unstable and rapidly underwent polymerization. Hence, we treated the cell extract with trimethylsilyl-diazomethane since the pigment was presumed to be derived from THN, which possesses phenolic hydroxyl groups that can be replaced by methyl ethers. The methyl-substituted pigment was identified as 4,9-dihydroxy-1,6,7,12-tetramethoxyperylene-3,10-quinone (Fig. 3A) by proton and carbon NMR spectra, with the aid of heteronuclear multiple bond correlation (HMBC) analysis, and HRESI−/TOF-MS. From the structure of the methylated compound, we deduced the structure of the green pigment to be HPQ (1,4,6,7,9,12-hexahydroxyperylene-3,10-quinone). This deduction was confirmed by observing a [M − H]− ion peak of HPQ at m/z 377.02980 (calculated for C20H9O8, 0.06 mmu error) by HRESI−/TOF-MS.
HPQ was also detected in the extract of S. griseus grown on agar medium (Fig. 2B), although its amount gradually decreased during prolonged cultivation (data not shown), perhaps due to polymerization of the HPQ produced. As described above, no HPQ formation was observed in the ΔrppA or ΔP-450mel mutant, whereas the ΔP-450mel mutant accumulated flaviolin as a shunt product. As described below, P-450mel did not form HPQ from flaviolin in vitro. These in vivo observations suggested that HPQ was an intermediate of melanin biosynthesis in S. griseus and that P-450mel was responsible for the synthesis of HPQ, catalyzing aryl coupling of the THN that had been produced from malonyl-CoA by RppA (Fig. 3A).
In vitro analysis of aryl coupling of THN by P-450mel.
For convenient purification of P-450mel, we placed its coding sequence under the control of the T7 promoter in pET26b and introduced it in E. coli BL21 (DE3). Plasmid pET26b-P-450mel thus constructed would direct the synthesis of P-450mel-Leu-Glu-His6. P-450mel purified from the soluble fraction using histidine-binding resin gave a single protein band of about 42 kDa, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. P-450mel produced in E. coli showed an absorbance at 450 nm on reduction by sodium dithionite and subsequent bubbling with CO, which suggested that the enzyme was active (results not shown).
For reconstitution of the P-450mel activity in vitro, we used the ferredoxin and ferredoxin-NADP+ reductase from spinach, as was successfully used for reconstitution of a soluble cytochrome P-450soy from S. griseus (20). Incubation of the reconstituted system with THN gave two products, as analyzed by HPLC (Fig. 4A). The product at retention time 9 min was identified as HPQ by its comigration with authentic HPQ, which was prepared from S. lividans harboring pNF1 (Fig. 4B). The product at retention time 6.2 min appeared to be pseudo-HPQ (Fig. 3A), a dimeric form of THN, because it had a molecular mass of 382, which is equal to [2Mw − 2] Da of THN (Mw 192), as revealed by liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry analysis (data not shown). The in vitro formation of HPQ from THN by the action of P-450mel was consistent with the in vivo observation that simultaneous overexpression of rppA and P-450mel in S. lividans led to accumulation of HPQ.
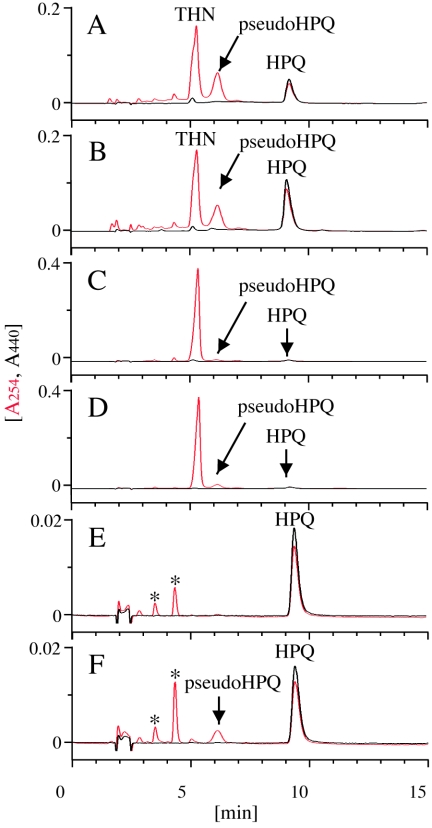
FIG. 4.
HPLC analysis of in vitro products. (A) The products from THN by the reconstituted P-450mel system were analyzed by simultaneously measuring the absorbances at 254 nm (in red) and 440 nm (in black). (B) HPQ prepared from S. lividans harboring pNF1 was coinjected with the products from the P-450 reaction on THN. (C) As a negative control, P-450mel was boiled before use for the reaction of P-450mel on THN. (D) As a negative control, ferredoxin and ferredoxin-NADP+ reductase were omitted from the reaction. (E) The products from pseudo-HPQ by the P-450mel reaction were analyzed. The peaks indicated with asterisks, derived spontaneously from pseudo-HPQ, were not identified. (F) As a negative control, P-450mel was boiled before use for the reaction on pseudo-HPQ.
