Abstract
The antimicrobial peptide MSI-99, an analog of magainin 2, was expressed via the chloroplast genome to obtain high levels of expression in transgenic tobacco (Nicotiana tabacum var. Petit Havana) plants. Polymerase chain reaction products and Southern blots confirmed integration of MSI-99 into the chloroplast genome and achievement of homoplasmy, whereas northern blots confirmed transcription. Contrary to previous predictions, accumulation of MSI-99 in transgenic chloroplasts did not affect normal growth and development of the transgenic plants. This may be due to differences in the lipid composition of plastid membranes compared with the membranes of susceptible target microbes. In vitro assays with protein extracts from T1 and T2 plants confirmed that MSI-99 was expressed at high levels to provide 88% (T1) and 96% (T2) inhibition of growth against Pseudomonas syringae pv tabaci, a major plant pathogen. When germinated in the absence of spectinomycin selection, leaf extracts from T2 generation plants showed 96% inhibition of growth against P. syringae pv tabaci. In addition, leaf extracts from transgenic plants (T1) inhibited the growth of pregerminated spores of three fungal species, Aspergillus flavus, Fusarium moniliforme, and Verticillium dahliae, by more than 95% compared with non-transformed control plant extracts. In planta assays with the bacterial pathogen P. syringae pv tabaci resulted in areas of necrosis around the point of inoculation in control leaves, whereas transformed leaves showed no signs of necrosis, demonstrating high-dose release of the peptide at the site of infection by chloroplast lysis. In planta assays with the fungal pathogen, Colletotrichum destructivum, showed necrotic anthracnose lesions in non-transformed control leaves, whereas transformed leaves showed no lesions. Genetically engineering crop plants for disease resistance via the chloroplast genome instead of the nuclear genome is desirable to achieve high levels of expression and to prevent pollen-mediated escape of transgenes.
Antimicrobial peptides (AMPs) with α-helical structures are ubiquitous and found in many organisms. They are a common component of innate defense mechanisms in the animal kingdom and help to control normal microbial flora and combat pathogens (Tossi et al., 2000). AMPs have been isolated from frogs, insects, and mammalian phagocytic vacuoles (Biggins and Sansom, 1999; Tossi et al., 2000). The AMP used in this study (MSI-99) is an analog of magainin 2, a defense peptide secreted from the skin of the African clawed frog (Xenopus laevis), first discovered by Zasloff (1987).
Magainins and their analogs have been studied as a broad-spectrum topical agent, a systemic antibiotic, a wound-healing stimulant, and an anticancer agent (Jacob and Zasloff, 1994). However, the possible agricultural use of magainin-type AMPs has received limited attention until recently. Li et al. (2001) have reported disease resistance, to both a fungal and a bacterial pathogen, conferred by expression of a magainin analog, Myp30, in transgenic tobacco (Nicotiana tabacum var. Petit Havana). Initial studies of AMPs including magainin were conducted using circular dichroism and solid-state NMR. These studies first reported the mechanism of AMPs on artificial membranes (Gesell et al., 1997; Shümann et al., 1997; Matsuzaki, 1998). The mode of action of these peptides has been more clearly defined recently. AMPs are selective for prokaryotic membranes over eukaryotic membrane due to the predominantly negatively charged phospholipids in the outer leaflet of the prokaryotic membrane (Biggin and Sansom, 1999; Huang, 2000; Tossi et al., 2000). Such preference is considered a regulatory function in target selectivity. Althought the overall charge of the peptide is important, it is known that other features play a role in potency and spectrum of the peptide. The size, sequence, structure (percent helical content), overall hydrophobicity, amphipathicity, and width of the hydrophobic and hydrophilic regions of the peptide have a function in the efficiency of the peptide (Tossi et al., 2000). The peptides initially lay parallel to the membrane surface in what is called a carpet affect. At this stage, the peptide has assumed a helical conformation. During this stage, the molar peptide-to-lipid ratio (P/L) is low. As the peptide concentration increases, the P/L reaches threshold. As the P/L continues to increase, the peptides become perpendicular to the membrane, where they begin to aggregate and disrupt the lipid composition by interacting with the phospholipids to form pores in the membrane (Huang, 2000). The membrane becomes depolarized and there is a loss of essential metabolites and secondary effects that disrupt respiration and signaling and triggers enzymes that cause peptidoglycan auto-lysis (Tossi et al., 2000). Because of the reported effectiveness of magainin and its analogs against pathogens (Jacob and Zasloff, 1994), we chose one of its analogs (MSI-99) to test against phytopathogenic bacteria and fungi.
Chloroplast transformation was selected because of several advantages over nuclear transformation (Daniell, 1999a, 1999b, 2000; Bogorad, 2000; Heifetz, 2000). Gene containment is possible when foreign genes are engineered via the chloroplast genome. This prevents pollen transmission of transgene in crops that maternally inherit the plastid genome. Although pollen from plants that exhibit maternal inheritance contain metabolically active plastids, the plastid DNA is lost during pollen maturation (Heifetz, 2000). Gene containment in transgenic plants is a serious concern when plants are genetically engineered for disease resistance because of the possibility of creating robust, disease-resistant weeds or passing on undesired traits to related crops. To prevent these consequences, it is desirable to genetically engineer crop plants for disease resistance via the chloroplast genome instead of the nuclear genome.
Because of the concentration dependent action of AMPs (Matsuzaki, 1998; Biggin and Sansom, 1999), we hypothesized that expression of MSI-99 via the chloroplast genome should accomplish high-dose release at the site of infection and prevent the spread of infection by microbes. Such compartmentalization of defense proteins is a natural occurrence in plants (Neuhas et al., 1991). Due to the high copy number associated with chloroplast expression, a larger amount of the AMP may be synthesized, provided the small peptide is not susceptible to extreme proteolytic degradation. The AMP should be released at the site of infection during the hypersensitive response that leads to cell and organelle lysis. Release of AMP at high concentrations should result in aggregation and formation of pores in the outer membrane of microbes and aid in preventing the spread of infection.
