Abstract
We report here the isolation of the Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1 (AtSERK1) gene and we demonstrate its role during establishment of somatic embryogenesis in culture. The AtSERK1 gene is highly expressed during embryogenic cell formation in culture and during early embryogenesis. The AtSERK1 gene is first expressed in planta during megasporogenesis in the nucleus of developing ovules, in the functional megaspore, and in all cells of the embryo sac up to fertilization. After fertilization, AtSERK1 expression is seen in all cells of the developing embryo until the heart stage. After this stage, AtSERK1 expression is no longer detectable in the embryo or in any part of the developing seed. Low expression is detected in adult vascular tissue. Ectopic expression of the full-length AtSERK1 cDNA under the control of the cauliflower mosaic virus 35S promoter did not result in any altered plant phenotype. However, seedlings that overexpressed the AtSERK1 mRNA exhibited a 3- to 4-fold increase in efficiency for initiation of somatic embryogenesis. Thus, an increased AtSERK1 level is sufficient to confer embryogenic competence in culture.
In flowering plants, zygotic embryos are formed as a result of the fusion of the male and female gametes. In Arabidopsis, where embryo development has been thoroughly characterized (for review, see Laux and Jürgens, 1997), the fertilized zygote elongates and divides once asymmetrically to give a basal and an apical cell. Further divisions of the basal cell result in the formation of the suspensor, the quiescent center, and the columella root cap of the root meristem. All other pattern elements of the zygotic embryo, including the shoot apical meristem (SAM), hypocotyl, and cotyledons derive from the apical cell. It is not clear whether the egg cell is competent to execute the embryo program by itself or whether fertilization is necessary for the acquisition of embryogenic competence.
In culture, a small proportion of single somatic cells can be induced to change fate toward embryogenesis by application of exogenous auxins. These cells are called “competent cells” and can give rise to embryogenic cells from which somatic embryos can develop (Toonen et al., 1993). The formation of competent cells in culture depends on the presence of certain arabinogalactan proteins produced by nonembryogenic cells in culture (McCabe et al., 1997; Toonen et al., 1997a). Therefore, some form of signaling between embryogenic and nonembryogenic cells appears to be required for embryo initiation.
One of the genes expressed in competent cells in carrot (Daucus carota) tissue culture is the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (DcSERK; GenBank accession no. A67796) gene, which encodes a Leu-rich repeat (LRR) transmembrane receptor-like kinase (RLK). Single competent cells destined to develop into somatic embryos expressed the luciferase reporter gene under the control of DcSERK regulatory elements. Therefore, DcSERK is considered to mark cells competent to form embryos in culture. During zygotic embryogenesis, expression of the DcSERK gene was found in globular zygotic embryos, and not in later embryo stages. Based on these observations, it was proposed that the same signal transduction pathway is activated during the acquisition of embryogenic competence by somatic cells and during zygotic embryogenesis after fertilization (Schmidt et al., 1997).
LRR-type cell surface RLKs possess a number of characteristic domains. These include an extracellular domain (EX) containing a variable number of LRR units immediately followed by a single transmembrane domain and an intracellular kinase domain responsible for phosphorylating downstream proteins. One example of this type of receptor is the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE 1 (Li and Chory, 1997), which is involved in perception of this plant growth regulator (He et al., 2000). Another example is the CLAVATA1 (CLV1) receptor that has a role in maintaining the proper balance between undifferentiated cells and cells destined to differentiate into organs in the SAM (Clark et al., 1997). Several components of the CLV1 signaling pathway have been identified. The kinase-associated protein phosphatase is a negative regulator of CLV1 (Williams et al., 1997; Stone et al., 1998). The small peptide CLAVATA3 (CLV3) is postulated to be the ligand of CLV1 (Clark et al., 1995; Fletcher et al., 1999; Trotochaud et al., 1999, 2000). A second LRR receptor kinase CLAVATA2 (CLV2) is required for the stability of the CLV1 receptor and may heterodimerize with it (Kayes and Clark, 1998; Jeong et al., 1999). Thus, RLKs appear to have a prominent role in cellular signaling in plants (Becraft, 1998; Lease et al., 1998).
The aim of the work presented here was to determine if the SERK-mediated signaling pathway is employed during zygotic and somatic embryogenesis in Arabidopsis. To achieve this, we first isolated the most closely related SERK gene from Arabidopsis, AtSERK1 (GenBank accession no. A67815). Several other SERK1-related sequences are present in the Arabidopsis genome database, indicating that AtSERK1 is part of a small family consisting of five members. The AtSERK1 expression pattern was determined by reverse transcriptase (RT)-PCR, promoter-reporter analysis, and in situ hybridization (ISH) during somatic and zygotic embryogenesis. Like DcSERK in carrot, AtSERK1 marks cells competent to form embryos in culture. The AtSERK1 gene is first expressed in the nuclear tissue of developing ovules including the megaspore mother cell (MMC). Furthermore, the embryogenic competence of callus derived from seedlings overexpressing AtSERK1 was 3 to 4 times higher when compared with wild-type callus. These results indicate that the AtSERK1 product is sufficient to confer embryogenic competence in culture. The possible acquisition of embryogenic competence by the egg cell mediated by this gene is discussed.
RESULTS
Molecular Cloning of AtSERK1
To isolate the orthologous SERK gene from Arabidopsis, we screened a genomic lambda phage library with the carrot cDNA clone as a probe and obtained one phage containing the entire AtSERK1 gene (GenBank accession no. A67815). A full-length SERK1 cDNA was isolated from an Arabidopsis cDNA library made from flower buds and opened flowers (Li and Thomas, 1998). This cDNA consisted of an open reading frame of 1,875 nucleotides and 194 nucleotides of 5′-untranslated region (GenBank accession no. A67827).
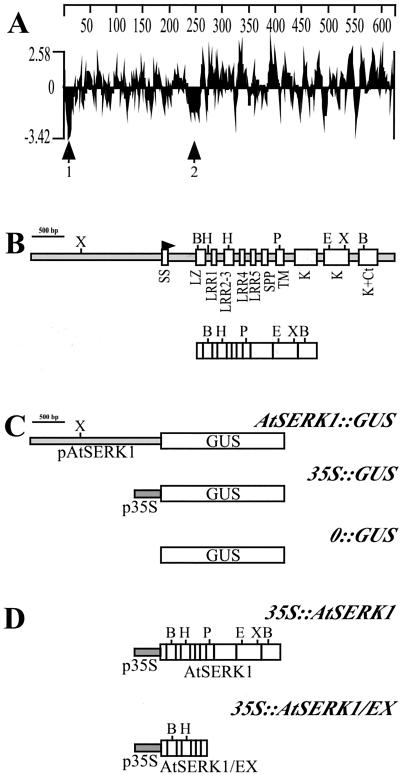
The predicted AtSERK1 protein of 625 amino acids has a calculated molecular mass of 69 kD, and is slightly acidic (predicted pI of 5.25). The amino acid sequence of AtSERK1 shows a high percentage of identity with DcSERK (92%) and shares all the characteristic features of that protein, including the five LRRs, the Pro-rich domain containing the so-called Ser-Pro-Pro (SPP) motif, containing two tandemly repeated SPP sequences, the transmembrane domain, and the kinase domain (Schmidt et al., 1997). Figure 1A shows the hydrophilicity plot for AtSERK1 containing two strongly hydrophobic regions. The first region, spanning residues 1 through 29, meets the conditions defining a signal peptide (von Heijne, 1986), with a potential signal peptidase cleavage site between positions 29 and 30. The second hydrophobic region, spanning residues 231 through 276, corresponds to the putative transmembrane domain separating the extracellular part of the protein and the intracellular kinase domain.
Figure 1.