Insignificant amounts of HPQ and pseudo-HPQ were observed in the control incubations with a boiled enzyme (Fig. 4C) and without the redox partners (Fig. 4D), indicating that these products were also formed nonenzymatically as a result of free radical-induced oxidation of THN, but at a lesser rate. We then performed the reaction using pseudo-HPQ which was recovered from the reaction of THN as a substrate to clarify whether both steps of sequential oxidation of THN leading to HPQ were catalyzed by P-450mel. P-450mel rapidly consumed pseudo-HPQ to yield HPQ (Fig. 4E), although pseudo-HPQ was extremely unstable and underwent spontaneous intramolecular aryl coupling to yield HPQ in a control reaction containing a boiled P-450mel enzyme (Fig. 4F). These results suggest that P-450mel catalyzes sequential oxidation of THN, which is intermolecular aryl coupling of THN and intramolecular aryl coupling of the resultant pseudo-HPQ, to yield HPQ (Fig. 3A).
Protective efficacy of HPQ melanin against UV radiation.
The melanin polymer has many interesting properties, the most conspicuous of which is its wide spectral absorbance, including the UV region, due to the high degree of conjugation in the molecule. We determined the viability of melanized and nonmelanized spores after irradiation with 254-nm UV light (Fig. 5). Although HPQ, which is dark green in a neutral to alkaline solution, irreversibly autopolymerized to yield a black amorphous solid when concentrated, the spores collected from the wild-type strain of S. griseus were greenish. This observation suggests that, in contrast to the fungal DHN melanin, which is black and extensively conjugated with fungal conidia (2), the amount of polymerized HPQ in the spore of S. griseus is rather small. The spores collected from the ΔrppA and ΔP-450mel mutants were colorless, reflecting the absence of HPQ, as described above (Fig. 2B).
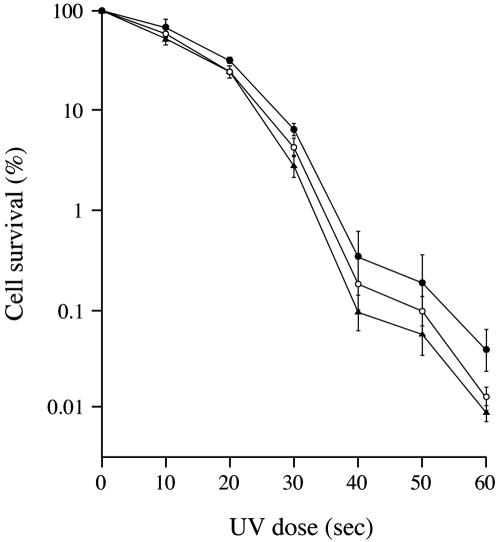
FIG. 5.
Effects of P-450mel and rppA mutations on spore survival after UV irradiation. The measure for UV resistance was the frequency of colony formation from spores after different doses of UV irradiation. The S. griseus strains examined were IFO13350 (solid circles), mutant ΔP-450mel (open circles), and ΔrppA (solid triangles). The values are means of the values obtained from four independent experiments.
The spores of the wild-type strain were the most resistant to UV among the three strains, whereas the ΔP-450mel mutant was slightly more resistant than mutant ΔrppA, probably due to the flaviolin produced by the ΔP-450mel mutant. The degree of the protection of HPQ from UV-irradiation was rather lower when compared to that of DHN melanin in fungi (18), which we assume is due to the low productivity of HPQ melanin in the Streptomyces spore. It is known that the degree of protection is proportional to the melanin concentration in spore walls (2). These observations show that HPQ melanin plays a role in protecting spores of S. griseus from UV irradiation, although DOPA melanin plays a major role, as is found in many other Streptomyces spp.
DISCUSSION
The present study has demonstrated that P-450mel mediates the biaryl coupling of THN without inserting an oxygen atom into the product, HPQ. The reaction mechanism is best explained by a radical coupling as follows. One-electron oxidation of THN affords a phenolate radical, and subsequent intermolecular radical coupling furnishes the aryl-aryl bond in a regiospecific manner. The resultant pseudo-HPQ is subjected to further intramolecular aryl coupling that elaborates HPQ (Fig. 3A). However, a cationic mechanism, which is an attack of an anion from THN on a cation of another THN molecule, cannot be excluded.