In addition, we hypothesized that chloroplast membranes may not be susceptible to MSI-99 because of the presence of neutral lipids in contrast to susceptible microbes. Therefore, transgenic expression of MSI-99 within plastids may not be harmful. It is known that the chloroplast envelope and thylakoid membranes consist of primarily glycolipids and galactolipids instead of phospholipids; monogalactosyldiacylglycerol makes up 50% of membrane lipid and digalactosyldiacylglycerol makes up 30% (Siegenthaler, 1998).
To date, chloroplast transformation has enabled generation of herbicide (Daniell et al., 1998), insect-resistant crops (McBride et al., 1995; Kota et al., 1999; DeCosa et al., 2001), and production of biopharmaceuticals (Guda et al., 2000; Staub et al., 2000; Daniell et al., 2001a). This work extends the chloroplast genetic engineering technology to confer pathogen resistance using a synthetic lytic peptide (MSI-99) against phytopathogenic bacteria and fungi.
RESULTS
Chloroplast Vectors and Plant Transformation
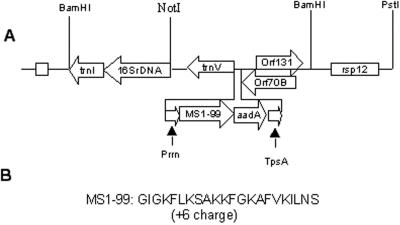
The synthetic peptide used in this study (MS1–99) is an analog of the naturally occurring 23-amino acid peptide, magainin 2. MSI-99 is a 22-amino acid sequence with an overall charge of +6 (Fig. 1). The gene cassette used for transformation consisted of the 16S rRNA promoter, the MS1–99 gene with a chloroplast preferred (GGAGG) ribosome-binding site, the aadA gene, which confers resistance to spectinomycin, and the psbA (photosynthetic binding protein) terminator for stabilizing the transcript. Flanking sequences are from the petunia chloroplast genome (Fig. 1A). Transformation efficiency (i.e. the percentage of spectinomycin resistant shoots that contained the transgene) was much lower (7%) than that routinely observed using the pLD vector (91%), which contains tobacco homologous flanking sequences. Out of 55 spectinomycin resistant shoots screened, only four contained the MSI-99 gene and the rest were mutants. Nuclear transgenic plants did not confer resistance to spectinomycin selection at 500 μg mL−1 (Daniell, 2000; Daniell et al., 2001b) and therefore were eliminated. The number of primary shoots observed per bombarded leaf does not give an accurate efficiency of chloroplast transformation because of the generation of mutants under spectinomycin selection. All transformants matured normally with no apparent morphological effects for T0, T1, and T2 generations (Fig. 2A). T1 seeds germinated in the presence of spectinomycin produced green seedlings, whereas control seedlings were bleached (Fig. 2B).
Figure 1.
Transformation vector and MSI-99 peptide sequence. A, Vector contains a selectable marker gene (aadA) that confers resistance to spectinomycin, 16S rrn promoter, psbA terminator, and petunia chloroplast DNA flanking sequences. B, Amino acid sequence of the AMP MSI-99.
Figure 2.
Phenotype of wild-type and transgenic plants. A, Plants 1 through 3 are T0 transgenic plants, whereas plant 4 is wild type. Plants 5 through 7 are T1 transgenic plants. Plants 8 and 9 are T2 transgenic plants. B, Seedlings germinated on MSO + 500 μg mL−1 spectinomycin. Three T1 transgenic lines (1–3) and wild type (4) are shown.
Foreign Gene Integration, Homoplasmy, and Copy Number
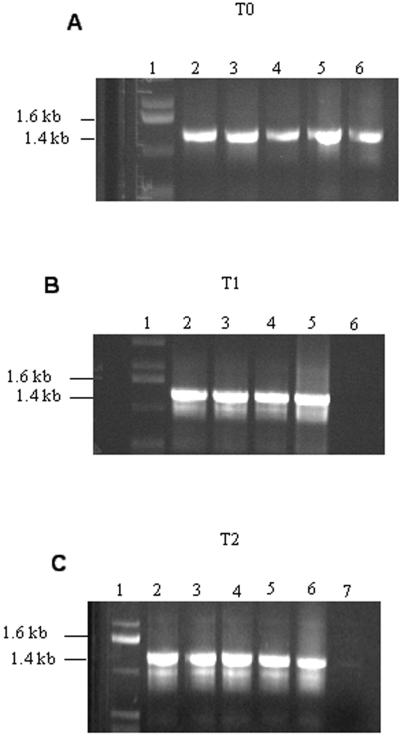
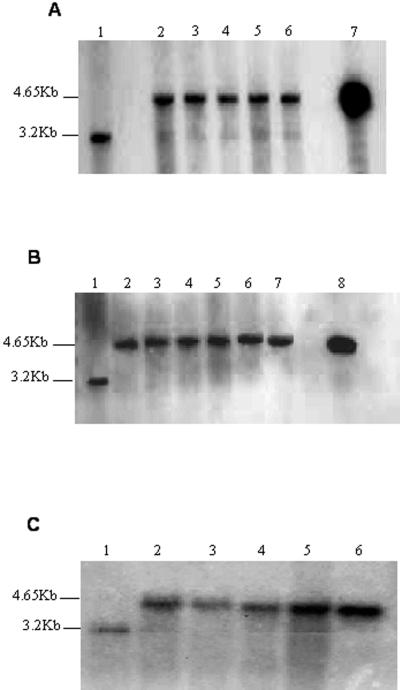
PCR was performed by landing one primer on the 5′ end of the aadA coding sequence, not present in native chloroplast and the 3′ end of the 16S rDNA. PCR products of T0, T1, and T2 generations yielded the same size product as the plasmid used for transformation (Fig. 3, A–C), confirming integration of transgenes. The probe used for the Southern analysis was a 2.3-kb fragment from the 5′ end of the trnI (BamHI) to the 3′ end of the 16SrDNA (NotI). The plant DNA was digested with BamHI. DNA from non-transformed plants produced a 3.2-kb fragment and transformed plant DNA produced a 4.6-kb fragment. Southern analysis confirmed integration of foreign genes for T0, T1, and T2 (Fig. 4, A–C). Untransformed DNA showed a 3.2-kb fragment, whereas the transformed contained a 4.6-kb fragment. Presence of some wild-type fragments in T0 transgenic samples indicated some heteroplasmy (Fig. 4A). However, DNA from T1 and T2 generation produced only the 4.6-kb fragment, confirming homoplasmy (Fig. 4, B and C). A cell is said to be homoplasmic when all of the chloroplast genomes are uniformly transformed. By confirming that the MSI-99-integrated genome is the only one present in transgenic plants (homoplasmy), one could estimate that the MSI-99 gene copy number could be as many as 10,000 per cell.
