Abstract
Phytosulfokine-α (PSK-α), a unique plant peptide growth factor, was originally isolated from conditioned medium of asparagus (Asparagus officinalis) mesophyll cell cultures. PSK-α has several biological activities including promoting plant cell proliferation. Four genes that encode precursors of PSK-α have been identified from Arabidopsis. Analysis of cDNAs for two of these, AtPSK2 and AtPSK3, shows that both of these genes consist of two exons and one intron. The predicted precursors have N-terminal signal peptides and only a single PSK-α sequence located close to their carboxyl termini. Both precursors contain dibasic processing sites flanking PSK, analogous to animal and yeast prohormones. Although the PSK domain including the sequence of PSK-α and three amino acids preceding it are perfectly conserved, the precursors bear very limited similarity among Arabidopsis and rice (Oryza sativa), suggesting a new level of diversity among polypeptides that are processed into the same signaling molecule in plants, a scenario not found in animals and yeast. Unnatural [serine-4]PSK-β was found to be secreted by transgenic Arabidopsis cells expressing a mutant of either AtPSK2 or AtPSK3 cDNAs, suggesting that both AtPSK2 and AtPSK3 encode PSK-α precursors. AtPSK2 and AtPSK3 were expressed demonstrably not only in cultured cells but also in intact plants, suggesting that PSK-α may be essential for plant cell proliferation in vivo as well as in vitro. Overexpression of either precursor gene allowed the transgenic calli to grow twice as large as the controls. However, the transgenic cells expressing either antisense cDNA did not dramatically decrease mitogenic activity, suggesting that these two genes may act redundantly.
The sporophytic phase of seed plants encompasses embryonic and postembryonic development. Following fertilization, the zygote undergoes cell division to produce the embryo. Higher plants elaborate much of their architecture postembryonically through development initiated at the tips of shoots and roots: Cell division is activated in the shoot and root apical cells, generating the shoot and root meristems. It is known that these apical meristematic cells can remain in the cell cycle to generate new cells for growth and development throughout the plant's life, but the molecular mechanisms are not yet well understood.
In animal systems, cells release and recognize extracellular signaling molecules such as hormones and growth factors. Oligopeptides play a major role in cell-cell communication, whereas mitogenic peptides contribute to cell cycle control, reactivation of the cell cycle being dependent on such extracellular signal molecules (Meyerowitz et al., 1998). Several peptide signals recently were isolated from plants. For example, systemin from tomato regulates the synthesis of defensive proteins in plant tissues (Pearce et al., 1991; McGurl et al., 1992). In Arabidopsis, CLAVATA3 (CLV3), a small extracellular protein, acts as a ligand in a stem cell-restricting signal transduction pathway that regulates the balance between cell proliferation and differentiation at the shoot meristem (Clark et al., 1997; Fletcher et al., 1999; Brand et al., 2000; Trotochaud et al., 2000). However, only one kind of mitogenic oligopeptide, the phytosulfokines (PSKs), has been identified so far.
PSKs were originally isolated from conditioned medium (CM) of asparagus (Asparagus officinalis) mesophyll cell cultures and their structures determined as a sulfated pentapeptide, PSK-α [Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln], and a C-terminal-truncated tetrapeptide, PSK-β [Tyr(SO3H)-Ile-Tyr(SO3H)-Thr] (Matsubayashi and Sakagami 1996). PSK-α is the most active compound although its amount in CM is less than 20% of that of PSK-β. PSK-β recently was shown to be an enzymatic derivative of PSK-α (Yang et al., 1999b). The latter strongly stimulates proliferation of plant cells in low-density cultures, the minimum concentration exhibiting mitogenic activity being 1.0 × 10−9 m. Although PSK-β contributes the major form in CM, it exhibits activity at a lower concentration of 1.0 × 10−8 m (Matsubayashi and Sakagami 1996).
PSK-α has several other biological activities in addition to promoting plant cell proliferation. For example, it enhances chlorophyll synthesis in etiolated cotyledons of cucumber (Yamakawa et al., 1998a), as well as the growth and chlorophyll content of Arabidopsis seedlings under high nighttime temperature conditions (Yamakawa et al., 1999). PSK-α also promotes adventitious root formation by hypocotyls of cucumber (Yamakawa et al., 1998b) and adventitious bud formation in snapdragon (Antirrhnum majus) (Yang et al., 1999a). Furthermore, it stimulates both tracheary element differentiation of isolated mesophyll cells from zinnia (Matsubayashi et al., 1999) and somatic embryogenesis in carrot cultures (Kobayashi et al., 1999; Hanai et al., 2000a).
We previously cloned the OsPSK gene encoding a precursor of PSK-α from monocot rice (Oryza sativa; Yang et al., 1999b, 2000a). Expression was found not only in cultured cell but also in intact plants (Yang et al., 1999b). In intact plants, transcripts of the OsPSK gene are concentrated in shoot and root meristems in which cells proliferate vigorously (Yang et al., 1999b). PSKs have also been found in dicots including Arabidopsis (Yang et al., 2000a), and the amino acid sequences of both PSK-α and PSK-β are perfectly conserved among different species (Yang et al., 2000a).
The flowering plant Arabidopsis is an important model system for identifying genes and determining their functions, because of its small body, short generation time, and small genome size. In the present article, we describe the functional and comparative analysis of two PSK precursor genes, AtPSK2 and AtPSK3, in Arabidopsis. Both precursors exhibit dibasic processing sites flanking PSK, analogous to animal prohormones. Although the PSK domain including the sequence of PSK-α is perfectly conserved, the two precursors bear very limited similarity. This structure suggests a new level of diversity among polypeptides that are processed into the same signaling molecule in plants, a scenario not found in animals and yeast.
RESULTS
Characterization of Arabidopsis Genes for Precursors to PSK
Although PSK peptides had been detected in Arabidopsis cell cultures (data not shown), our attempts to clone Arabidopsis PSK precursor gene(s) using a rice PSK precursor cDNA (OsPSK) as a probe were unsuccessful, suggesting that Arabidopsis PSK precursor gene(s) may share very low similarity with the rice OsPSK gene. To identify OsPSK homologous gene(s), the Arabidopsis expressed sequence tags (EST) were searched with the BLAST program using the amino acid sequence of PSK-α. Several putative homologs were found to encode proteins that contain the PSK sequence. Their sequences were compared and showed to correspond to two genes that we designated as AtPSK2 and AtPSK3, respectively, based on their chromosomal location.