Although the aryl-aryl bond formation by a P-450 catalyst was first demonstrated in the biosynthesis of a benzylisoquinoline alkaloid at the cell-free level (26), P-450 enzymes catalyzing aryl-aryl coupling have not been characterized until the recent discovery of OxyC, which is perhaps involved in the last oxidative phenol coupling in vancomycin biosynthesis (17). However, no in vitro analysis of OxyC, which shares 32% identity in amino acid sequence to P-450mel, was conducted because its physiological substrate was unknown and unavailable. Very recently, Zhao et al. (27) solved the crystal structure of CYP158A2 from S. coelicolor A3(2) and showed that it catalyzes aryl coupling of two molecules of flaviolin (Fig. 3A), as predicted by Cortés et al. (4). Therefore, these two enzymes are the same in their catalytic properties, although CYP158A2 and P-450mel share only 43% identity in amino acid sequence. In fact, we used flaviolin as a substrate for P-450mel and detected a probable dimer of flaviolin as the product (data not shown). Since disruption of the CYP158A2 gene or an rppA homologue in S. coelicolor A3(2) (10) or in S. lividans TK21 (our unpublished data) caused no effects on pigmentation, these genes appear to be involved in the biosynthesis of a secondary metabolite other than HPQ melanin.
Perylenequinones form a relatively small but expanding group of biologically active pigments obtainable from natural sources (23). Although HPQ was chemically synthesized for evaluation of its antiviral activity (22), this is the first report of isolation of HPQ from natural sources. We have shown that HPQ melanin in S. griseus is synthesized by the condensation of malonyl-CoA to yield THN by the action of RppA and the subsequent aryl coupling of THN to yield unstable HPQ by the action of P-450mel. HPQ then autopolymerizes to form HPQ melanin (Fig. 3A). On the other hand, a wide variety of fungi synthesize melanin via the 1,8-dihydroxynaphthalene (DHN) pathway (Fig. 3B). The first intermediate in the pathway is THN, which is synthesized by PKS1, a type I PKS (6). Thereafter, a series of reductions and dehydrations take place, leading to DHN (2). The last step is polymerization of DHN, yielding DHN melanin.
It is interesting that 4,9-dihydroxy-3,10-perylenequinone, which is derived from 4,5,4′,5′-tetrahydroxy-1,1′-dinaphthyl, was isolated as a precursor of perylenequinone-related melanin from the ascomycete Daldinia concentrica (1). The only difference in the biosynthetic pathway between S. griseus and D. concentrica is that the ascomycete synthesizes the perylenequinone by symmetrical oxidative coupling of two molecules of DHN instead of THN. This analogy is remarkable because the analogous pathway is conserved between two distinct organisms, prokaryotic streptomycetes and eukaryotic ascomycetes. It is noteworthy that DHN melanin is associated with virulence and pathogenicity on infection of animal and plant hosts with DHN melanin producers (15). We showed that the HPQ melanin of S. griseus enhances protection from UV irradiation, which supports the idea that HPQ melanin plays a role in protection of the host cell from an environmental stress.
Acknowledgments
We thank Akira Arisawa for advice on production of P-450mel in E. coli. We also thank Hirofumi Shoun and Akinori Ohta for technical support in measuring the CO difference spectra of P-450mel.
This work was supported by a research grant from the Noda Institute for Scientific Research, by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan, and by a Grant-in-Aid for Scientific Research on Priority Areas from Monkasho.
REFERENCES
- 1.Allport, D. C., and J. D. Bu'Lock. 1958. The pigmentation and cell-wall material of Daldinia sp. J. Chem. Soc. 1958:4090-4094. [Google Scholar]
- 2.Bell, A. A., and M. H. Wheeler. 1986. Biosynthesis and function of fungal melanins. Annu. Rev. Phytopathol. 24:411-451. [Google Scholar]
- 3.Challis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100:14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortés, J., J. Velasco, G. Foster, A. P. Blackaby, B. A. Rudd, and B. Wilkinson. 2002. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 44:1213-1224. [DOI] [PubMed] [Google Scholar]
- 5.Endo, K., K. Kamo, K. Hosono, T. Beppu, and K. Ueda. 2001. Characterization of mutants defective in melanogenesis and a gene for tyrosinase of Streptomyces griseus. J. Antibiot. 54:789-796. [DOI] [PubMed] [Google Scholar]