Figure 3.
PCR analysis of plant DNA. DNA extracted from T0 (A), T1 (B), and T2 (C) plants were run on a 0.8% (w/v) agarose gel. A, T0, lane 1, 1-kb ladder; lanes 2 through 5, transgenic lines; lane 6, MSI-99 plasmid. B, T1, lane 1, 1-kb ladder; lanes 2 through 4, transgenic; lane 5, plasmid control; lane 6, wild-type plant DNA. C, T2, lane 1, 1-kb ladder; lanes 2 through 5, transgenic; lane 6, plasmid control; lane 7, wild-type plant DNA.
Figure 4.
Southern analysis of T0, T1, and T2 generation plants. A, Lanes 2 through 6, T0 transgenic lines; lane 1, wild type; lane 7, plasmid DNA. B, Lanes 2 through 7, T1 transgenic lines; lane 1, wild type; lane 8, plasmid DNA. C, Lanes 2 through 5, T2 transgenic lines; lane 1, wild type; lane 6, plasmid DNA.
Northern Analysis
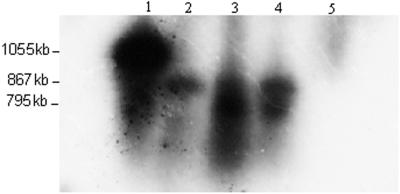
Northern analysis was done to detect transcription of the foreign genes (Fig. 5). Eight to 10 ng of RNA was loaded in each well. Transcripts of 867 bp are present in lane 2, whereas transcripts of 867 and 795 bp in lanes 3 and 4 indicate dicistronic transcripts (lanes 2–4 are transgenic). Lane 5, which is from a non-transformed plant, did not produce any transcripts. Lane 1 is the 1,055-bp probe containing the MSI-99 gene, promoter, and the aadA gene. The monocistronic transcript for MSI-99 is too small (77 bp) to be detected on this gel.
Figure 5.
Northern analysis of T2 generation. Eight to 10 ng of RNA was loaded in each well. Lane 1 is positive control consisting of the MSI-99 and promoter sequences and the aadA gene (1,055 kb). Lanes 2 through 4 are transgenic lines and lane 5 is non-transformed control plant.
In Vitro Antibacterial Activity of Plant Extracts to Pseudomonas syringae pv tabaci (Pst)
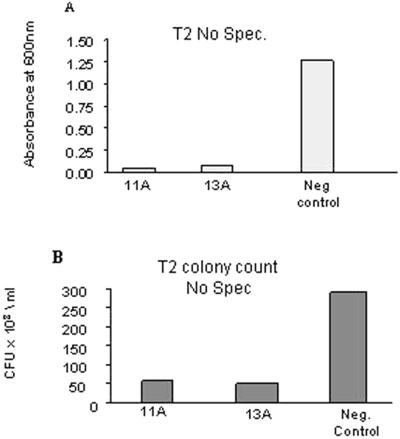
Cell-free extracts of T1 and T2 transgenic plants inhibited growth of Pst in vitro by 86% and 96% compared with wild-type plants (Fig. 6). Cell-free extracts from transgenic plants germinated in the absence of spectinomycin displayed an equivalent ability to inhibit growth of P. syringae pv tabaci, indicating that the growth inhibition observed was not caused by spectinomycin that might be present in the plant tissue (Fig. 7). The control plant extracts inhibited bacterial growth more than the buffer only. This is most probably due to natural defense peptides such as defensins and thionins produced by plants (Mourgues et al., 1998). Differences in bacterial growth inhibition observed using plate assays versus liquid culture assays (e.g. Fig. 7, A versus B) can be explained by the differences in their environment. Although the plated bacteria were no longer exposed to AMP, bacteria in the liquid media were constantly surrounded by active peptides.
Figure 6.
In vitro bioassays for T1 and T2 generations of three transgenic lines (10A, 11A, and 13A). Five microliters of bacterial cells from an overnight culture was diluted to A600 0.1 to 0.3 and incubated for 2 h at 25°C with 100 μg of total plant protein extract. One milliliter of Luria broth (LB) was added to each sample. Samples were incubated overnight at temperature appropriate for the specific bacteria. A600 was recorded. Negative control was non-transformed plant extract. Buffer only was added as a control and stock culture was used as a reference point.
Figure 7.
In vitro bioassays of plants grown in the absence of spectinomycin. Five microliters of an overnight culture of P. syringae pv tabaci diluted to A600 0.1 to 0.3 was mixed with 100 μg of total protein extract from T2 lines 11A and 13A (germinated in the absence of spectinomycin). After 2 h incubation, 50 μL of each mix was plated onto LB plates and incubated overnight at 27°C. A, A600 was recorded the next morning using a spectrophotometer. B, Count of viable colony forming units (CFUs) were made using the Gell Dock (Bio-Rad, Hercules, CA).