Description of the Arabidopsis SERK1 gene and protein. A, Kyte-Doolitle hydrophylicity plot of AtSERK1 protein. Arrows 1 and 2 indicate the two hydrophobic regions of the protein. B, Genomic organization of AtSERK1 gene in Arabidopsis. Up, Genomic DNA; down, cDNA. White boxes indicate exons and gray boxes indicate non-coding regions. SS, Signal sequence; SPP, Pro-rich domain containing the SPP motif; TM, transmembrane domain; K, kinase domain; Ct, C terminus. C, Constructs used for expression analysis. The AtSERK1 2-kb promoter was fused to the GUS reporter gene in AtSERK1::GUS construct. As positive control, the cauliflower mosaic virus (CaMV) 35S promoter was fused to GUS 35S::GUS, and as negative control, the GUS gene was promoterless 0::GUS. D, Constructs used for overexpression analysis. 35S::AtSERK1 and 35S::AtSERK1-EX are fusions between AtSERK1 complete or partial cDNA to the 35S promoter. Restriction sites: B, BamHI; E, EcoRI; H, HindIII; P, PstI; and X, XbaI.
Directly adjacent to the cleavage site of the putative signal peptide, the AtSERK1 protein contains a Leu-rich domain of 45 amino acids fitting the Leu-zipper (LZ) pattern Lx6Lx6Lx6L (Landschulz et al., 1988). It is surprising that these two domains are not present in DcSERK, which instead contains 28 amino acids that are absent in AtSERK1. The substantial similarity between the two proteins (92%) only begins at position 99 of AtSERK1. The LRR domain of AtSERK1 extends from positions 75 through 194 and is composed of five units. In most LRR receptor kinases, the transmembrane domain immediately follows the LRR domain. However, in AtSERK1, as in DcSERK, a Pro-rich region containing a repeated SPP motif separates these domains. We consider the SPP motif to be one of the hallmarks of the SERK-like RLKs. This motif has been suggested to act as a hinge providing flexibility to the extracellular part of the receptor or as a region for interaction with the cell wall. The intracellular region of AtSERK1 is also similar to DcSERK, containing the 11 subdomains characteristic of the catalytic core of Ser/Thr protein kinases (Hanks et al., 1988; Stone and Walker, 1995) and a C-terminal Leu-rich domain suggested to be involved in protein-protein interactions (Schmidt et al., 1997). Both intracellular domains from AtSERK1 and DcSERK have been shown to be active Ser/Thr kinases (Shah et al., 2001a, 2001b).
Comparison of cDNA and genomic sequences of the AtSERK1 gene shows the presence of 11 exons in the coding region (Fig. 1B). The intron splice site consensus sequences fit the canonical GT/AG-U2-dependent borders (Brown et al., 1996). The overall structure of the AtSERK1 gene is such that the putative protein domains described above are all located in separate exons. In particular, each LRR unit is encoded by a different exon, with the exception of LRR2 and 3, which are encoded by exon 4. The separation of individual LRR units in different exons has been described previously for other LRR protein-encoding genes such as the LRP (Leu-rich protein) gene (Tornero et al., 1996) and the ZmSERK genes (Baudino et al., 2001). This phenomenon also occurs in some RLK genes such as ERECTA, which contains 21 LRRs that are all encoded in separate exons (Torii et al., 1996). These instances of similarity in the genomic organization of LRRs suggest an exon-based definition of a LRR unit as aLxxNNLSGxaPxxLxxLxxLxxL, which differs in frame from the LRR consensus sequence of xLxxLaLxxNNLSGxaPxxLxxLx previously proposed (Kobe and Deisenhofer, 1994).
We also obtained the full genomic sequence of DcSERK (GenBank accession no. U93048). Comparison with the DcSERK cDNA revealed the presence of nine introns in the coding region. Of the 11 AtSERK1 exons, the last eight correspond closely to the last eight of DcSERK, encoding exactly the same amino acid regions of the predicted proteins. The first two exons of DcSERK are not represented in the AtSERK1 sequence. Sequences highly homologous to the first three exons of AtSERK1 are present in the DcSERK genomic sequence, about 2.3 kb upstream of the predicted DcSERK translation initiation site.
Arabidopsis Contains Five SERK-Related Genes
A tBLASTn search (Altschul et al., 1990) identified a large number of sequences related to AtSERK1. However, only four sequences (on three bacteria artificial chromosomes [BACs]; GenBank accession nos. AC07454, AL035678, and AC06436) contained the characteristic structure of AtSERK1, including the LZ domain, five LRRs, SPP motif, and transmembrane and kinase domains; therefore, we designated them AtSERK2, AtSERK3, AtSERK4, and AtSERK5 (Baudino et al., 2001). Complete cDNA clones and sequences from AtSERK2 and AtSERK3 were obtained from the cDNA library described above (GenBank accession nos. AF384969 and AF384970). The amino acid identity with AtSERK1 ranges from 90% for AtSERK2 to 67% for AtSERK5. Identity within the kinase domain is 95% to 85% for all five sequences and all contain the core sequences characteristic of Ser/Thr kinases. Identity is also high in the LRR region (89%–66%) and transmembrane domain (82%–54%). The greatest divergence is seen for AtSERK3, AtSERK4, and AtSERK5 in the SPP (47%, 38%, and 31% identity, respectively) and C-terminal domains (44%, 38%, and 38% identity, respectively).
The AtSERK1 gene is the most similar to the DcSERK gene at the nucleic acid level (74%). The genomic structure of all homologs is also similar to AtSERK1, with 11 predicted exons, each encoding a different domain of the protein. Because the Arabidopsis Genome Initiative sequencing project is completed (The Arabidopsis Genome Initiative, 2000), we believe that these five genes constitute the entire SERK family in Arabidopsis.
The chromosomal location of the AtSERK1 gene was determined by hybridization to the physically ordered Centre d'Etude du Polymorphisme Humain, Institut National de la Recherche Agronomique, Centre National de la Recherche Scientifique (CIC) Yeast Artificial Chromosome library (Creusot et al., 1995; Meinke et al., 1998). Alignment of the seven hybridizing clones (3F2, 7E8, 9D6, 11B11, 12A5, 12G10, and 12H9) provided a map position for AtSERK1 between markers g4552 and nga111 on chromosome 1, between genetic markers CLV2 and CLV1. Fluorescence ISH on pachytene chromosomes using the AtSERK1 lambda phage clone and the Yeast Artificial Chromosome clones CIC3F2 and CIC12H9 confirmed the location on chromosome 1 of all three clones (V. Hecht and P. Fransz, unpublished data). The BAC clone F14O23 sequence (GenBank accession no. AC12654) was recently released that contains the full AtSERK1 gene. The map position of this BAC clone confirms the location of AtSERK1 on chromosome 1. The three BAC clones containing the other Arabidopsis SERK homologs are located on chromosome 1 (AtSERK2), 2 (AtSERK4 and AtSERK5), and 4 (AtSERK3; http://www.Arabidopsis.org/).
AtSERK1 Is Expressed Postembryonically and Marks Embryogenic Competent Cells
To examine the expression pattern of AtSERK1, a transcriptional fusion between the AtSERK1 promoter and the Escherichia coli β-glucuronidase (GUS) gene was constructed. The CaMV 35S promoter was used as a positive control to evaluate the strength and the specificity of the AtSERK1 promoter. A promoterless GUS construct was used as a negative control to evaluate the background GUS activity. The constructs used are detailed in Figure 1C. These constructs were stably transformed into the Arabidopsis genome via Agrobacterium tumefaciens using vacuum infiltration. Several independent transformants were obtained for each construct. No GUS activity was detected in lines containing the 0::GUS construct. Lines containing the construct 35S::GUS showed GUS activity throughout the whole plant life cycle (data not shown). Three independent transformant lines containing the AtSERK1::GUS construct were obtained. All three showed exactly the same GUS expression pattern.
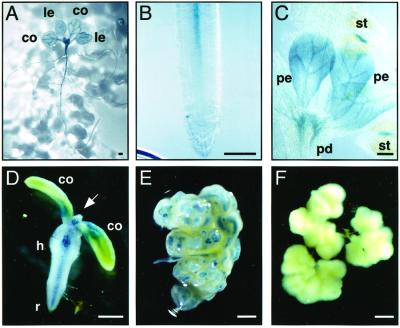
We first analyzed the AtSERK1::GUS expression pattern during postembryonic development. GUS staining was found in vascular tissue of seedlings and adult plants after long incubation times and is shown in Figure 2, A through C. Germinating seedlings show GUS expression in vascular bundles of the cotyledons, primary leaves, hypocotyl, and roots (Fig. 2, A and B). This vascular expression pattern is also found in other organs of the mature plant, such as the pedicel and the petals (Fig. 2C).