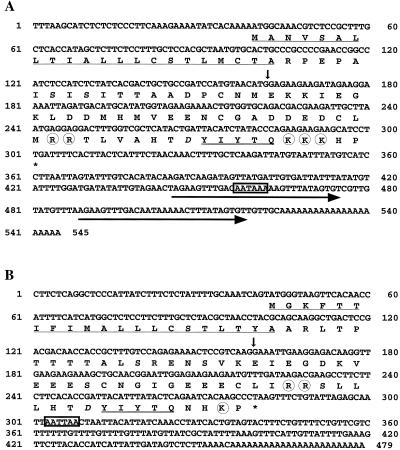
AtPSK2 cDNA (EST no. 120C15T7) is 524 bp in length with polyadenylation motifs (AATAAA) that present 19 and 62 bp upstream from the polyadenyl tail (Fig. 1A). Two imperfect 35-bp repeats were noted in the 3′-untranslated region (Fig. 1A). The AtPSK2 cDNA contains an open reading frame of 261 bp that can code for a precursor to PSK with 87 amino acids (Fig. 1A) and a predicated molecular mass of 9.6 kD. In contrast, AtPSK3 cDNA (EST no. 91K2T7) has no repeats and is shorter than AtPSK2 cDNA. The precursor deduced from AtPSK3 cDNA (79 amino acids with a predicated molecular mass of 8.9 kD) is also smaller than that of AtPSK2. (Fig. 1B).
Figure 1.
Nucleotide and deduced amino acid sequences of AtPSK2 and AtPSK3 cDNAs. The deduced amino acid sequences with single-letter abbreviations are shown below the nucleotide sequences of AtPSK2 cDNA (A) and AtPSK3 cDNA (B). The positions of introns are shown with vertical arrows. The repeats are indicated with horizontal arrows. The most likely sequences for a polyadenylation signal are boxed. The potential N-terminal signal sequences are underlined, and the PSK-α sequence is underscored with double lines. The Asp residues near PSK-α are printed in italic, and the putative bordering processing sites are circled. The nucleotide sequences reported in this paper have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases (accession nos. AB029344, AB052752, AB029820, and AB050627).
To analyze the genomic structure of the AtPSK genes, we isolated clones containing the entire AtPSK2 or AtPSK3 genes. The AtPSK2 gene was shown to consist of two exons (162 and 362 bp) that perfectly match the cDNA sequence interrupted by an intron of 148 bp with a well-conserved GT-AG intron border sequence. The first exon consists of the 5′-non-coding region and a coding region for the 22-amino acid N-terminal signal peptide. The second exon was found to consist of the 3′ non-coding region and a coding region for the remaining residues of the precursor including the 5-amino acid PSK-α sequence. AtPSK3, like AtPSK2, consists of two exons and one intron, but the intron (265 bp) of AtPSK3 is longer than that of AtPSK2.
Structures of the PSK Precursors Encoded by AtPSK2 and AtPSK3
As expected, the AtPSK2 has a potential hydrophobic N-terminal signal sequence (residues 1–22), and the mature precursor is predicted to be 65 amino acids in length (Fig. 1A). The 5-amino acid PSK-α sequence occurs only once (amino acids 78–82) within the AtPSK2 close to its C terminus, as shown in Figure 1A. There is an Asp residue immediately N terminal to the first Tyr of PSK-α in the −1 position (Fig. 1A), suggesting the Tyr residues in the PSK-α sequence can be sulfated (Hanai et al., 2000b). It is interesting that two Arg (amino acids 69–70) and three Lys (amino acids 83–85) residues exist (Fig. 1A), suggesting that putative processing sites bordering PSK-α of the Arabidopsis PSK precursor conform to the consensus sequence for endoproteolytic processing sites flanking bioactive peptides in animal prohormone precursors (Harris, 1989), and that PSK-α can be proteolytically processed from the Arabidopsis PSK precursor.
The AtPSK3 also was found to contain a potential N-terminal signal sequence (amino acids 1–21) and one copy of the PSK-α sequence (amino acids 71–75) at its C terminus (Fig. 1B). The Asp residue is conserved in the −1 position relative to the first Tyr of PSK-α. Also, two Arg (amino acids 62–63) and one Lys (amino acid 28) residues were found to border the PSK-α sequence (Fig. 1B).
A search for homologies to AtPSK2 and AtPSK3 genes in the Arabidopsis Database (AtDB) indicated that the AtPSK2 gene is located on chromosome II, within the region contained in bacterial artificial chromosome clone T20K9, and the closest genetic marker is mi238, a recombinant inbred marker that exists at 39.92 cM (Liu et al., 1996). The bacterial artificial chromosome clone T16K5 harbored the AtPSK3 gene indicating that AtPSK3 is located on chromosome III close to MS–3–1 (Reiter et al., 1992), a recombinant inbred marker that exists at 72.75 cM.
Similarity among Arabidopsis and Rice Precursors to PSK
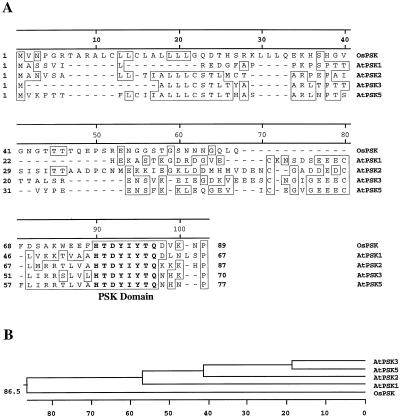
Two more PSK precursor genes recently were found in the AtDB by a BLAST homology search using the sequence of the PSK domains. These genes were designated as AtPSK1 (accession no. AC027656.4) and AtPSK5 (accession no. AB018108.1), respectively. The AtPSK1 gene is located on chromosomes I, close to mi348 (23.7 cM), whereas the AtPSK5 gene is present in the south part of chromosome V, close to mi335 (131.1 cM). Both the AtPSK1 and AtPSK5 genes also have a single intron and the predicted amino acid sequences contain N-terminal signal peptides. Therefore, the Arabidopsis genome appears to possess a gene family encoding PSK precursors.