- 6.Fujii, I., Y. Mori, A. Watanabe, Y. Kubo, G. Tsuji, and Y. Ebizuka. 2000. Enzymatic synthesis of 1,3,6,8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase PKS1. Biochemistry 39:8853-8858. [DOI] [PubMed] [Google Scholar]
- 7.Funa, N., M. Funabashi, E. Yoshimura, and S. Horinouchi. 2005. A novel quinone-forming monooxygenase family involved in modification of aromatic polyketides. J. Biol. Chem. 280:14514-14523. [DOI] [PubMed] [Google Scholar]
- 8.Funa, N., Y. Ohnishi, I. Fujii, M. Shibuya, Y. Ebizuka, and S. Horinouchi. 1999. A new pathway for polyketide synthesis in microorganisms. Nature 400:897-899. [DOI] [PubMed] [Google Scholar]
- 9.Ichinose, K., Y. Ebizuka, and U. Sankawa. 2001. Mechanistic studies on the biomimetic reduction of tetrahydroxynaphthalene, a key intermediate in melanin biosynthesis. Chem. Pharm. Bull. 49:192-196. [DOI] [PubMed] [Google Scholar]
- 10.Izumikawa, M., P. R. Shipley, J. N. Hopke, T. O'Hare, L. Xiang, J. P. Noel, and B. S. Moore. 2003. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 30:510-515. [DOI] [PubMed] [Google Scholar]
- 11.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 12.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Montellano, P. R., and J. J. De Voss. 2002. Oxidizing species in the mechanism of cytochrome P450. Nat. Prod. Rep. 19:477-493. [DOI] [PubMed] [Google Scholar]
- 14.Munro, A. W., and J. G. Lindsay. 1996. Bacterial cytochromes P-450. Mol. Microbiol. 20:1115-1125. [DOI] [PubMed] [Google Scholar]
- 15.Noasanchuk, J. D., and A. Casadevall. 2003. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5:203-223. [DOI] [PubMed] [Google Scholar]
- 16.Omura, T., and R. Sato. 1964. The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 239:2370-2385. [PubMed] [Google Scholar]
- 17.Pylypenko, O., F. Vitali, K. Zerbe, J. A. Robinson, and I. Schlichting. 2003. Crystal structure of OxyC, a cytochrome P450 implicated in an oxidative C-C coupling reaction during vancomycin biosynthesis. J. Biol. Chem. 278:46727-46733. [DOI] [PubMed] [Google Scholar]
- 18.Romero-Martinez, R., M. Wheeler, A. Guerrero-Plata, G. Rico, and H. Torres-Guerrero. 2000. Biosynthesis and function of melanin in Sporothrix schenckii. Infect. Immun. 68:3696-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takano, E., J. White, C. J. Thompson, and M. J. Bibb. 1995. Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133-137. [DOI] [PubMed] [Google Scholar]
- 20.Trower, M. K., S. F. Sariaslani, and D. P. O'Keefe. 1989. Purification and characterization of a soybean flour-induced cytochrome P-450 from Streptomyces griseus. J. Bacteriol. 171:1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda, K., K.-M. Kim, T. Beppu, and S. Horinouchi. 1995. Overexpression of a gene cluster encoding a chalcone synthase-like protein confers redbrown pigment production in Streptomyces griseus. J. Antibiot. 48:638-646. [DOI] [PubMed] [Google Scholar]
- 22.Wang, H.-K., J.-X. Xie, J.-J. Chang, K.-M. Hwang, S.-Y. Liu, L.-M. Ballas, J. B. Jiang, and K.-H. Lee. 1992. Antitumor agents. 134. New Shiraiachrome-A- and Calphostin-C-related perylene derivatives as cytotoxic and antiviral agents and inhibitors of protein kinase C. J. Med. Chem. 35:2717-2721. [DOI] [PubMed] [Google Scholar]
- 23.Wiess, U., L. Merlini, and G. Nasini. 1987. Naturally occurring perylenequinones. Prog. Chem. Org. Nat. Prod. 52:1-71. [DOI] [PubMed] [Google Scholar]
- 24.Williams, S. T., M. Goodfellow, G. Alderson, E. M. Wellington, P. H. Sneath, and M. J. Sackin. 1983. Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129:1743-1813. [DOI] [PubMed] [Google Scholar]
- 25.Williams, S. T., M. Goodfellow, E. M. Wellington, J. C. Vickers, G. Alderson, P. H. Sneath, M. J. Sackin, and A. M. Mortimer. 1983. A probability matrix for identification of some streptomycetes. J. Gen. Microbiol. 129:1815-1830. [DOI] [PubMed] [Google Scholar]
- 26.Zenk, M. H., R. Gerardy, and R. Stadler. 1989. Phenol oxidative coupling of benzylisoquinoline alkaloids catalysed by regio- and stereo-selective cytochrome P-450 linked plant enzymes: salutaridine and berbamunine. J. Chem. Soc. Chem. Commun. 1989:1725-1727. [Google Scholar]
- 27.Zhao, B., P. Guengerich, A. Bellamine, D. C. Lamb, M. Izumikawa, L. Lei, L. M. Podust, M. Sundaramoorthy, J. A. Kalaitzis, L. M. Reddy, S. L. Kelly, B. S. Moore, D. Stec, M. Voehler, J. R. Falck, T. Shimada, and M. R. Waterman. 2005. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J. Biol. Chem. 280:11597-11607. [DOI] [PubMed] [Google Scholar]