Figure 2.
AtSERK1 expression pattern postembryonically and in embryogenic cultures. GUS expression of plants containing the AtSERK1::GUS construct was followed throughout plant life (A–C) and during induction of embryogenic cultures in amp1 seedlings (D–F). A, Seedling 15 d after germination (DAG). B, Root tip of the seedling shown in A. C, Flower of an adult plant. D, 4 DAG seedlings germinated in presence of 2, 4-D; arrow indicates the SAM. E, Embryogenic callus at 28 DAG. F, Nonembryogenic callus after 40 DAG. Bar, 1 mm; co, cotyledon; h, hypocotyl; le, primary leaves; pd, pedicel; pe, petal; r, root; st, stamen.
A line containing the AtSERK1::GUS construct was used to examine if the AtSERK1 gene was expressed during the early stages of somatic embryogenesis. In the altered meristem program 1 (amp1) or primordia timing (pt) mutant seedling protocol (Mordhorst et al., 1998), amp1 seeds are germinated in inducing medium containing the growth regulator 2,4-dichlorophenoxyacetic acid (2,4-D). This leads to the formation of embryogenic callus, predominantly from the SAM region. Somatic embryos form on primary callus and on callus cultured in liquid medium. Plants containing the AtSERK1::GUS construct were crossed with the amp1 mutant. The F2 progeny of this cross was screened for the presence of the construct and the amp1 phenotype.
The AtSERK1::GUS expression pattern was determined in embryogenic and nonembryogenic cultures from F3 seeds germinated in inducing medium is shown in Figure 2, D through F. Seedlings germinated in inducing medium exhibited a strong AtSERK1::GUS expression in the SAM region and in all vascular tissue (Fig. 2D), indicating a possible promoter effect of auxin on AtSERK1 promoter activity. Outer cell layers of the cotyledons, hypocotyl, and radicle did not show GUS expression. Soon after culture initiation in inducing medium, embryogenic structures appeared from the SAM region. These structures show strong AtSERK1::GUS expression (Fig. 2E). Regular subculture of the embryogenic callus gave rise to high numbers of somatic embryos, which appeared to originate from the GUS-expressing cell clusters. Nonembryogenic callus, characterized by its white color and the absence of somatic embryo formation, was gradually obtained from embryogenic callus via selective subculturing. During the first weeks of subculturing, some AtSERK1::GUS expression remained in isolated groups of cells (data not shown). With continued subculture and selection, the culture became nonembryogenic and AtSERK1::GUS expression decreased to zero within 40 DAG (Fig. 2F). A more detailed analysis of AtSERK1::GUS expression during initiation of embryogenic cultures in Arabidopsis will be described elsewhere.
From these observations it appears that AtSERK1::GUS expression is found in cells as they acquire embryogenic competence. This supports the previous conclusion from studies in carrot (Schmidt et al., 1997) and in Dactylis glomerata (Somleva et al., 2000) that AtSERK1 is a marker for competent cells to form embryos in culture.
The AtSERK1 Gene Is Expressed in Developing Ovules and Early Embryos
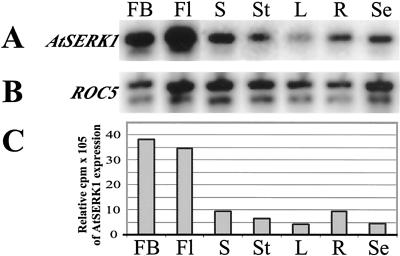
To identify which cells express AtSERK1 during ovule development and zygotic embryogenesis, we investigated the expression pattern of AtSERK1in planta. We first analyzed the AtSERK1 expression at the organ level using semiquantitative RT-PCR analysis. Total RNA was isolated from various tissues including flower buds, fertilized flowers, siliques, stems, leaves, roots, and seedlings. After reverse transcription, AtSERK1 cDNA (accession no. A67827) was amplified and the cyclophilin ROC5 mRNA (Chou and Gasser, 1997) was used as internal control. No amplification of mRNAs with specific primers was observed when the RT step was omitted. The results are shown in Figure 3, A and B. AtSERK1 mRNA was most abundant in closed flower buds before fertilization and in flowers 3 d after pollination, which contained developing seeds with embryos from stages 1 through 7 (Jürgens and Mayer, 1994). A low level of AtSERK1 mRNA was also detected in all other organs tested. Quantitative analysis showed that AtSERK1 mRNA was almost 10 times higher in flower buds relative to leaf tissue (Fig. 3C).
Figure 3.
AtSERK1 expression during plant development determined by semiquantitative RT-PCR. A, AtSERK1 expression pattern at 30 cycles. B, ROC5 expression pattern at 30 cycles. C, Relative expression of AtSERK1. FB, Flower buds; Fl, opened flowers containing embryos from stages 0 through 7; S, siliques containing embryos from stages 7 through 20; St, stems; L, rosette leaves; R, roots; Se, seedlings 7 DAG.
The AtSERK1 expression pattern at the cellular level was examined using mRNA ISH and GUS staining. The results are presented in Figure 4. For ISH experiments, a partial AtSERK1 cDNA fragment containing the 5′-untranslated region and the first two exons was used as an AtSERK1-specific probe. For GUS expression analysis, the three independent transformant lines containing the AtSERK1::GUS construct were used.
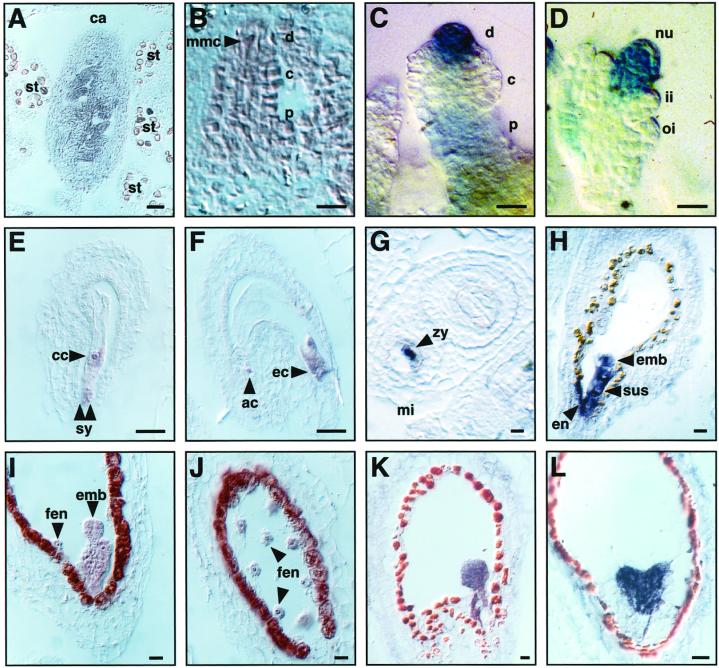
Figure 4.
Expression of the AtSERK1 in developing ovules and seeds. Expression pattern determined by ISH or by GUS staining (GUS). A, Transversal section of a flower bud containing young ovule primordia (ISH). B, Ovule primordia at stage 2-II, arrow indicates dividing MMC (ISH). C, Ovule primordia at stage 2-IV (GUS). D, Ovule primordia at stage 3-I (GUS). E, Mature embryo sac showing expression in the egg cell and the antipodal cells (ISH). F, Mature embryo sac showing expression in the synergids and the central cell (ISH). G, Transversal section of a developing seed containing an embryo at stage 2 (ISH). H, Longitudinal section of a developing seed containing an embryo at stage 4 (ISH). I, Longitudinal section of a developing seed containing an embryo at stage 5 (ISH). J, Longitudinal section of the same developing seed as in (I) showing the free endosperm nuclei (ISH). K, Longitudinal section of a developing seed containing an embryo at stage 8 (ISH). L, Longitudinal section of a developing seed containing an embryo at heart stage (ISH). Bars: A through D, 10 μm; E through L, 20 μm. ac, Antipodal cell; c, central; d, distal; cc, central cell; ec, egg cell; emb, embryo proper; en, endothelium; fen, free endosperm nuclei; ii, inner integument; mi, micropylar pole; nu, nucleus; oi, outer integument; p, proximal; st, stamen; su, suspensor; sy, synergids; zy, zygote.