The amino acid sequences of the Arabidopsis and rice PSK precursors were aligned using the Clustal W (1.7) Multiple Sequence Alignment provided by the GenomeNet CLUSTALW Server (Thompson et al., 1994). It is quite unexpected that these precursors do not share significant similarity throughout the whole length, although the PSK-α sequence and three amino acid residues immediately N terminal to it are perfectly conserved (Fig. 2). We refer to this eight-amino acid region as the PSK domain. In addition, a very limited similarity was found for N-terminal signal peptides.
Figure 2.
Amino acid sequence comparison of Arabidopsis and rice PSK precursors. The alignment (A) and phylogenetic tree (B) were generated using the MegAlign program. Single-letter abbreviations for the amino acid residues are used. Gaps are shown as dashes (−). Identical amino acid residues are boxed. The amino acid sequence of the PSK domain is in bold.
Copy Numbers of the PSK Precursor Genes
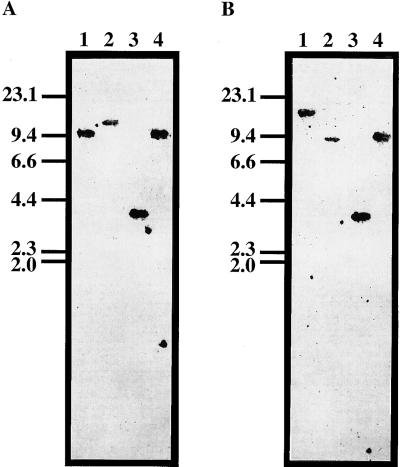
Copy numbers of the AtPSK2 and AtPSK3 genes in the Arabidopsis genome were investigated by genomic Southern-blot analyses. Wild-type DNA was probed with the full-length cDNAs and hybridizations were performed under low- and high- stringency conditions. As results, only one band was detected under either low or highly stringent conditions in the DNA fragments derived from single digestion of the four different restriction enzymes (Fig. 3, A and B), indicating that both AtPSK2 and AtPSK3 are single-copy genes.
Figure 3.
DNA gel blot showing the presence of AtPSK2 and AtPSK3 genes. Total DNA (10 μg) isolated from Arabidopsis culture cells was digested with BamHI (lane 1), EcoRI (lane 2), HindIII (lane 3), or XbaI (lane 4) and subjected to DNA gel-blot analysis using 32P-labeled AtPSK2 (A) or AtPSK3 cDNA (B) at 50°C. Marker lengths are indicated on the left in kilobases.
Expression of PSK Precursor cDNAs in Transgenic Cells
To examine whether AtPSK2 and AtPSK3 code for precursors to PSK, we transformed 28-d-old Arabidopsis roots with artificially mutated AtPSK2 or AtPSK3 cDNAs in sense orientation, which were designed to produce [Ser-4]PSK-α [Tyr(SO3H)-Ile-Tyr(SO3H)-Ser-Gln] instead of PSK-α. The chimeric genes genes harboring the mutated cDNAs in frame were placed under the control of the constitutive cauliflower mosaic virus 35S promoter incorporated within the binary vector pMAT037 (Matsuoka and Nakamura, 1991). B5 medium (Gamborg et al., 1968) supplemented with 0.5 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.05 mg L−1 kinetin (KIN) was used to induce calli; transgenic calli were selected on basis of kanamycin resistance. Transgenic calli were obtained at a frequency of 79%. DNA gel-blot hybridization analyses confirmed the presence of transgenes; around 70% of the transformants had a single locus insertion (data not shown).
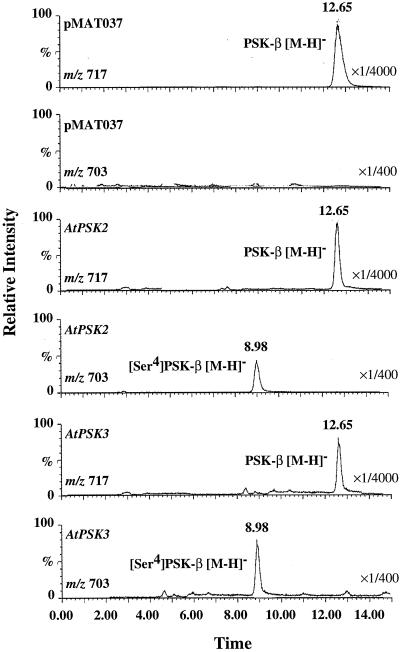
Transgenic cells were transferred to liquid callus-inducing medium after 1 month. Two weeks after the onset of subculture, PSK-α and its analogs in the CM by the transgenic cells were purified and analyzed by liquid chromatography/mass spectrometry (LC/MS). Although only PSK-β was detected with transgenic cells containing the pMAT037 vector alone, both PSK-β and [Ser-4]PSK-β-α [Tyr(SO3H)-Ile-Tyr(SO3H)-Ser] peaks were present in eluates derived from the CM of transgenic cells harboring either the mutated AtPSK2 or AtPSK3 cDNAs (Fig. 4).
Figure 4.
Mass chromatograph-obtained LC/MS analyses. The transgenic cells harboring the empty vector, the mutated AtPSK2 cDNA, or the mutated AtPSK3 cDNA were cultured in B5 liquid medium for 2 weeks to prepare CM. PSK-α and its analogs containing in the CM were concentrated by two steps of column chromatograph and subjected to LC/MS analysis with selected ion monitoring at m/z 703 ([M-H]− of [Ser-4]PSK-β) and m/z 717 ([M-H]− of PSK-β). The peaks eluting at 8.9 and 12.65 were [Ser-4]PSK-β and PSK-β, respectively.
Transgenic cells harboring the mutated cDNAs were quantified for their secretion of PSK analogs into CM. Amounts of the PSKs in CM were measured by LC/MS based on peak heights without an internal standard. Transgenic cells containing the binary vector alone served as controls. The amount of [Ser-4]PSK-β produced by the transgenic cells with the mutated cDNA was one order of magnitude higher than that of PSK-β derived from the endogenous gene (Table I).
Table I.