In Vitro Antifungal Activity of Plant Extracts to Aspergillus flavus, Fusarium moniliforme, and Verticillium dahliae
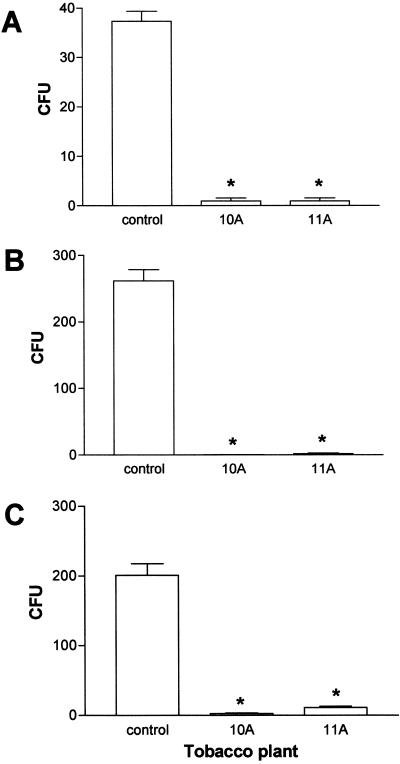
Plant extracts from each transformed tobacco plant significantly reduced (P < 0.001) the number of fungal colonies arising from germinating conidia of A. flavus, F. moniliforme, and V. dahliae compared with the extracts from non-transformed controls (Fig. 8, A–C). Germinating conidia of A. flavus were susceptible to the extracts from transformed plants resulting in a greater than 95% reduction in the number of colonies. Extracts from all the transformants significantly reduced (P < 0.001) the number of colonies compared with the control (Fig. 8A). Extracts from T1 transgenic plants 10A and 11A inhibited almost 100% of germinating conidia of F. moniliforme (Fig. 8B). Germinating conidia of V. dahliae were also susceptible to extracts from the transformed plants. Extracts from the transformed plants reduced the number of germinating conidia of V. dahliae by 99% compared with extracts from non-transformed controls (Fig. 8C).
Figure 8.
In vitro antifungal bioassays in T1 transgenic lines. A, Inhibition of germinated conidia of A. flavus by exposure to leaf extracts from tobacco plants expressing the antifungal peptide MSI-99 for 1 h. Asterisk, Denotes a significant reduction (P < 0.001) in the number of A. flavus colonies compared to extracts of non-transformed control. B, Inhibition of germinated conidia of F. moniliforme by exposure to leaf extracts from tobacco plants expressing the antifungal peptide MSI-99 for 1 h. Asterisk, Denotes a significant reduction (P < 0.001) in the number of F. moniliforme colonies compared with extracts of non-transformed control. C, Inhibition of germinated conidia of V. dahliae by exposure to leaf extracts from tobacco plants expressing the antifungal peptide MSI-99 for 1 h. Asterisk, Denotes a significant reduction (P < 0.001) in the number of V. dahliae colonies compared with extracts of non-transformed control. Error bars indicate se of means. Mean separation was performed using the method of Tukey.
In Planta Resistance to P. syringae pv tabaci
Inoculation of T0 potted plants (6–7 months old) with P. syringae pv tabaci using a sandpaper technique (see “Materials and Methods”) resulted in areas of necrosis surrounding the point of inoculation in wild-type control for all cell densities, whereas transgenic mature leaves showed no areas of necrosis (Fig. 9). Even inoculation of 8 × 105 cells resulted in no necrosis in mature transgenic leaves (Fig. 9A), suggesting the local concentration of the AMP to be very high. However, non-transformed plants inoculated with 8 × 103 cells displayed necrosis (Fig. 9B). Similar results were obtained with bacteria inoculated using a syringe (Fig. 9, C and D). Transgenic mature leaves injected with P. syringae pv tabaci showed a mild discoloration at the site of inoculation of 8 × 105 cells (Fig. 9C), whereas the wild-type plants displayed necrosis even when inoculated with 8 × 103 cells (Fig. 9D).
Figure 9.
In planta bioassays. Five- to 7-mm areas of T0 transformants and non-transformed tobacco cv Petit Havana leaves were scraped with fine-grain sandpaper. Ten microliters of 8 × 105, 8 × 104, 8 × 103, and 8 × 102 cells from an overnight culture of P. syringae pv tabaci were added to each prepared area. Photos were taken 5 d after inoculation. A, Transgenic leaf; B, wild-type leaf; C, transgenic; D, wild type were injected with 25 μL of 8 × 103 cells of Pst. Pictures were taken 5 d after inoculation. Transformed leaf show only slight discoloration, whereas the wild-type leaf shows necrosis.
Inoculation of T1 transgenic plants 10A and 11A with P. syringae pv tabaci using the syringe method produced results essentially the same as for the T0 plants; no necrotic lesions formed. To assess whether this lack of symptoms reflected a reduction in bacterial growth, we quantified the number of bacteria in the injected region. The starting population of bacteria right after inoculation was determined to be about 400 to 450 CFU per disc. After 4 d, the bacterial population increased to 13,750 ± 750 CFU per disc in control plants, whereas a lower number of colonies were counted in T1 transgenic plants (4,650 ± 125 CFU for 10A and 5,150 ± 350 for 11A).
In Planta Anthracnose Resistance
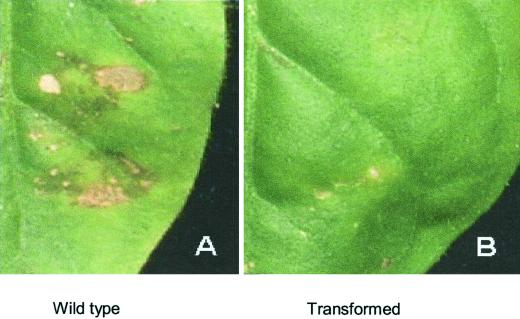
Leaves inoculated with Colletotrichum destructivum developed anthracnose lesions within 48 to 72 h after inoculation on non-transformed controls, whereas T1 progeny plants of MSI-99 transformants (10A and 11A) did not develop lesions even after 1 week of inoculation (Fig. 10).