The proximal-distal polarity of the ovule is established prior to meiosis. It divides the ovule primordia into three distinct parts: The proximal part is the precursor of the funiculus, the central part is the precursor of the integuments, and the distal part is the precursors of the nucleus and embryo sac. Expression of AtSERK1 was first detected in the whole ovule primordium at stage 1 (Schneitz et al., 1995). At this stage, no expression was detected in the placental tissue or in the developing carpel (Fig. 4A). AtSERK1 expression in the ovule persisted throughout MMC differentiation and meiosis (stage 2-I; Fig. 4B), and appeared to increase by the time of functional megaspore differentiation. In agreement with the ISH data, GUS expression was also found in developing ovules. Strong AtSERK1::GUS expression was found in the distal side and weaker expression in the proximal part of ovule primordia (stage 2-IV; Fig. 4C). At this stage, the nucleus has begun to divide and enlarge (Schneitz et al., 1995). In contrast, the central area of the ovule primordia, which gives rise to the integuments, was devoid of GUS expression. AtSERK1 expression was detectable in all distal cells of the ovule primordia, including the epidermis, cortical cells, and the dividing megaspore. GUS activity persisted in the functional megaspore and in the nucleus during subsequent stages of ovule development. During megagametogenesis, the GUS staining continued to be restricted to the nucleus, and was not present in the inner or outer integuments (stages 2-IV; Fig. 4D). It is interesting that AtSERK1 expression was detected in all constituents of the embryo sac, including the synergids and the central cell (Fig. 4E), as well as the egg cell and the antipodal cells (Fig. 4F). After fertilization, AtSERK1 expression appeared to decrease rapidly and was only detected in a few cells at the micropylar pole of the developing seed corresponding to the fertilized zygote (Fig. 4, G and H). Later in embryo development, a hybridization signal was found in all cells derived from the fusion of the gametes, including the embryo proper and suspensor (Fig. 4, H and I), as well as in the free endosperm nuclei (Fig. 4J). AtSERK1 expression persisted in all cells of the embryo until the heart stage (stage 14; Fig. 4, K and L; Jürgens and Mayer, 1994). At later stages of embryo development, no ISH signal or GUS expression was detectable even after long incubation times (data not shown).
Taken together, these results show that AtSERK1 is expressed prior to fertilization in ovules and transiently during early embryo development. The low level of AtSERK1 expression in other organs found by RT-PCR is likely to reflect the expression in vascular tissues seen in the AtSERK1::GUS lines. However, expression in these tissues was never confirmed by ISH, possibly due to low steady-state AtSERK1 mRNA levels.
AtSERK1 Increases Embryogenic Competence
We next investigated the effect of ectopic expression of the AtSERK1 gene using two different constructs containing AtSERK1 under the control of the CaMV 35S constitutive promoter. The first construct contained the full AtSERK1 cDNA (35S::AtSERK1), whereas the second only contained the EX of the protein (35S::AtSERK1/EX). The constructs used are depicted in Figure 1D. These constructs were transferred into Arabidopsis by vacuum infiltration in two different transformation series for each construct. Several independent transformants containing single insertions were identified and allowed to self-fertilize to obtain homozygous lines. These lines showed normal fertility and seed set.
Several of these lines were tested for embryogenic cell formation using the in vitro seedling assay (Mordhorst et al., 1998). Representative calli are shown in Figure 5. In this assay, seeds are germinated in medium with 2,4-D. After 3 weeks, the percentage of embryogenic structures is scored. Embryogenic cultures are established after 7 weeks and are then scored for embryogenic capacity, which is assessed as the number of somatic embryos developed per individual cell cluster. This is a more reproducible measure of embryogenic potential than the percentage of embryogenic structures that initially developed because this latter measure is more variable across different seed batches (M.V. Hartog, A.P. Mordhorst, and S.C. de Vries, unpublished data).
Figure 5.
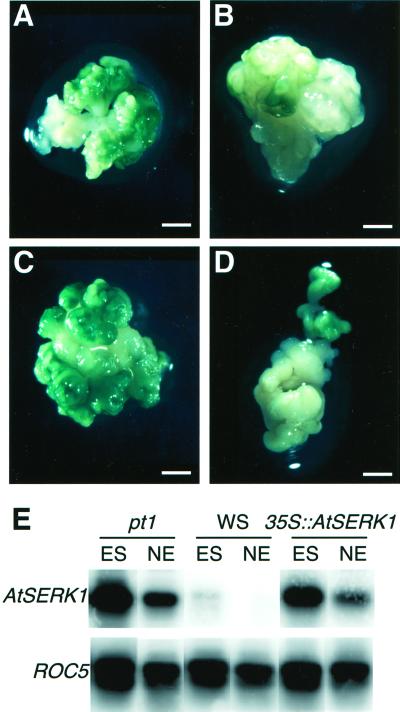
The effect of ectopic AtSERK1 expression on embryogenic potential of seedlings. A through D, Embryogenic callus 4 weeks after initiation. A, Embryogenic callus of a amp1 culture. B, Nonembryogenic callus of a Wassilewskija (WS) wild-type culture. C, Embryogenic callus of a 35S::AtSERK1culture. D, Embryogenic callus of a 35S::AtSERK1-EX culture. E, AtSERK1 expression determined by RT-PCR after 30 cycles of callus from amp1, WS wild type, and 35S::AtSERK1 4 weeks after embryogenic culture initiation as shown in Figure 4, I through K. ROC5 expression is shown as internal control. ES, Embryogenic structures; NE, nonembryogenic structures. Bar, A through D, 1 mm.
Using this assay, it was shown previously that somatic embryogenesis is facilitated by mutations in genes repressing meristematic cell divisions such as amp1 and the clavata mutants (Mordhorst et al., 1998). Therefore, we included cultures obtained from amp1 mutant seedlings as a positive control in our analysis (Fig. 5A), in addition to cultures from wild-type WS seedlings as negative controls (Fig. 5B). Calli obtained from seedlings homozygous for the 35S::AtSERK1 construct 4 weeks after induction (Fig. 5C) were highly embryogenic when compared with the positive control. Similar calli obtained from seedlings homozygous for the 35S::AtSERK1/EX construct were almost nonembryogenic (Fig. 5D) when compared with the negative control. A more quantitative analysis is presented in Table I. Out of 1,727 seedlings tested that contained the 35S::AtSERK1 construct, 16% developed embryogenic structures after 3 weeks. Scored after 7 weeks, the cultures developed from 35S::AtSERK1 lines had an embryogenic capacity score of 1.4 on an arbitrary scale ranging from 0 (wild-type WS) to 3 (amp1). Out of 1,258 tested seedlings containing the 35S::AtSERK/EX construct, 13% developed embryogenic cultures after 3 weeks, but the embryogenic capacity scored after 7 weeks was only 0.1.
Table I.
Effect of ectopic AtSERK1 expression on embryogenic potential of seedlings in culture
| Linea | No. of Experimentsb (No. of Lines Tested) | Total No. of Seedlings Tested | Percentage of ESc | Embryogenic Capacity after 7 Weeksd |
|---|---|---|---|---|
| amp1 (Landsberg erecta) | 7 | 363 | 18 | 3 |
| WS (wild type) | 9 | 403 | 5 | 0 |
| 35S∷AtSERK1 | 44 (9) | 1,727 | 16 | 1.4 |
| 35S∷AtSERK1-EX | 19 (6) | 1,258 | 13 | 0.1 |
35S∷AtSERK constructs were introduced by vacuum infiltration. Transformants were selected on kanamycin and were maintained by selfing. T2 through T4 generations were selected for 100% kanamycin-resistant progeny and were tested for embryogenic induction using the amp1 seedling assay (Mordhorst et al., 1998).
Data are pooled from several generations including two that were obtained via somatic embryogenesis of a T2 plant. Individual experiments were performed over a period of 1.5 years. No significant differences were found between lines obtained via normal propagation and via somatic embryogenesis.