PSK production by transgenic Arabidopsis cells
| Construct | PSK-β | [Ser4]PSK-β |
|---|---|---|
| PMAT037 | 413 ± 30 | 0 |
| [Ser4]PSK AtPSK2 cDNA | 387 ± 41 | 3,750 ± 40 |
| [Ser4]PSK AtPSK3 cDNA | 350 ± 36 | 4,785 ± 50 |
Transgenic cells (0.8 g) harboring pMAT037 alone or mutated cDNAs were incubated in 100 mL of fresh B5 liquid medium supplemented with 0.5 mg L−1 of 2,4-D and 0.05 mg L−1 of KIN at 25°C in the light with rotary shaking at 120 rpm. After 2 weeks of culture, PSK-β and its analog concentrations in the CM were quantified by LC/MS. PSK concentrations are presented in pm. Averages of three independent experiments are given with the sd. PSK-α was not detectable by this assay because its amount was beyond the detection limit.
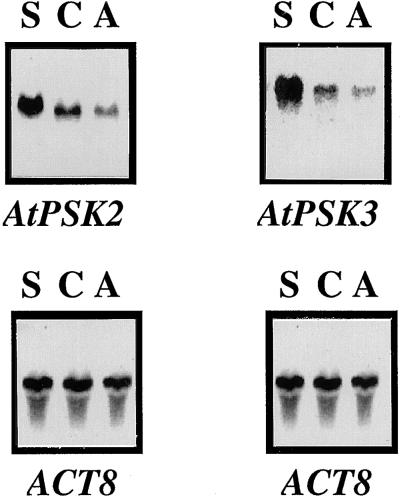
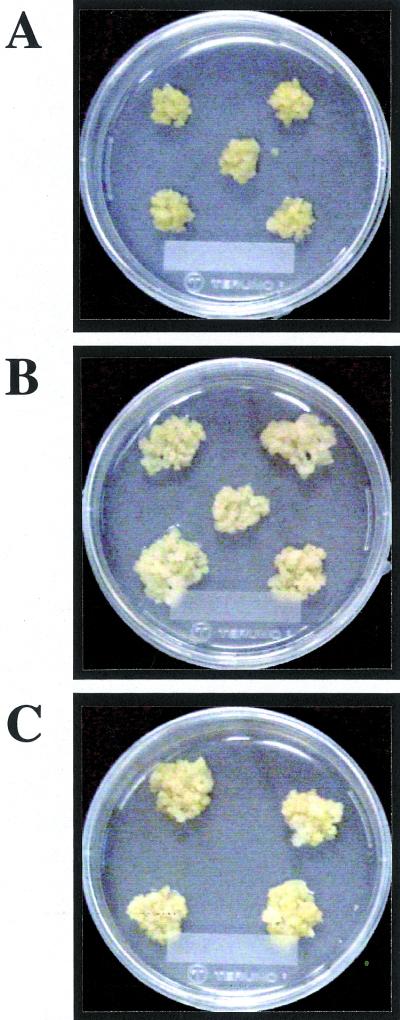
We also introduced wild-type cDNAs in the sense and antisense orientations into Arabidopsis cells to examine the effects of supernumerary or suppressed expression of the PSK precursor genes on cell proliferation. Amounts of PSK precursor mRNA were higher in the sense but lower in the antisense transgenic cells than in the controls (Fig. 5). Overexpression of the PSK precursor genes allowed the transgenic calli to grow twice as large as the controls (Fig. 6). However, the transgenic cells in which expression of the PSK precursor genes was suppressed by the antisense cDNA did not dramatically decrease mitogenic activity (data not shown), suggesting that these two genes may act redundantly.
Figure 5.
Changes in AtPSK2 and AtPSK3 transcripts in transgenic cells. Twenty-microgram aliquots of total RNA extracted from sense transgenic cells (lane S), control (lane C), or antisense transgenic cells (lane A) cultured for 14 d were separated on 1.2% (w/v) agarose gels, transferred to nylon membranes, and allowed to hybridize with 32P-labeled full-length AtPSK2 or AtPSK3 cDNAs, as described in “Materials and Methods.” The blots were reprobed with an Arabidopsis actin cDNA to indicate equal loading of RNA.
Figure 6.
Comparison of growth of control and transgenic calli. Control or transgenic cells were transplanted into fresh B5 medium containing 0.5 mg L−1 of 2,4-D, 0.05 mg L−1 of KIN, and 0.8% (w/v) agar. Photographs were taken 2 weeks after culture at 25°C under 16-h-light/8-h-dark cycle. This experiment was repeated for three times and four or five lines of each construct were shown. A, Transgenic calli harboring empty pMAT037 alone. B, Transgenic calli harboring the sense AtPSK2 chimeric gene. C, Transgenic calli harboring the sense AtPSK3 chimeric gene.
Expression of the AtPSK2 and AtPSK3 Genes
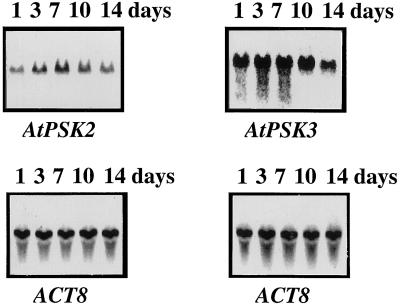
RNA-blot analysis with RNA extracted from suspension culture cells revealed the AtPSK2 gene to be continuously expressed, suggesting provision of a supply of PSK-α, which allows the cells to proliferate rapidly. The transcripts increased gradually after cells were transformed to fresh medium and maximal expression values were observed after 7 to 10 d, followed by a decrease (Fig. 7). AtPSK3 showed an expression pattern similar to that of AtPSK2, although maximal expression started earlier (Fig. 7).
Figure 7.
Persistent expression of AtPSK2 and AtPSK3 in cultured Arabidopsis cells. Twenty micrograms of total RNA extracted from Arabidopsis cells cultured for 1 (lane 1), 3 (lane 3), 7 (lane 7), 10 (lane 10), or 14 (lane 14) d were separated on 1.2% (w/v) agarose gels, transferred to nylon membranes, and allowed to hybridize with 32P-labeled full-length AtPSK2 or AtPSK3 cDNAs, as described in “Materials and Methods.” The blots were reprobed with an Arabidopsis actin cDNA to indicate equal loading of RNA.