Figure 10.
Tobacco leaf cv Petit Havana from A, a non-transformed control tobacco plant showing anthracnose symptoms 7 d after inoculation with the fungal pathogen C. destructivum. B, The leaf from the T1transgenic plant 11A expressing MSI-99 turned slightly chlorotic at the site of inoculation but did not develop lesions.
DISCUSSION
This study shows that AMPs can be expressed in tobacco chloroplasts and maintain their biological activity without harmful effects to the transgenic plants or chloroplasts. T0, T1, and T2 transgenic plants were healthy and showed no morphological or developmental abnormalities. The initial low rate of transformation in this study was most likely due to less than 100% homology between the petunia flanking sequences and the tobacco chloroplast genome. This is not surprising because very low transformation efficiency was also observed when tobacco chloroplast flanking sequences were used to transform potato chloroplast genome (Sidorov et al., 1999). In addition, other projects in our lab that use the pLD vector (has tobacco flanking sequences) obtained transformation efficiency of 91% (ratio of transformants to mutants). Retention of biological activity was evident in the sharp decrease in cell viability during in vitro bioassays. When comparing Southern blots with biological activity, antimicrobial activity increased as homoplasmy was achieved. Equal lytic activity was also observed in transgenic plants germinated in the presence or absence of spectinomycin. Transgenic plants transferred to potting soil for 5 to 6 months after being removed from spectinomycin selection displayed similar antimicrobial properties against inoculations of P. syringae pv tabaci. These observations eliminate the possibility that spectinomycin absorbed into the plant tissue during germination of seeds may be responsible for the growth inhibition in the in vitro and in situ bioassays. In addition, the observation that MSI-99 was equally active in transgenic plants germinated in the presence or absence of spectinomycin shows the stability of the introduced trait in the absence of any selection pressure.
MSI-99 is an analog of a naturally occurring peptide (magainin 2) found in the skin of the African frog. Changes have been made to the amino acid sequence to enhance its lytic abilities. It has been speculated previously that AMPs with a high antibacterial activity might also have a high potential for toxic activity against the chloroplast (Everett, 1994). However, as reported in this study, the transgenic plants grew normally, flowered, and set seeds like the wild-type control. In contrast to prokaryotic membranes, the chloroplast envelope and thylakoid membranes consist of primarily glycolipids and galactolipids instead of phospholipids. It has been shown that monogalactosyldiacylglycerol makes up 50% of membrane lipid and digalactosyldiacylglycerol makes up 30% (Siegenthaler, 1998). Both of these lipids are neutral and this may explain why chloroplasts were not affected by MSI-99.
Key features of cationic peptides such as MSI-99 are a net positive charge, an affinity for negatively charged prokaryotic membrane phospholipids over neutral-charged eukaryotic membranes, and the ability to form aggregates that disrupt the bacterial membrane (Houston et al., 1997; Biggin and Sansom, 1999). Given the fact that the outer membrane is an essential and highly conserved part of all bacterial cells, it would seem highly unlikely that bacteria would be able to adapt (as they have against aminoglycosides or other types of antibiotics) to resist the lytic activity of these peptides.
Western blots could not be performed because there were no antibodies available for this peptide. Northern analysis confirmed the transcription of MSI-99 in the transgenic plants but not in the control plants. A high level of AMP expression can be expected due to the following reasons. The nature of plastids to move from a somatically unstable heteroplasmic state to a state of homoplasmy itself lends to high expression (Bock and Hagemann, 2000). The A + T percent of MSI-99 is 51.39%, which is compatible with the tobacco chloroplast 61% A + T content (Shimada and Sugiura, 1991). Also, published reports from our lab report expression of Cry2A operon (A + T content of 65%) at levels as high as 46% total soluble protein (DeCosa et al., 2001). Although these facts point to high expression, it must be noted that when protein extracts from the transgenic and control plants were run on a 16% (w/v) Gly gel, we were unable to detect any peptides below 14 kD. MSI-99 is approximately 2.7 kD and was probably lost due to diffusion.
The minimum inhibitory concentration of MSI-99 was investigated in this study (Table I). Based on total inhibition of 1,000 P. syringae pv tabaci cells mL−1, MSI-99 was most effective against Pst, requiring only 1 μg mL−1 of MSI-99. The overall charge of MSI-99 is more positive than magainin 2. However, although the overall charge of the peptide is important, it is known that other features play a role in potency and spectrum of the peptide. The size, sequence, structure (percent helical content), overall hydrophobicity, amphipathicity, and width of the hydrophobic and hydrophilic regions of the peptide have a function in the efficiency of the peptide (Tossi et al., 2000). Further study is needed to investigate other changes that occurred in MSI-99 as a result of increased positive charge. The results of this investigation provide yet another option in the ongoing battle against phytopathogens to reduce yield losses and prevent mycotoxin contamination caused by some fungi in food and feed crops.
Table I.
Minimum Inhibitory Concentrationsa (μg mL−1) of magainin 2 and MSI-99
| Phytopathogens | MSI-99 | Magainin 2 |
|---|---|---|
| P. syringae | 1 | 32 |
| Erwinia carotovora | 1 | 32 |
| Phytophthora parasitica | 64 | >256 |
| Fusarium solani | 2 | 4 |
| Fusarium graminearum | 4 | 4 |
| Thielaviopsis basicola | 4 | 8 |
| Botrytis cinerea | 8 | 16 |
Minimum Inhibitory Concentrations required to inhibit growth after 24 h. Based on 1,000 fungal conidiospores or 1,000 bacterial cells.