The percentage of embryogenic structures was determined by counting the seedlings that developed bright-green compact callus at their shoot apices after 3 weeks in culture, indicative of embryogenic capacity (Mordhorst et al., 1998).
The ability to develop into a stable embryogenic suspension culture was determined on a scale between 0 (WS wild-type plants) and 3 (amp1 plants in Landsberg erecta background), based on the frequency and the quality of somatic embryos.
The AtSERK1 expression levels in embryogenic and nonembryogenic cultures obtained from amp1-, WS-, and 35S::AtSERK1-containing seedlings were analyzed by RT-PCR. After reverse transcription, AtSERK1 cDNA was amplified and the cyclophilin ROC5 mRNA (Chou and Gasser, 1997) was used as an internal control (Fig. 5E). No amplification of mRNAs with specific primers was observed when the RT step was omitted. AtSERK1 expression was detectable in all culture samples tested, although it was fairly weak in WS wild-type cultures, consistent with the weak embryogenic capacity of these cultures. In all cases, the AtSERK1 mRNA was more abundant in embryogenic than in nonembryogenic cultures of each line. It is of interest to note that AtSERK1 expression is considerably up-regulated in cultures derived from the amp1 mutant (Fig. 5E). This may be a reflection of the natural tendency of this mutant to regenerate (Chaudhury et al., 1993; Mordhorst et al., 1998). Taken together, these results suggest that overexpression of AtSERK1 is sufficient to confer sustained post-germination embryogenic competence in culture.
DISCUSSION
We describe here the isolation of the AtSERK1 gene, the Arabidopsis gene most closely related to the carrot DcSERK gene. AtSERK1 expression is found in cells acquiring embryogenic competence, in embryogenic cells, and in early somatic embryos. AtSERK1 is also expressed in ovules prior to fertilization and transiently during zygotic embryo development. Low expression was found in vascular tissue. Ectopic expression of AtSERK1 confers sustained embryogenic competence to seedlings under in vitro conditions. These results suggest that the AtSERK1 gene plays an essential role in determining embryogenic competence.
SERK Is Represented by a Multigene Family in Arabidopsis
The carrot SERK gene was previously reported to be a marker for single cells competent to form embryos in suspension cultures (Schmidt et al., 1997). DcSERK encodes an LRR-containing RLK and belongs to a large and diverse family of receptor kinases in plants (Becraft, 1998; Lease et al., 1998). Limitations of the carrot system for functional analysis prompted us to initiate a search for SERK homologs in Arabidopsis.
The predicted primary structure of the AtSERK1 protein consists of a signal peptide, an LZ domain, five LRR units, a Pro-rich domain containing the SPP motif, a single transmembrane domain, and the 11 conserved subdomains of a Ser-Thr kinase. At the carboxy-terminal end of the protein, there is a region rich in Leu residues, which may possibly be involved in interaction with other proteins. Although the LZ motif was described as a characteristic feature of DNA-binding proteins (Landschulz et al., 1988), it is also found in plant LRR-containing proteins, such as LRP from tomato (Lycopersicon esculentum), which is involved in pathogen resistance (Tornero et al., 1996).
The main feature distinguishing SERK proteins from other RLKs is the Pro-rich domain containing the SPP motif, located between the LRRs and the transmembrane domain. The presence of an SPP motif was used as a criterion for the identification of four other SERK genes (AtSERK2 to AtSERK5) among numerous LRR-RLK encoding genes in the Arabidopsis database. Each of these genes has all the other characteristic features of SERK proteins outlined above. AtSERK2 is the most closely related to AtSERK1, with AtSERK3, 4, and 5 comprising a separate subfamily. One of the most striking features of the SERK gene family is the highly conserved genomic structure. In all of the predicted SERK proteins, each of the EXs and each individual LRR unit is encoded in a separate exon. This genomic organization suggests a composition of a single LRR unit that is different from the one previously proposed by Kobe and Deisenhofer (1994). A similar genomic organization was previously described for other LRR encoding genes (Torii et al., 1996; Tornero et al., 1996) and suggests that genes of this type may have evolved by exon duplication from a prototypic gene containing one LRR unit.
The presence of a SERK multiple gene family in Arabidopsis and also in maize (Zea mays; Baudino et al., 2001) implies that carrot is also likely to contain more than one SERK gene. The precise phylogenetic relationship between SERK genes in Arabidopsis and DcSERK, therefore, cannot yet be clearly defined. Nevertheless, AtSERK1 is somewhat more similar to DcSERK at the nucleic acid level than the other Arabidopsis SERK genes, and was the only sequence identified in a genomic library using the DcSERK cDNA as a probe. Although we chose to first investigate the function of the AtSERK1 gene due to higher DNA identity, it is possible that AtSERK2 has a function similar to AtSERK1, due to the high degree of identity at the amino acid level with DcSERK.
The main difference between the DcSERK and AtSERK1 mRNAs lies in their 5′ regions. The first three exons in the Arabidopsis SERK1 mRNA are absent in the carrot SERK mRNA and are replaced by two different exons, resulting in a slightly shorter mRNA. Although the three exons used in Arabidopsis are present in the genomic sequence of DcSERK, the predicted transcript could not be identified, despite extensive RT-PCR experiments (E.D.L. Schmidt, unpublished data). Furthermore, the first two exons present in the DcSERK mRNA are not present in any of the five Arabidopsis SERK genomic sequences. Therefore, we conclude that no alternatively spliced precursor corresponding to the DcSERK mRNA is produced in Arabidopsis.
The AtSERK1 Gene Marks Embryogenic Competence in Culture
During initiation and maintenance of embryogenic carrot cell cultures, DcSERK expression was detected in a small number of cells attached to the explant, in a small subpopulation of cells in culture, in embryogenic cell clusters, and in somatic embryos up to the globular stage. Analysis of luciferase expression under the control of the carrot SERK regulatory elements showed a tight quantitative correlation between the ability of single cells to develop an embryo and DcSERK expression, demonstrating that this gene is a marker for single competent cells (Schmidt et al., 1997). The expression analysis of the AtSERK1 gene confirms and extends these observations.
When seedlings are grown in the presence of auxin during the initiation of Arabidopsis embryogenic cultures, AtSERK1 promoter activity is detected in the SAM and at the base of the cotyledons. Both are sites at which embryogenic callus emerges in Arabidopsis (Mordhorst et al., 1998; von Recklinghausen et al., 2000). It is of interest to note that weak AtSERK1 promoter activity is also observed in the vascular tissue of seedlings grown in the absence of auxin. With a few exceptions, cells within the vascular tissue are among the first to reinitiate cell division in response to hormonal treatments (Guzzo et al., 1994, 1995). Cells competent to form embryos are derived from such dividing cells (Schmidt et al., 1997; Somleva et al., 2000). However, SERK expression was never seen in cells of the vascular tissue without 2,4-D treatment in D. glomerata leaf explants, and in carrot hypocotyl explants. This may be due to low steady-state AtSERK1 mRNA levels, or may point to a difference between Arabidopsis and these two other plant species. If the AtSERK1 expression as seen in cells of the vascular tissue marks embryogenic potential, the intriguing possibility exists that plants contain a small population of cells that retain embryogenic competence, reminiscent of stem cells in animals (van der Kooy and Weiss, 2000; Watt and Hogan, 2000).
Embryogenic cultures are routinely established using persistent auxins as an inducing agent. In these cultures, only a small proportion of cells is competent to form embryos (Toonen and de Vries, 1997). Although the AtSERK1 expression is clearly enhanced by application of 2,4-D, this expression does not remain constitutive in all cells grown in 2,4-D-containing media. Therefore, the SERK-mediated signaling pathway may interact at some point with the auxin pathway, but SERK is certainly not an integral part of it. In contrast, it is believed that acquisition of embryogenic competence in tissue culture requires the presence of nonembryogenic cells that produce and secrete molecules into the culture medium. These molecules can then be perceived by other cells that in turn express their competence and develop into embryos (Pennell et al., 1992; de Jong et al., 1993). Several plant-produced molecules that may have a role in cell-to-cell signaling have been identified, including chitinases (de Jong et al., 1992) and arabinogalactan proteins (McCabe et al., 1997; Toonen et al., 1997a).