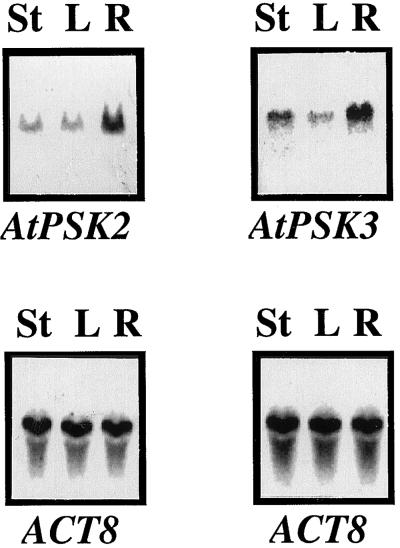
AtPSK2 transcripts also accumulated in 28-d-old Arabidopsis plants and were most abundant in roots (Fig. 8). AtPSK3 was also expressed most abundantly in roots, but transcripts in stems including shoot apexes were greater in this case than with AtPSK2 (Fig. 8). These findings suggest that the PSK-α molecule is also produced and has physiological functions in intact plants.
Figure 8.
Tissue specificity of AtPSK2 and AtPSK3 expression. Twenty micrograms of total RNA prepared from stems (lane S), leaves (lane L), and roots (lane R) of 28-d-old Arabidopsis plants were separated on 1.2% (w/v) agarose gels, transferred to nylon membranes, and allowed to hybridize with 32P-labeled full-length AtPSK2 or AtPSK3 cDNAs, as described in “Materials and Methods.” The blots were reprobed with an Arabidopsis actin cDNA to indicate equal loading of RNA.
DISCUSSION
PSK-α, the first sulfated peptide found in plants, is a unique signal strongly promoting proliferation of plant cells at low concentrations that may be universally distributed in the plant kingdom (Yang et al., 2000a). We previously identified OsPSK, a PSK-α precursor gene in rice (Yang et al., 1999b, 2000b), and in this article have described the molecular cloning and characterization of AtPSK2 and AtPSK3 genes that encode PSK precursors in Arabidopsis.
AtPSK2 or AtPSK3 are found as single copies with two exons and one intron, on chromosomes II and III, respectively. The first exon of AtPSK2 contains a coding region for about half of the PSK precursor including the N-terminal region that might serve as a signal peptide involved in secretion, a common feature of animal active peptide hormones, and the second exon has a coding region within which the 5-amino acid PSK-α sequence occurs only once, close to the C terminus, as reported for rice OsPSK (Yang et al., 2000b). AtPSK3 also shares these characteristics. Although the GT-AG intron border sequences are well conserved in both Arabidopsis and rice PSK precursors, the introns of the Arabidopsis PSK precursor genes (148 bp for AtPSK2 and 256 bp for AtPSK3) are only 13% to 22% identical to the OsPSK intron (1,150 bp). This difference may reflect characteristics of Arabidopsis: The size of the genome is the smallest of those for higher plants. Although OsPSK cDNA has 16 GA repeats in the 5′-untranslated region (Yang et al., 1999b), and AtPSK2 cDNA contains two 35-bp repeats in the 3′-untranslated region, AtPSK3 cDNA has no such repeats (Fig. 1, A and B), and these possible functions remain to be clarified. AtPSK2 consists of 87 amino acids with a predicated molecular mass of 9.6 kD, similar to those of rice PSK precursor (89 amino acids and 9.8 kD; Yang et al., 1999b). On the other hand, AtPSK3 is composed of 79 amino acids with a predicated molecular mass of 8.9 kD and thus is smaller than both the rice PSK precursor and AtPSK2.
A comparison of Arabidopsis and rice PSK precursors at the amino acid sequence level uncovered little similarity between AtPSK2 and AtPSK3 and only limited resemblance to rice PSK precursor. However, it is noteworthy that an eight-amino acid (HTDYIYTQ) region is perfectly conserved, and some similarity was also observed among the three N-terminal peptides (Fig. 2). The conserved eight-amino acid region includes the PSK-α sequence itself and the Asp residue (Fig. 2) that has been proved to be of importance for Tyr sulfation at −1 to the first Tyr of PSK (Hanai et al., 2000b). Therefore, this region, which we have termed the “PSK domain,” may be the most important for posttranslational modification of the PSK precursors and the production of PSK-α.
The similarity at the nucleic acid level among OsPSK, AtPSK2, and AtPSK3 genes is so low that they do not cross-hybridize even under low-stringency conditions. AtPSK1 and AtPSK5 genes recently were found in the AtDB by a BLAST homology search using the sequence of the PSK domains. The AtPSK1 gene is located on chromosome I, whereas the AtPSK5 gene is present in the south part of chromosome V. It is interesting that these genes bear very low similarity. Our data presented here suggest a new level of diversity among polypeptides that are processed into same signaling molecules in plants, a scenario not found in animals and yeast. Furthermore, several rice EST clones (C743, D15509, and S15186) may also encode proteins that contain the PSK domain but share very limited similarity, suggesting that the divergence observed outside the PSK domain is a more general feature of (at least two) plant PSK precursor gene families.
Polyploidy occurs widely in plants and it is proposed to be a key factor in plant evolution (Wendel, 2000). The evolution of Arabidopsis apparently involved a whole-genome duplication, followed by subsequent gene loss and extensive local gene duplications, giving rise to a dynamic genome (The Arabidopsis Genome Initiative, 2000). It is interesting that AtPSK2 and AtPSK3 genes are located in the large duplicated segments of chromosomes II and III, suggesting a derivation from the same ancestor but divergence after the duplication event. However, the long period of time over which genome stabilization has occurred has provided ample opportunity for divergence of functions of genes during through duplication. In many cases, the number of copies of a gene and its counterpart differ (for example, one copy on one chromosome and multiple copies on the other). As described above, Arabidopsis PSK precursor genes are separately located on chromosomes I, II, III, and V.
Protein Tyr O-sulfation is one of the posttranslational modifications that occurs with many secretory and membrane proteins in animal cells (Huttner, 1984). It has been shown to affect the biological activity of tyrosylproteins (Niehrs and Huttner, 1990) and deletion of the sulfate groups of Tyr-1 and Tyr-3 results in compounds with 0.6% and 4% of the activity of PSK-α (Matsubayashi et al., 1996). Unsulfated PSK-α has neither competitive ability for ligand binding nor mitogenic activity (Matsubayashi et al., 1997). Sulfated tyrosines are located within acidic regions of tyrosylproteins and all sites that have been characterized in animals have associated aspartic or Glu, with three or more acidic residues usually found between −5 and +5 in tyrosylproteins (Dorner and Kaufman, 1990). We previously demonstrated the existence of an Asp at −1, a Glu at −5, and an Asp at position +5 relative to the first Tyr, as well as aspartic acids at −3 and +5 positions around the second Tyr residue (Yang et al., 1999b). Amino acid substitution tests confirmed that the acidic amino acid residues adjacent to the Tyr residues of rice PSK precursor to be essential for Tyr sulfation, and that the Asp at −1 to the first Tyr is the most important determinant in rice PSK precursor (Hanai et al., 2000b). In Arabidopsis PSK precursors, the PSK-α sequence also exists within the acidic region, and an Asp at −1 to the first Tyr is conserved in the PSK domain (Fig. 2), suggesting that the Tyr residues in Arabidopsis PSK precursors could be similarly sulfated.