MATERIALS AND METHODS
Plant Materials
With the exception of T0, all wild-type and transgenic plants were germinated from seeds at the same time. The only difference in growth media was the presence of spectinomycin for selection. All plants and leaves were of the same age. Although T0 plants originated from tissue culture, the T0 controls were germinated on Murashige and Skoog medium with no hormones (MSO; Murashige and Skoog, 1962; Daniell, 1997). With this exception, all experimental procedures and plant materials were identical in both transgenic and control plants.
Plant Transformation
For plant transformation, tobacco (Nicotiana tabacum var. Petit Havana) seeds were germinated aseptically on MSO media at 26°C with photoperiods of 16 h light and 8 h dark. Sterile intact leaves, about 2 to 3 weeks old, were placed abaxial side up on Whatman (Clifton, NJ) No. 1 filter papers laying on regeneration medium of plants (RMOP; Daniell, 1993) in standard petri plates (100 × 15 mm) for bombardment. Gold microprojectiles were used for bombardment using the Bio-Rad helium-driven PDS-1000/He System (Daniell, 1997). After bombardment, the petri dishes were sealed with Parafilm and incubated in the dark for 48 h at 26°C. Leaves were then cut into 1-cm2 squares and placed on a petri dish containing RMOP medium with 500 μg mL−1 spectinomycin with photoperiods of 16 h light and 8 h dark (first round of selection). Four to 6 weeks later, green intact shoots were transferred to fresh RMOP containing spectinomycin (500 μg mL−1; second round of selection). Green shoots that appeared during the second selection were transferred to sterile bottles containing MSO and spectinomycin (500 μg mL−1). Plants were screened via PCR to verify chloroplast integration of transgenes. Those that were PCR positive for the presence of the MSI-99 gene were transferred to pots and grown in chambers at 26°C with photoperiods of 16 h light and 8 h dark. After flowering, seeds were harvested and sterilized with a solution of 1 part 15% (v/v) bleach and 2 parts water with 1 drop of Tween 20. Seeds were vortexed for 5 min, washed six times with 500 μL of distilled water and dried in speed vac. T1 and T2 seeds were germinated on MSO + 500 μg mL−1 spectinomycin unless indicated otherwise. Non-transformed tobacco cv Petit Havana seeds were germinated on the same media as a control to ensure that spectinomycin was active.
PCR Analysis
Plant DNA extraction on T0, T1, and T2 was performed using the DNeasy Mini Kit (Qiagen, Valencia, CA) on putative transgenic samples and non-transformed plants. PCR primers were designed using Primer Premier software (Premier Biosoft International, Palo Alto, CA) and synthesized by GIBCO BRL (Carlsbad, CA). Primer (8P: 5′-ATCACCGCTTCCCTCAT-AAATCCCTCCC-3′) anneals with the 5′ end of the aadA and primer (8 m:5′-CCACCTACAGA CGCTTTACGCCCAATCA-3′) anneals with the 3′ end of 16SrDNA (Fig. 3). PCR was carried out using the Gene Amp PCR system 2400 (Perkin-Elmer, Santa Clara, CA). Samples were run for 29 cycles in the following sequence: 94°C for 1 min, 65°C for 1 min, and 72°C for 3 min The cycles were proceeded by a 94°C denaturation period and followed by a 72°C final extension period. A 4°C hold followed the cycles. PCR products were separated on 0.8% (w/v) agarose gels.
Southern Analysis
Integration of foreign genes for T0 and T1 was determined by Southern-blot analysis. DNA from transformed and wild-type plants was isolated and digested with BamHI and run on a 0.7% (w/v) agarose gel. The DNA was then transferred to a nylon membrane by capillary action. The probe was digested with BamHI and NotI and was labeled with 32P using the Probe Quant G-50 Micro Columns and protocol provided with the kit (Amersham, Piscataway, NJ). Labeled probe was hybridized with the nylon membrane using the Stratagene (La Jolla, CA) QUICK-HYB hybridization solution and protocol. Membrane was exposed to film and developed.
Northern Analysis
RNA was extracted using the Rneasy Mini Kit (Qiagen) and protocol. Probes for the aadA and the MSI-99 genes were digested with XbaI and NotI. Eight to 10 μg RNA was loaded in each well. Plant RNA was transferred to a nylon membrane by capillary action. The probe was labeled with 32P using the Probe Quant G-50 Micro Columns and protocol (Amersham). Labeled probe was hybridized with the nylon membrane using the Stratagene QUICK-HYB hybridization solution and protocol. Membrane was exposed to film and developed.
In Vitro Bioassays
To determine minimum inhibitory concentration of MSI-99, conidiospores of Fusarium solani, Thielaviopsis basicola, and Botrytis cinerea were collected from potato dextrose agar (PDA) plates by flooding a 2-week-old culture with a solution of 0.01% (v/v) Tween 20 and rubbing the surface. The spore suspension was filtered through glass wool to remove mycelial fragments. The spore concentration (per milliliter) was determined using a hemocytometer. For Phytophthora parasitica, zoospores grown in liquid culture were use for the assay. Bacteria were harvested from log phase-grown cultures and the concentrations were determined based on optical density readings and plating of serial dilutions of the cultures.
Known amounts of pathogen (1,000 spores or 1,000 bacteria) were added to serial dilutions of chemically synthesized MSI-99 (Cornell University, Ithaca, NY) ranging from 0 to 256 μg mL−1 in individual wells of a 96-well microtiter plate. An equivalent amount of growth medium (LB for bacteria and potato dextrose broth [PDB] for fungi) was added to each well, bringing the total volume to 50 μL per well. Plates were incubated overnight at 25°C with gentle shaking. The following day, wells were scored for the presence or absence of growth. The lowest concentration of peptide, which inhibited all cell growth, was recorded as the minimum inhibitory concentration (microgram per milliliter) value.