Are All Cells of the Embryo Sac Competent to Form Embryos?
Zygotic embryo development commences at fertilization and the zygote could therefore be considered to be the first “embryogenic cell” of the ovule. In line with this idea, DcSERK expression was not detected prior to fertilization in developing carrot flowers, suggesting that embryogenic competence is not acquired before formation of the zygote in carrot (Schmidt et al., 1997). Experiments with isolated egg cells have also shown that fertilization is an absolute requirement for embryogenesis to proceed (Dumas and Mogensen, 1993; Faure et al., 1994). However, AtSERK1 is clearly expressed before meiosis during ovule development in gametophytic and sporophytic tissue. At ovule maturity, all cells of the embryo sac express AtSERK1. Assuming that AtSERK1 expression strictly correlates with embryogenic potential, these results suggest that all cells of the embryo sac are competent for embryogenesis. Rare cases where non-gametic cells of the embryo sac such as the synergids and antipodals develop autonomously into an embryo have been described (for review, see Maheswari, 1950; Kamelina, 1995; Solntseva, 1995), but their existence remains controversial (Czapik, 1999; Shishkinskaya and Yudakova, 1999).
Somatic (sporophytic) cells surrounding the embryo sac can develop into an embryo, as occurs in apomixis (Koltunow, 1993; Koltunow et al., 1995), and reduced egg cells can initiate embryogenesis parthenogenetically in the absence of fertilization (Matzk et al., 1995; Matzk, 1996). Apomixis refers to asexual reproduction through seeds that involves the avoidance of meiosis and fertilization-independent embryo development (Grossniklaus, 2001). Different mechanisms for apomixis have been distinguished according to the identity of the initial cell that gives rise to the embryo sac or to the embryo. Gametophytic apomixis requires the formation of a non-reduced embryo sac and autonomous embryo development. In diplospory, the MMC gives rise to non-reduced spores, which in turn form a non-reduced embryo sac, whereas in apospory, a somatic cell develops into an non-reduced embryo sac (Koltunow, 1993). Adventitious embryony corresponds to the direct development of an embryo from cells outside of the sexual embryo sac. Depending on the plant species analyzed, it is considered that apomixis is controlled by one or two major loci (Nogler, 1995; Savidan, 1982; Koltunow et al., 1998; van Baarlen et al., 1999). It has been shown recently that in addition to these major loci, other genetic factors or modifiers are also important for the efficient expression of apomixis (Koltunow et al., 2000). These results suggest the existence of a complex pathway controlling apomixis. None of the components of the apomictic pathway have been identified yet, and a role for SERK in apomictic reproduction remains a possibility.
Somatic and gametophytic cells are competent to form embryos. This suggests that embryogenesis does not rely on specific information stored in the unfertilized egg cell or that other embryogenic cells have an egg cell-like character. However, the nature of the stimuli that induce embryo development in different situations remains unknown. Although all cells of the embryo sac express AtSERK1 and could be competent to form embryos, only the egg cell develops into an embryo after fertilization. Thus, the interaction between male and female gametes or even components delivered into the egg cell during fertilization might be the “inducers” of embryo development. Some of the genetically defined modifiers of apomixis (Koltunow et al., 2000) may also contribute to this induction.
The AtSERK1 Gene Determines Embryogenic Competence in Culture
Embryogenic competence in plant tissue culture is an operational definition (Toonen and de Vries, 1997). The processes that govern the property of embryogenic competence in plant cells remain largely unknown (Mordhorst et al., 1997). Our results indicate that AtSERK1 not only marks cells competent to form embryos, but is also involved in conferring this embryogenic competence. Thus, it appears that AtSERK1-activating ligands are present during embryogenic culture and are not the rate-limiting step in activation of the AtSERK1-mediated signal transduction cascade.
Mutations in the AMP1, CLV1, and CLV3 genes result in similar increase in embryogenic competence. It was proposed previously that the enhanced embryogenic capacity of these mutants is an indirect effect, resulting from an increased number of undifferentiated cells in the SAM of these mutants (Mordhorst et al., 1998). The higher AtSERK1 expression in amp1 cultures in comparison with wild type may correlate with the embryogenic competence of such undifferentiated cells. Therefore, one of the effects of AMP1 activity could be to suppress the expression of AtSERK1 after germination. It is interesting to note that the loss of function of AMP1 has a stronger effect on embryogenic potential and plant development than the 35S promoter-driven AtSERK1 expression. Artificial increase of AtSERK1 expression level is apparently not sufficient to overcome possible inhibition by AMP1. AtSERK1 RNA levels are higher in calli derived from amp1 mutants than in calli derived from 35S::AtSERK1 plants, consistent with the higher embryogenic potential of the amp1 cultures.
Induction of embryo development can also occur on leaves of plants ectopically expressing LEAFY COTYLEDON 1 (LEC1; Lotan et al., 1998) and on roots in the pickle (pkl) mutant (Ogas et al., 1997, 1999). The loss of function mutant lec1 shows trichome development on cotyledons, suggesting that early vegetative development is occurring during late embryogenesis. One explanation could be that the LEC1 transcription factor represses vegetative development, and as an unexpected side effect, its ectopic expression results in spontaneous somatic embryo formation. In a scenario similar to the one we propose for continued AtSERK1 expression in amp1 seedlings, the chromatin-remodeling factor PKL is suggested to repress the transcription of LEC1 (Ogas et al., 1997, 1999). As has been discussed (de Vries, 1998; Harada et al., 1998), PKL and LEC may be involved in repressing certain aspects of postembryonic development. Because none of the genes mentioned above have been shown to interact genetically, it appears therefore that several different independent pathways influence embryogenic competence.
SERK Signaling Pathway during Embryogenesis
Our results indicate that the SERK-mediated signaling pathway, as it occurs during somatic embryogenesis, is recruited from a pathway that operates normally during ovule development. Therefore, we propose that AtSERK1 could be a component of an embryogenesis-signaling pathway. Competent cells may contain an inactive receptor, which is activated by the presence of the proper ligand to switch on the embryogenesis program. In the near future, it would be of great importance to identify the components of the SERK signaling pathway, such as the activating ligand and downstream targets, and to determine whether other ovule-expressed genes are involved in this pathway.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype WS seeds were sown on filter paper incubated at 4°C for 48 h and were germinated on soil in a growth chamber at 22°C with 16-h-light/8-h-night periods.
Library Screening, Subcloning, and DNA Sequencing
The screening procedure is described in Sambrook et al. (1989). For the first screening, autoradiography was done at −80°C using x-ray films (XR-omat, Eastman-Kodak, Rochester, NY) and hyperscreen intensifying screens (Amersham, Buckinghamshire, UK). For the second and third screening, membranes were hybridized with the enhanced chemiluminescence direct nucleic acid labeling and detection kit (Amersham).
Independent clones (2 × 105) of an Arabidopsis ecotype Landsberg erecta genomic library in Lambda-FIXII were screened using the carrot (Daucus carota) cDNA clone 31-50 (Schmidt et al., 1997) as a probe. Six lambda phages were recovered, purified, and one of them was used for subcloning. The entire AtSERK1 gene, including the promoter, was included in this phage, subcloned in pBLUESCRIPT (Stratagene, La Jolla, CA), and completely sequenced.
Phages (1.5 × 106) from an amplified Lambda-ZAPII (Stratagene) cDNA library made from flower buds and open flowers (Li and Thomas, 1998) were plated according to the manufacturer's protocol. The plaques were hybridized with a 4-kb XbaI fragment of genomic AtSERK1 gene containing most of the coding sequence, and five positives were recovered and characterized by end sequencing.
DNA sequences were determined by the dideoxy-chain terminator method on double-stranded DNA templates (Sanger et al., 1977; Chen and Seeburg, 1985) using an ABI 373A (Applied Biosystems, Foster City, CA) automatic sequencer.