Peptide hormones and other biologically active peptides are generally synthesized as inactive higher Mr precursors that must undergo a variety of posttranslational processing steps to yield the active forms (Harris, 1989). It is considered that the primary processing recognition sequence for endoproteolysis of animal prohormone precursor proteins is a pair of basic amino acid residues (Lys-Arg, Lys-Lys, or Arg-Arg) that bracket the peptide hormone (Harris, 1989). It is interesting that a doublet of basic amino acids (Arg-Arg) was conserved in the Arabidopsis PSK precursors close to the N terminus of PSK-α (Fig. 1), suggesting that the initial cleavage event with Arabidopsis PSK precursors may be similar to that of animal prohormones. Consensus sequences for Trypsin were found at amino acid residues 69 to 70 and 83 to 85 in the AtPSK2, and 62 to 63 as well as 78 in the AtPSK3, flanking PSK-α on both sides (Fig. 1, A and B). However, there is no such recognition site in the amino acid sequence of rice PSK precursor (Yang et al., 1999b), suggesting that the endoproteolytic enzymes may differ between monocots and dicots. Details of the processing procedure are now under investigation.
Our transgenic experiments demonstrated that the AtPSK2 and AtPSK3 cDNAs could produce PSK-α in transformed Arabidopsis cells. The finding of [Ser-4]PSK-β in the CM prepared from transgenic cells harboring the mutated AtPSK2 or AtPSK3 cDNAs simultaneously confirmed that PSK-β is an enzymatic derivative of PSK-α (Yang et al., 1999b). The fact that the amount of [Ser-4]PSK-β produced by mutated AtPSK2 or AtPSK3 cDNAs was more than PSK-β produced by the endogenous gene might result from a higher level of expression of foreign genes and no influence to the modification and/or processing efficiency by the amino acid substitution. It is also plausible that the degradation speed of [Ser-4]PSK-β is slower than that of natural PSK-β in Arabidopsis cultures.
Our previous data indicated that rice Oc cells produce more PSKs than any other plant cells tested. We could clearly detect PSK-α as well as PSK-β from only 40 mL of CM derived from Oc cells by LC/MS analysis (Yang et al., 1999b). PSK-α is the active principle of cell proliferation activity in CM; however, the amount of PSK-β is much more than that of PSK-α in all investigated cultures. The total amount of PSKs in Arabidopsis cultures were found to be almost two orders less than that produced by rice Oc cells, explaining why PSK-α was not detectable by LC/MS in the present study. It is likely that the degradation of PSK-α in the Arabidopsis culture is much faster than in rice Oc culture or alternatively, the PSK degradation system in rice Oc culture is unusually weak.
We found that AtPSK2 and AtPSK3 transcripts remained at a high level in cultured Arabidopsis cells with maximum values observed 7 to 10 d after transplanting (Fig. 7). Hitherto, the PSK-α peptide has not been detected in any intact plants with chemical and immunological methods. We previously detected OsPSK transcripts in rice seedlings by reverse transcriptase-PCR (Yang et al., 1999b), and here, we could detect transcripts of both the AtPSK2 and AtPSK3 genes in Arabidopsis plants by northern hybridization (Fig. 8), indicating that these PSK precursor genes may also function in whole plants.
AtPSK2 and AtPSK3 transcripts accumulated in all tissues tested (Fig. 8), suggesting that these two genes may act redundantly. AtPSK1 and AtPSK5 genes are also expressed in a similar pattern (data not shown). Such pairs of genes allow functional stabilization in case one becomes switched off. To clarify the mode of action of AtPSK2 or AtPSK3 genes in intact plants, it is of importance to make transgenic plants in which they are inactivated, both individually and in combination.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis ecotype Columbia (collection no. Col-0) were purchased from Lehle Seeds (Round Rock, TX), sown on vermiculite, and grown in a greenhouse at 25°C under continuous illumination from daylight fluorescent tubes providing 150 mmol m−2 s−1 (FL40SBRN, Toshiba, Tokyo). Plants 28 d after germination were used as the source of RNA for northern-blot analysis.
Establishment of Arabidopsis Cell Suspensions
Arabidopsis seeds (collection no. Col-0) were surface sterilized for 5 min in 70% (v/v) ethanol, washed three times with sterile distilled water, transferred to 5% (w/v) NaOCl for 10 min, rinsed five times with sterile distilled water, and placed on 150- × 25-mm petri dishes containing Murashige and Skoog medium (Murashige and Skoog, 1962) to germinate. Plants were grown at 25°C in a 16-h-light/8-h-dark cycle. The same growth room conditions were used for tissue culture procedures. Roots were collected from 28-d-old plants and cultured on B5 medium (Gamborg et al., 1968) supplemented with 0.5 mg L−1 2,4-D and 0.05 mg L−1 KIN. Calli obtained from the explants were subcultured on the same medium at regular intervals of 2 weeks.
Genomic DNA-Blot Analysis
Genomic DNA was extracted from the calli using the cetyl-trimethyl-ammonium bromide method (Murry and Thompson, 1980). Aliquots of 10 μg of DNA were digested with restriction endonucleases, separated on 0.8% (w/v) agarose gels, and transferred to Biodyne nylon filters (Pall, New York) in 20× SSC. Hybridization was executed in a solution of 5× SSC, 0.5% (w/v) SDS, 5× Denhardt's solution, and 500 μg mL−1 of salmon sperm DNA at 50°C and 65°C using the 32P-labeled full-length cDNA prepared with a Random Primed DNA Labeling Kit (Takara, Shiga, Japan). Washing after hybridization was performed with 2× SSC at 25°C for 15 min, three times, and then 2× SSC containing 0.1% (w/v) SDS at 50°C and 65°C for 15 min, three times.