Pseudomonas syringae pv tabaci (ATCC 17914) (Pst) was cultured overnight prior to the assay. Plants were grown on MSO medium in sterile bottles. Fifty milligrams of leaf tissue (minus mid-rib) from the second or third leaf of young (2- to 3-week-old) plants in bottles, were ground in a microcentrifuge containing 150 μL of phosphate buffer (pH 5.5) with 5 mm phenylmethylsulfonyl fluoride and 5 mm EDTA using a plastic pestle. Samples were centrifuged for 5 min at 10,000g at 4°C. Supernatant was transferred to a fresh tube and kept on ice. Protein concentration was determined by Bradford assay. One hundred micrograms of total plant protein (volumes ranged from 50–100 μL) from each plant was mixed with 5 μL of Pst from overnight culture or buffer alone, in a Falcon tube. Initial absorbance of the culture ranged from 0.1 to 0.3 (A600). Tubes were incubated for 2 h at 25°C on a rotary shaker at 125 rpm. Next, 1 mL of LB was added and the tubes were allowed to incubate for 18 h at 27°C on a rotary shaker at 125 rpm. Absorbance (A600) was read for each tube at the end of the incubation period. The mean and se were determined using GraphPad Prism software (San Diego).
To rule out spectinomycin as the cause of growth inhibition in the in vitro experiments, the same experiment with Pst was repeated using T2 plants that were germinated on MSO with no spectinomycin. To confirm the absorbance readings, a serial dilution was made of samples after the initial 2-h incubation. Dilutions of 10−3 to 10−5 were plated onto LB plates and incubated overnight at 27°C. The next morning, a count of viable CFUs were made using the Gel Dock 2000 (Bio-Rad). The optical density readings were then compared with the CFU counts.
In Vitro Analysis of Antifungal Activity of Plant Extracts to Aspergillus flavus, Fusarium moniliforme, and Verticillium dahliae
The inhibitory activity of extracts from tobacco plants transformed with MSI-99 was assessed in vitro following the method of Cary et al. (2000). In brief, conidial suspensions were prepared from cultures grown on PDA (Difco, Detroit, MI) slants for 7 d at 30°C (A. flavus and F. moniliforme) or 22°C (V. dahliae). Conidial suspensions in 1% (w/v) PDB (pH 6.0) were adjusted to a density of 105 conidia mL−1 and were germinated in PDB for 8 h at 30°C (A. flavus, F. moniliforme) or overnight at 22°C (V. dahliae) prior to assay.
Plant homogenates were prepared by directly grinding tobacco leaves into a fine powder in liquid nitrogen with no buffer added. Ground tissues were then centrifuged at 8,200g for 10 min at room temperature and extract collected from each sample. Conidial suspensions (25 μL) were then added to 225 μL of plant extract, mixed, and incubated for 1 h at 30°C (A. flavus and F. moniliforme) or 22°C (V. dahliae). Three 50-μL aliquots from each sample were then spread onto PDA plates and incubated at 30°C or 22°C for 24 to 48 h and fungal colonies enumerated. One-way ANOVA was used to determine the significance of the effect of transgenic plant extracts on germinating conidia. Mean separations were performed using the method of Tukey (Sokal and Rohlf, 1981).
In Planta Assay for Anthracnose Resistance
Seven days prior to plant inoculation, Czapek yeast autolysate agar plates were inoculated with mycelium of a virulent isolate of Colletotrichum destructivum (ATCC 42492) and incubated at 24°C. A single Czapek yeast autolysate agar plate was flooded with 9 mL of sterile distilled water and spores were aseptically removed to yield a final inoculum density of approximately 1 × 106 spores mL−1. T1 plants expressing MSI-99 (10A and 11A) were inoculated by placing eight drops of 10 μL each onto the adaxial surface of three young, expanding tobacco leaves according to previously published procedures (Cary et al., 2000). Two leaves each of three non-transformed tobacco plants were also inoculated to serve as controls.
In Planta Assay for Resistance to Wildfire Disease Caused by P. syringae pv tabaci
P. syringae pv tabaci (ATCC 17914) was cultured overnight prior to the assay. Five- to 7-mm areas of T0 transformants and non-transformed tobacco cv Petit Havana leaves were scraped with fine-grain sandpaper. Ten microliters of 8 × 105, 8 × 104, 8 × 103, and 8 × 102 cells from an overnight culture of Pst were added to each prepared area. Photos were taken 5 d after inoculation. In another assay, the same serial dilution was made with Pst from an overnight culture. Twenty-five microliters of each sample was injected into leaves using a needle, followed by a syringe, of both transgenic and non-transformed plants. Pictures were taken 5 d after infection. T0 transgenic plants 10A, 11A, and 13A were tested. A total of 12 transgenic (four per plant) and control leaves were tested.
In planta assay on T1 progeny plants to evaluate resistance to Pst was adapted from Huang et al. (1997). Pst inoculum from a fresh culture was grown in liquid nutrient broth (Difco) overnight. The culture was centrifuged and pellet resuspended in 50 mL of phosphate buffer (0.01 m, pH = 7.0). The suspension was diluted in phosphate buffer to about 8×103 CFU mL−1. At least three seedlings from each transgenic line were inoculated by infiltrating Pst inoculum into the lamina using a syringe through a needle hole and the water-soaked area was outlined with a marker. Starting immediately after inoculation and after 4 d, two leaf discs were collected from within a single inoculated area (i.e. within the outlined area) from each plant and were quickly ground in 300 μL of 10 mm MgCl2. The homogenates were transferred to 5 mL PO4 buffer. Both the undiluted and 1:100 diluted homogenates were plated on Pseudomonas Agar F (Difco) using a Spiral Plater (Spiral Systems Inc., Cincinnati). Pst colonies were enumerated after incubation at 28°C for 24 to 48 h.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Steven Hutcheson (University of Maryland, College Park) for the Pst culture and the monitoring editor Dr. Roger Innes (Indiana University, Bloomington) for his invaluable suggestions toward improvement of this manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010233.