Primers Used
AtSERK1-specific primers used were: S1 (5′-TAAGTTTGTCAGATTTCCAAGATTACTAGG-3′), V1 (5′-TTGGAAATCTGACAAACTTAGTGAGTTTGG-3′), S2 (5′-TCGTCGCCACCAAGCAAAGGCTATTGCAGG-3′), V2 (5′-GCTGCTCCTGCAATAGCCTTTGCTTGGTGG-3′), S3 (5′-AGAGATATTCTGGAGCGATGTGACCGATGG-3′), V3 (5′-CGTGACAACAGCAGTCCGTGGCACCATCGG-3′), S4 (5′-TGCAGACACTAAAGATAGCGATTCACCTCC-3′), V4 (5′-TGGAGGTGAATCGCTATCTTTAGTGTCTGC-3′), S5 (5′-CACATTATGCTTACCCCATGTGGTGGATGG-3′), V5 (5′-ATGAAAATAAAGAGTCCATCCACCACATGG-3′), S6 (5′-ACCCTCAAAGTATGCAAAGC-3′), V6 (5′-ATGCTTTG-CATACTTTGAGG-3′), and V7 (5′-GACGACGACGAGAACGCGG-3′).
The cyclophilin constitutively expressed ROC5-specific primers used were: ROC5-5 (5′-TCTCTCTTCCAAATCTCC-3′) and ROC5-3 (5′-AAGTCTCTCACTTTCTCACT-3′).
AtSERK1 Gene Constructs and Transformation of Arabidopsis
The AtSERK1 promoter region of 2 kb was cloned into pGPTV-KAN (Becker et al., 1992) by directional cloning with a blunt end and SalI restriction enzyme sites. AtSERK1 full-length cDNA was cloned as an SacI-KpnI fragment in pRT105 (Töpfer et al., 1993) containing the CaMV 35S promoter. The 35S::AtSERK1 fragment was then transferred to the pMOG800-based binary vector (Toonen et al., 1997b) by HincII and HindIII digestion. The 35S::AtSERK1-EX construct was cloned in the binary vector pMOG800 by HindIII digestion. All constructs were verified by sequencing using the specific AtSERK1 primers and were electroporated in Agrobacterium tumefaciens strain C58C1 containing a disarmed C58 Ti plasmid (Koncz et al., 1989).
Arabidopsis ecotype WS plants were transformed by vacuum infiltration (Bechtold et al., 1993) with the different constructs. Between 24 and 36 plants were used for each transformation experiment. T1 seeds were selected on one-half-strength Murashige and Skoog salt medium (Murashige and Skoog, 1962; DUCHEFA, Haarlem, The Netherlands) supplemented with 10% (w/v) Suc and 50 mg L−1 kanamycin for 10 d. The kanamycin-resistant seedlings were transferred in soil and used for amplification of seeds, and each T1 plant was the mother plant of independent lines. The T2 seeds were selected on kanamycin, transferred to soil, and analyzed.
Initiation of Embryogenic Cultures
Embryogenic cultures were initiated using the amp1 seedling assay as described by (Mordhorst et al., 1998). In brief, around 30 seeds were surface sterilized and incubated in 20 mL of liquid medium. The induction medium was Murashige and Skoog salts containing 2% (w/v) Suc, 4.5 μm 2,4-D, and 10 mm MES [2-(N-morpholino)-ethanesulonic acid] medium at pH 5.8 (MS-4). After a cold treatment of 2 d at 4°C, cultures were kept on a rotary shaker (100 rpm) at 25°C in the light (3,000 lux for 16 h of light and 8 h of darkness). Each germinated seedling developed a callus aggregate. After 2 weeks of culture, and subsequently every week, medium was replaced with fresh MS-4 medium. After 3 weeks, seedlings developing embryogenic green clusters with a smooth surface and/or yellowish nonembryogenic callus were subcultured independently and gave rise to embryogenic and nonembryogenic cultures, respectively. Somatic embryos were obtained after culturing embryogenic clusters in absence of 2,4-D for 1 week.
GUS Assays
Plant tissues were immersed in ice-cold 90% (w/v) acetone and placed for 1 h at −20°C. The tissues were then washed three times for 20 min in 0.1 m sodium phosphate buffer (pH 7) containing 2.5 mm potassium ferry- and ferrocyanide. GUS staining was performed in 50 mm sodium phosphate buffer, pH 7, 10 mm EDTA, 0.1% (w/v) Triton X-100, 10% (w/v) dimethyl sulfoxide, 2.5 mm potassium ferri- and ferrocyanide, and 1 mg mL−1 of 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (DUCHEFA) for 2 to 4 h at 37°C for ovules and developing seeds and for more than 16 h for vascular tissues. Unfertilized carpels and ovules were cleared in 20% (w/v) lactic acid and 20% (w/v) glycerol in 1× phosphate-buffered saline, whereas fertilized carpels, developing seeds, and other organs were cleared in Hoyer's clearing solution (100 g of chloral hydrate, 5 mL of glycerol, and 30 mL of water). Observations were performed using an Optiphot microscope (Nikon, Tokyo) equipped with Nomarski optics. Pictures were taken with Ectachrom 320ASA (Eastman-Kodak) and were processed with Adobe Photoshop 5.0.2 (Adobe Systems, Mountain View, CA).
RT-PCR Analysis
Total RNA was extracted as described previously (Kay et al., 1987) from 2 to 4 g of different tissues (flower buds, opened flowers, siliques at different developmental stages, leaves, stems, roots, seedlings, embryogenic structures, and nonembryogenic structures). DNAseI treatment (Promega, Madison, WI), reverse transcription, and PCR reactions were performed according to Albrecht et al. (1998). For semiquantitative RT-PCR analysis, PCR products were collected after 24, 26, 28, 30, 32, 34, and 36 cycles to determine the linearity of the PCR. The linearity of the PCR was determined for AtSERK1 and ROC5 genes between 28 and 32 cycles. The amplified fragments were separated on 1.5% (w/v) agarose gels, blotted, and hybridized with the corresponding probe. The AtSERK1-specific primers used were V1 and S2, resulting in a fragment of 460 bp. The ROC5-specific primers used were ROC5-5 and ROC5-3 and resulted in a fragment of 568 bp (Chou and Gasser, 1997). The intensity of each band was measured by Image Quant for Macintosh (Molecular Dynamics, Sunnyvale, CA) and values were processed by Excel (Microsoft Office 98, Microsoft Corporation, Redmond, WA). AtSERK1 expression levels were calculated after normalization relative to ROC5 expression.
ISH
A BamHI fragment of 350 bp of AtSERK1 cDNA containing the 5′-untranslated region and the first two exons was subcloned in pBLUESCRIPT (Stratagene). Sense and antisense probes were obtained from this partial AtSERK1. ISH was performed according to Vielle-Calzada et al. (1999).
ACKNOWLEDGMENTS
We are grateful to Carlos Alonso Blanco and Ton Peeters for giving us the genomic DNA library, to Terry Thomas for providing us with the cDNA library, and to Chuck Gasser for the primer sequences for the amplification of ROC5. We thank Tony van Kampen for DNA sequencing and David Bouchez for mapping results. We are very grateful to Boudewijn van Veen for his help in the artwork in Figures 4 and 5. We especially thank Jim Weller for critical comments on the manuscript.
Footnotes
This work was supported through a collaboration with Novartis (grant nos. PL96662282 and ERBIO4–CT96–0689 from the European Union Biotechnology program to S.C.d.V.), by Pioneer Hi-Bred International (competitive research award to U.G. and J.-P.V.-C.), and by the Swiss National Science Foundation (fellowship to J.-P.V.-C.).