Construction and Screening of Genomic Library
Genomic DNA (50 μg) was partially restricted with Sau3AI to generate BamHI-compatible fragments ranging in size from 9 to 23 kb. The genomic fragments (0.3 μg) were inserted into the BamHI sites of EMBL3 vectors that had been digested with BamHI and EcoRI (Stratagene, La Jolla, CA) and packaged with Gigapack III Gold Packaging Extract (Stratagene) to construct a genomic library. Two independent cDNAs encoding PSK precursors were found in the Arabidopsis EST database in June 1999 by a BLAST homology search using the sequence of the PSK domains. The EST clones (accession nos. 120C15T7 and 91K2T7) were ordered from the DNA Stock Center of the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The inserts were purified, sequenced, and then used to screen the genomic library by plaque hybridization. Hybridization was performed as described above.
DNA Sequence Analysis
The inserts of positive phages were subcloned into pBS SK− plasmids (Stratagene). Escherichia coli strain JM109 was used as the host for the plasmids. Deletion clones were generated for DNA sequencing with a Kilo Sequencing Kit (Takara) according to the protocol recommended by the manufacturer. The plasmid DNA templates were amplified with a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), and sequenced with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) in accordance with the manufacturer's protocols.
Construction of Chimeric Genes
A 22-mer primer (5′-TCTTCTGGGAATAGATGTAATC-3′) based on the nucleic acid sequence of the AtPSK2 cDNA and a 24-mer primer (5′-CTGAGAATAAATGTAATCGGTGTG-3′) based on the nucleic acid sequence of the AtPSK3 cDNA were synthesized and used to obtain the mutated cDNAs that were designed to produce unnatural [Ser-4]PSK-α instead of PSK-α. Mutations were introduced with an LA PCR In Vitro Mutagenesis Kit (Takara) and confirmed by sequencing. The pMAT037 (Matsuoka and Nakamura, 1991) was employed as a binary vector for Arabidopsis root transformation. Original or mutated cDNAs were digested with SacI and HindIII or KpnI and inserted in frame into the same sites of the vectors to construct chimeric genes harboring the cDNA in sense or antisense orientations, respectively. Expression of the chimeric genes was driven by the promoter of the cauliflower mosaic virus 35S transcript.
Transformation of Arabidopsis Roots
Constructs were transformed into Agrobacterium tumefaciens (C58C1RifR) by triparental mating (Van Haute et al., 1983), and A. tumefaciens-mediated transformation of roots of Arabidopsis Columbia ecotype was essentially as described (Valvekens et al., 1988). Transgenic microcalli were generated by incubating cocultivated root explants for 3 weeks on the callus induction medium (B5 + 0.5 mg L−1 2,4-D and 0.05 mg L−1 KIN) supplemented with 50 mg L−1 kanamycin sulfate (Life Technologies, Grand Island, NY) and 750 mg L−1 vancomycin hydrochloride (Wako, Osaka). The individual regenerating microcalli were transferred to fresh medium to ensure that all transformants were due to independent T-DNA insertions. To detect the introduced genes, genomic DNA was isolated from transgenic calli, digested with BamHI, and allowed to hybridize with the labeled cDNA as described above. Arabidopsis cells transformed with the binary vector alone served as controls.
Purification of PSKs from CM
CM was prepared from 14-d-cultured calli by filtration (Advantec no. 2) and stored at −20°C until use. Eighty-milliliter aliquots of each CM were buffered by adding Tris to a final concentration of 20 mm, adjusted to pH 8.0 with 6.0 n HCl, and then applied to a DEAE Sephadex A-25 column (1.7 × 8 cm, Pharmacia, Piscataway, NJ) which was first equilibrated with 20 mm Tris-HCl buffer at pH 8.0. The column was washed with 50 mL of equilibration buffer and eluted successively with 50 mL of buffer containing 0, 400, 800, then 1,200 mm KCl at a flow rate of 60 mL h−1. Trifluoroacetic acid (TFA) was added to a final concentration of 0.1% (v/v) into the last two fractions containing PSKs, and then the samples were applied to a Sep-Pak Vac column (12 cc, C18, Millipore, Tokyo) after equilibration with 0.1% (v/v) TFA. The column was washed with 30 mL of the same buffer at a flow rate of 60 mL h−1 and eluted with 30 mL of 30% (v/v) acetonitrile containing 0.1% (v/v) TFA. The fraction containing PSKs was collected and lyophilized for subsequent analyses.
LC/MS Analysis
Mass spectra were obtained using a Fisons VG platform quadruple mass spectrometer with electrospray ionization interfaced to a Jasco PU 980 HPLC system. The fraction containing PSKs was dissolved in 200 μL of water and separated on a reverse-phase HPLC column (Develosil ODS-HG-5, 4.6 × 250 mm, Nomura Chemicals, Seto, Japan) with 10% (v/v) acetonitrile containing 0.1% (v/v) TFA at 1.0 mL min−1. The HPLC eluate was split 1:9 (v/v) so that 100 μL min−1 flowed to the mass spectrometer during the separation. The pseudomolecular ions of PSKs were scanned every 1.9 s with selected ion monitoring at m/z 703 ([M-H]− of [Ser-4]PSK-β) and m/z 717 ([M-H]− of PSK-β). The semiquantitative amounts of the PSK-β and [Ser-4]PSK-β in CM were measured based on the peak heights without an internal standard.
Expression Analysis of the AtPSK2 and AtPSK3 Genes
Total RNAs were isolated from Arabidopsis roots, leaves, shoots, or culture cells incubated for 1, 3, 7, 10, or 14 d as reported (Chomczynski, 1993). Twenty-microgram aliquots were denatured at 65°C for 5 min in 50% (v/v) formamide, 1× MOPS [3-(N-morpholino)-propanesulfonic acid] (200 mm MOPS, 10 mm EDTA, and 50 mm NaOAc, pH 7.0) and 1.5% (v/v) formaldehyde. The RNAs were then fractionated by electrophoresis on a 1.2% (v/v) agarose gel containing 2.2 m formaldehyde, and subsequently transferred to Biodyne nylon membranes (Pall) in 20× SSC and then allowed to hybridize with 32P-labeled cDNA as described above.
ACKNOWLEDGMENT
We thank the Arabidopsis Biological Resource Center for providing the EST clones.