LITERATURE CITED
- Biggin P, Sansom M. Interactions of α-helices with lipid bilayers: a review of simulation studies. Biophys Chem. 1999;76:161–183. doi: 10.1016/s0301-4622(98)00233-6. [DOI] [PubMed] [Google Scholar]
- Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- Bock R, Hagemann R. Extracellular inheritance: plastid genomics: manipulation of plastid genomes and biotechnological applications. Prog Bot. 2000;6:76–90. [Google Scholar]
- Cary J, Rajasekaran K, Jaynes J, Cleveland T. Transgenic expression of a gene encoding a synthetic antimicrobial peptide results in inhibition of fungal growth in vitro and in planta. Plant Sci. 2000;154:171–181. doi: 10.1016/s0168-9452(00)00189-8. [DOI] [PubMed] [Google Scholar]
- Daniell H. Foreign gene expression in chloroplast of higher plants mediated by tungsten particle bombardment. Methods Enzymol. 1993;217:536–556. doi: 10.1016/0076-6879(93)17088-m. [DOI] [PubMed] [Google Scholar]
- Daniell H. Transformation and foreign expression in plants mediated by microprojectile bombardment. Methods Mol Biol. 1997;62:463–489. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- Daniell H. New tools for chloroplast genetic engineering. Nat Biotechnol. 1999a;17:855–856. doi: 10.1038/12841. [DOI] [PubMed] [Google Scholar]
- Daniell H. GM crops: public perception and scientific solutions. Trends Plant Sci. 1999b;4:467–469. doi: 10.1016/s1360-1385(99)01503-4. [DOI] [PubMed] [Google Scholar]
- Daniell H. Genetically modified food crops: current concerns and solutions for next generation crops. Biotechnol Genet Eng Rev. 2000;17:327–352. doi: 10.1080/02648725.2000.10647997. [DOI] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression and assembly of the native cholera toxin B subunit gene as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001a;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB. Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Curr Genet. 2001b;39:109–116. doi: 10.1007/s002940100185. [DOI] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H. Hyper-expression of the Cry2Aa2 operon in chloroplasts leads to the formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett NP. Design of antifungal peptides for agricultural applications. In: Hedin PA, Menn JJ, Hollingworth RM, editors. Natural and Engineered Pest Management Agents. Washington, DC: American Chemical Society; 1994. pp. 278–292. [Google Scholar]
- Gesell J, Zasloff M, Opella S. Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an α-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. J Biochem NMR. 1997;9:127–135. doi: 10.1023/a:1018698002314. [DOI] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Heifetz P. Genetic engineering of the chloroplast. Biochimie. 2000;82:655–666. doi: 10.1016/s0300-9084(00)00608-8. [DOI] [PubMed] [Google Scholar]
- Houston ME, Jr, Kondejewski L, Gough M, Fidai S, Hodges RS, Hancock R. Influence of performed α-helix and α-helix induction on the activity of cationic antimicrobial peptides. J Peptide Res. 1997;52:81–88. doi: 10.1111/j.1399-3011.1998.tb01361.x. [DOI] [PubMed] [Google Scholar]
- Huang H. Action of antimicrobial peptides: two state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- Huang Y, Nordeen RO, Di M, Owens LD, MacBeath JH. Expression of an engineered cecropin gene cassette in transgenic tobacco plants confers disease resistance to Pseudomonas syringae pv. tabaci. Phytopathology. 1997;87:494–499. doi: 10.1094/PHYTO.1997.87.5.494. [DOI] [PubMed] [Google Scholar]
- Jacob L, Zasloff M. Potential therapeutic applications of magainins and other antimicrobial; agents of animal origin: antimicrobial Peptides. Ciba Found Symp. 1994;186:197–223. doi: 10.1002/9780470514658.ch12. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski F, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis Cry2A protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lawrence CB, Xing H-Y, Babbitt RA, Bass WT, Maiti IB, Everett NP. Enhanced disease resistance conferred by expression of an antimicrobial magainin analog in transgenic tobacco. Planta. 2001;212:635–639. doi: 10.1007/s004250000480. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogen PS, Stalker DM, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to extraordinary level of an insecticidal protein in tobacco. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- Mourgues F, Brisset MN, Cheveau E. Strategies to improve plant resistance to bacterial diseases through genetic engineering. Trends Biotechnol. 1998;16:203–210. doi: 10.1016/s0167-7799(98)01189-5. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 1962;15:473–497. [Google Scholar]
- Neuhas J, Sticher L, Meins F, Jr, Boller T. A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA. 1991;88:10362–10366. doi: 10.1073/pnas.88.22.10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shümann M, Dathe M, Wieprecht T, Beyermann M, Bienert M. The tendency of magainin to associate upon binding to phospholipids bilayers. Biochemistry. 1997;36:4345–4351. doi: 10.1021/bi962304x. [DOI] [PubMed] [Google Scholar]
- Sidorov V, Kasten D, Pang S, Hajdukiewicz P, Saub J, Nehra N. Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Siegenthaler PA. Molecular organization of acyl lipids in photosynthetic membranes of higher plants. In: Siegenthaler P-A, Murata N, editors. Lipids in Photosynthesis: Structure, Function and Genetics: Advances in Photosynthesis. Vol. 6. Boston: Kluwer Academic Publishers; 1998. pp. 120–144. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: W.H. Freeman and Co.; 1981. [Google Scholar]
- Staub J, Garcia B, Graves J, Hajdukiewicz P, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll J, Saptola L. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nature Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Tossi A, Sandri L, Giangaspero A. Amphipathic, α-helical antimicrobial peptides. Biopolymer. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]