LITERATURE CITED
- Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 1998;15:605–614. doi: 10.1046/j.1365-313x.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baudino S, Hansen S, Brettschneider R, Hecht V, Dresselhaus T, Lörz H, Dumas C, Rogowsky P. Molecular characterization of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta. 2001;213:1–10. doi: 10.1007/s004250000471. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Becraft PW. Receptor kinases in plant development. Trends Plant Sci. 1998;3:384–388. [Google Scholar]
- Brown JWS, Smith P, Simpson CG. Arabidopsis consensus intron sequences. Plant Mol Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES. amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 1993;4:907–916. [Google Scholar]
- Chen Y, Seeburg PY. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–190. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chou IT, Gasser CS. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol. 1997;35:873–892. doi: 10.1023/a:1005930024796. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Le Paslier D, Cohen D, Chaboue ME, Durr A. The CIC library: a large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Czapik R. Enigma of apogamety. Protoplasma. 1999;208:206–210. [Google Scholar]
- de Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, van Kammen A, de Vries SC. A carrot somatic embryo mutant is rescued by chitinase. Plant Cell. 1992;4:425–433. doi: 10.1105/tpc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AJ, Schmidt EDL, de Vries SC. Early events in higher-plant embryogenesis. Plant Mol Biol. 1993;22:367–377. doi: 10.1007/BF00014943. [DOI] [PubMed] [Google Scholar]
- de Vries SC. Making embryos in plants. Trends Plant Sci. 1998;3:451–452. [Google Scholar]
- Dumas C, Mogensen HL. Gametes and fertilization: maize as a model system for experimental embryogenesis in flowering plants. Plant Cell. 1993;5:1337–1348. doi: 10.1105/tpc.5.10.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J-E, Digonnet C, Dumas C. An in vitro system for adhesion and fusion of maize gametes. Science. 1994;263:1598–1600. doi: 10.1126/science.263.5153.1598. [DOI] [PubMed] [Google Scholar]
- Fletcher LC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U. From sexuality to apomixis: molecular and genetic approaches. In: Savidan Y, Carman J, Dresselhaus T, editors. Advances in Apomixis Research. CYMMIT Press, Texcoco, Mexico. 2001. pp. 168–211. [Google Scholar]
- Guzzo F, Baldan B, Levi M, Sparvoli E, Lo Schiavo F, Terzi M, Mariani P. Early cellular events during induction of carrot explants with 2,4-D. Protoplasma. 1995;185:28–36. [Google Scholar]
- Guzzo F, Baldan B, Mariani P, Loschiavo F, Terzi M. Studies on the origin of totipotent cells in explants of Daucus carota L. J Exp Bot. 1994;45:1427–1432. [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;242:42–51. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Harada JJ, Lotan T, Fisher RL. … response: embryos without sex. Trends Plant Sci. 1998;3:452–453. [Google Scholar]
- He Z, Wang Z, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1933. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard JBL, editor. Embryos. Color Atlas of Development. London: Wolf Publishers; 1994. pp. 7–21. [Google Scholar]
- Kamelina O. Synergid apogamety in the genus Tetradiclis Stev. (Tetradiclidiaceae) and occurrence of this phenomenon in flowering plants. Apomixis Newslett. 1995;8:32–33. [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of the CaMV 35S promoter sequences create a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biol Sci. 1994;19:415–420. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Koltunow AM. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell. 1993;5:1425–1437. doi: 10.1105/tpc.5.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Bicknell RA, Chaudhurry AM. Apomixis: molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiol. 1995;108:1345–1352. doi: 10.1104/pp.108.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. Sexual and apomictic development in Hieracium. Sex Plant Reprod. 1998;11:213–230. [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sex Plant Reprod. 2000;12:253–266. [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Korber H, Redei GP, Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA. 1989;86:8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Laux T, Jürgens G. Embryogenesis: a new start in life. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease K, Ingham E, Walker JC. Challenges in understanding RLK function. Curr Opin Plant Biol. 1998;1:388–392. doi: 10.1016/s1369-5266(98)80261-6. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li Z, Thomas TL. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell. 1998;10:383–398. doi: 10.1105/tpc.10.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee MK, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON 1 is sufficient to induce embryo development in vegetative tissue. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Maheswari P. An Introduction to the Embryology of Angiosperms. New York: McGraw-Hill; 1950. [Google Scholar]
- Matzk F. The Salmon system of wheat: a suitable model for apomixis research. Hereditas. 1996;125:299–301. [Google Scholar]
- Matzk F, Meyer HM, Baumlein H, Balzer HJ, Schubert I. A novel approach to the analysis of the initiation of embryo development in Gramineae. Sex Plant Reprod. 1995;8:266–272. [Google Scholar]
- McCabe PF, Valentine TA, Forsberg LS, Pennell RI. Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell. 1997;9:2225–2241. doi: 10.1105/tpc.9.12.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopis thaliana: a model plant for genome analysis. Science. 1998;282:679–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Toonen MAJ, de Vries SC. Plant embryogenesis. Crit Rev Plant Sci. 1997;16:535–576. [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics. 1998;149:549–563. doi: 10.1093/genetics/149.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nogler GA. Genetics of apomixis in Ranonculus auricomus: VI. Epilogue Bot Helv. 1995;105:111–115. [Google Scholar]
- Ogas J, Cheng J-C, Sung ZR, Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryogenic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Scofield GN, Booij H, de Vries SC, Roberts K. Identification of a transitional cell state in the developmental pathway to carrot somatic embryogenesis. J Cell Biol. 1992;119:1371–1380. doi: 10.1083/jcb.119.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis R. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Niklen S, Coulson AR. Sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidan Y. Nature et hérédité de l'apomixie chez Panicum maximum Jacq. Trav Doc ORSTOM. 1982;153:1–159. [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hülskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 1995;7:731–749. [Google Scholar]
- Shah K, Schmidt EDL, Vlak JM, de Vries SC. Expression of the Daucus carota somatic embryogenesis receptor kinase (DcSERK) protein in insect cells. Biochimie. 2001a;83:415–421. doi: 10.1016/s0300-9084(01)01257-3. [DOI] [PubMed] [Google Scholar]
- Shah K, Vervoort J, de Vries SC (2001b). Role of threonines in the AtSERK1 activation loop in auto- and transphosphorylation. J Biol Chem (in press) [DOI] [PubMed]
- Solntseva PM. Letters. Apomixis Newslett. 1995;8:55–57. [Google Scholar]
- Shishkinskaya NA, Yudakova OI. The problem of apogamety in plants. Apomixis Newslett. 1999;11:12–13. [Google Scholar]
- Somleva MN, Schmidt EDL, de Vries SC. Embryonic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep. 2000;19:718–726. doi: 10.1007/s002999900169. [DOI] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interaction. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:795–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Toonen M, Schmidt E, Hendriks T, Verhoeven H, de Vries S. Identification of single embryo-forming cells in carrot suspension cultures. Acta Bot Neerl. 1993;42:518–519. [Google Scholar]
- Toonen MAJ, Schmidt EDL, van Kammen A, de Vries SC. Promotive and inhibitory effects of diverse arabinogalactan proteins on Daucus carota L. somatic embryogenesis. Planta. 1997a;203:188–195. [Google Scholar]
- Toonen MAJ, Verhees JA, Schmidt EDL, van Kammen A, de Vries SC. AtLTP1 luciferase expression during carrot somatic embryogenesis. Plant J. 1997b;12:1213–1221. doi: 10.1046/j.1365-313x.1997.12051213.x. [DOI] [PubMed] [Google Scholar]
- Töpfer R, Maas C, Höricke-Grandpierre C, Schell J, Steinbiss HH. Expression vectors for high-level gene expression in dicotyledonous and monocotyledonous plants. Methods Enzymol. 1993;217:66–78. doi: 10.1016/0076-6879(93)17056-b. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Mayda E, Gómez MD, Cañas L, Conjero V, Vera P. Characterization of LRP, a leucine-rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J. 1996;10:315–330. doi: 10.1046/j.1365-313x.1996.10020315.x. [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang ZB, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–405. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science. 2000;289:613–617. doi: 10.1126/science.289.5479.613. [DOI] [PubMed] [Google Scholar]
- van Baarlen P, Verduijn M, van Dijk PJ. What can we learn from natural apomicts? Trends Plant Sci. 1999;4:43–44. [Google Scholar]
- van der Kooy D, Weiss S. Why stem cells. Science. 2000;287:1439–1441. doi: 10.1126/science.287.5457.1439. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J-P, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage site. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Recklinghausen IR, Iwanowska A, Kieft H, Mordhorst AP, Schel JHN, van Lammeren AAM. Structure and development of somatic embryos formed in Arabidopsis thaliana pt callus cultures derived from seedlings. Protoplasma. 2000;211:217–224. [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]