Footnotes
This research was supported in part by the Japan Society for the Promotion of Science (Grant-in-Aid no. JSPS–RFTF00L01601 from the Research for the Future Program).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010452.
LITERATURE CITED
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15:532–536. [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Dorner AJ, Kaufman RJ. Analysis of synthesis, processing, and secretion of proteins expressed in mammalian cells. Methods Enzymol. 1990;185:577–598. doi: 10.1016/0076-6879(90)85046-q. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Hanai H, Matsuno T, Yamamoto M, Matsubayashi Y, Kamada H, Sakagami Y. A secreted peptide growth factor, phytosulfokine, acting as a stimulatory factor of carrot somatic embryo formation. Plant Cell Physiol. 2000a;41:27–32. doi: 10.1093/pcp/41.1.27. [DOI] [PubMed] [Google Scholar]
- Hanai H, Nakayama D, Yang H, Matsubayashi Y, Hirota Y, Sakagami Y. Existence of a plant tyrosylprotein sulfotransferase: novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine variants in vitro. FEBS Lett. 2000b;470:97–101. doi: 10.1016/s0014-5793(00)01299-0. [DOI] [PubMed] [Google Scholar]
- Harris RB. Processing of pro-hormone precursor proteins. Arch Biochem Biophys. 1989;275:315–333. doi: 10.1016/0003-9861(89)90379-2. [DOI] [PubMed] [Google Scholar]
- Huttner WB. Determination and occurrence of tyrosine O-Sulfate in protein. Methods Enzymol. 1984;107:200–223. doi: 10.1016/0076-6879(84)07013-0. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Eun C-H, Hanai H, Matsubayashi Y, Sakagami Y, Kamada H. Phytosulfokine-α, a peptidyl plant growth factor, stimulates cell division that leads to somatic embryogenesis in carrot. J Exp Bot. 1999;50:1123–1128. [Google Scholar]
- Liu YG, Mitsukawa N, Lister N, Dean C, Whittier RF. Isolation and mapping of a new set of 129 RFLP markers in Arabidopsis thaliana using recombinant inbred lines. Plant J. 1996;10:733–736. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Hanai H, Hara O, Sakagami Y. Active fragments and analogs of the plant growth factor, Phytosulfokine: structure-activity relationships. Biochem Biophys Res Commun. 1996;225:209–214. doi: 10.1006/bbrc.1996.1155. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induced the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Takagi L, Omura N, Morita A, Sakagami Y. The endogenous sulfated pentapeptide, phytosulfokine-α, stimulates tracheary element differentiation of isolated mesophyll cells of Zinnia elegans. Plant Physiol. 1999;120:1043–1048. doi: 10.1104/pp.120.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Takagi L, Sakagami Y. Phytosulfokine-α, a sulfated pentapeptide, stimulates the proliferation of rice cells by means of specific high- and low-affinity binding sites. Proc Natl Acad Sci USA. 1997;94:13357–13362. doi: 10.1073/pnas.94.24.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Nakamura K. Prepeptide of a precursor to a plant vacuolar protein required for vacuolar targeting. Proc Natl Acad Sci USA. 1991;88:834–838. doi: 10.1073/pnas.88.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. Structure, expression, and antisense inhibition of the systemin precursor gene. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM, Running MP, Sakai H, Williams RW. Multiple modes of cell division control in Arabidopsis flower development. Symp Soc Exp Biol. 1998;51:19–26. [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Murry MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Huttner WB. Purification and characterization of tyrosylprotein sulfotransferase. EMBO J. 1990;9:35–42. doi: 10.1002/j.1460-2075.1990.tb08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Reiter RS, Williams JG, Feldmann KA, Rafalski JA, Tingey SV, Scolinik PA. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci USA. 1992;89:1477–1481. doi: 10.1073/pnas.89.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Thompson JP, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science. 2000;289:613–617. doi: 10.1126/science.289.5479.613. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebttens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute E, Joos H, Maes S, Warren G, Van Montagu M, Schell J. Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J. 1983;2:411–418. doi: 10.1002/j.1460-2075.1983.tb01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Yamakawa S, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S. Promotion by a peptidyl plant growth factor, phytosulfokine-α, of chlorophyll formation in etiolated cotyledons of cucumber. Biosci Biotechnol Biochem. 1998a;62:2441–2443. doi: 10.1271/bbb.62.2441. [DOI] [PubMed] [Google Scholar]
- Yamakawa S, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S. Promotive effects of the peptidyl plant growth factor, phytosulfokine-α, on the growth and chlorophyll content of Arabidopsis seedlings under high night-time temperature conditions. Biosci Biotechnol Biochem. 1999;63:2240–2243. doi: 10.1271/bbb.63.2240. [DOI] [PubMed] [Google Scholar]
- Yamakawa S, Sakurai C, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S. The promotive effects of a peptidyl plant growth factor, phytosulfokine, on the formation of adventitious roots and expression of a gene for a root-specific cystatin in cucumber hypocotyls. J Plant Res. 1998b;111:453–458. [Google Scholar]
- Yang G, Shen S, Kobayashi T, Matsubayashi Y, Sakagami Y, Kamada H. Stimulatory effects of a novel peptidyl plant growth factor, phytosulfokine-α, on adventitious bud formation in Antirrhinum majus. Plant Biotechnol. 1999a;16:231–234. [Google Scholar]
- Yang H, Matsubayashi Y, Hanai H, Sakagami Y. Phytosulfokine-α, a peptide growth factor found in higher plants: its structure, functions, precursor and receptors. Plant Cell Physiol. 2000a;41:825–830. doi: 10.1093/pcp/pcd009. [DOI] [PubMed] [Google Scholar]
- Yang H, Matsubayashi Y, Hanai H, Nakamura K, Sakagami Y. Molecular cloning and characterization of OsPSK, a gene encoding a precursor for phytosulfokine-α, required for rice cell proliferation. Plant Mol Biol. 2000b;44:635–647. doi: 10.1023/a:1026576423870. [DOI] [PubMed] [Google Scholar]
- Yang H, Matsubayashi Y, Nakamura K, Sakagami Y. Oryza sativa PSK encodes a precursor of phytosulfokine-α, a sulfated peptide growth factor found in plants. Proc Natl Acad Sci USA. 1999b;96:13560–13565. doi: 10.1073/pnas.96.23.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]