Abstract
Many plants increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Cold acclimation in Arabidopsis involves rapid cold-induced expression of the C-repeat/dehydration-responsive element binding factor (CBF) transcriptional activators followed by expression of CBF-targeted genes that increase freezing tolerance. Here, we present evidence for a CBF cold-response pathway in Brassica napus. We show that B. napus encodes CBF-like genes and that transcripts for these genes accumulate rapidly in response to low temperature followed closely by expression of the cold-regulated Bn115 gene, an ortholog of the Arabidopsis CBF-targeted COR15a gene. Moreover, we show that constitutive overexpression of the Arabidopsis CBF genes in transgenic B. napus plants induces expression of orthologs of Arabidopsis CBF-targeted genes and increases the freezing tolerance of both nonacclimated and cold-acclimated plants. Transcripts encoding CBF-like proteins were also found to accumulate rapidly in response to low temperature in wheat (Triticum aestivum L. cv Norstar) and rye (Secale cereale L. cv Puma), which cold acclimate, as well as in tomato (Lycopersicon esculentum var. Bonny Best, Castle Mart, Micro-Tom, and D Huang), a freezing-sensitive plant that does not cold acclimate. An alignment of the CBF proteins from Arabidopsis, B. napus, wheat, rye, and tomato revealed the presence of conserved amino acid sequences, PKK/RPAGRxKFxETRHP and DSAWR, that bracket the AP2/EREBP DNA binding domains of the proteins and distinguish them from other members of the AP2/EREBP protein family. We conclude that components of the CBF cold-response pathway are highly conserved in flowering plants and not limited to those that cold acclimate.
Plants vary greatly in their abilities to survive freezing temperatures (Sakai and Larcher, 1987). Whereas plants from tropical regions have essentially no capacity to withstand freezing, herbaceous plants from temperate regions can survive freezing at temperatures ranging from −5 to −30°C, depending on the species. It is significant that the maximum freezing tolerance of plants is not constitutive, but is induced in response to low temperatures (below approximately 10°C), a phenomenon known as “cold acclimation” (Hughes and Dunn, 1996; Thomashow, 1999). Nonacclimated wheat (Triticum aestivum L. cv Norstar) plants, for instance, are killed at freezing temperatures of about −5°C, but after cold acclimation, can survive temperatures down to about −20°C. Determining what accounts for the differences in freezing tolerance between plant species and the molecular basis of cold acclimation is of basic scientific interest and has the potential to provide new approaches to improve the freezing tolerance of plants, an important agronomic trait.
A recent advance in understanding cold acclimation in Arabidopsis was the discovery of the C-repeat/dehydration-responsive element binding factor (CBF) cold-response pathway (see Thomashow, 2001). Arabidopsis encodes a small family of cold-responsive transcriptional activators known either as CBF1, CBF2, and CBF3 (Stockinger et al., 1997; Gilmour et al., 1998) or DREB1b, DREB1c, and DREB1a (Liu et al., 1998; Kasuga et al., 1999), respectively. The CBF transcription factors, which are members of the AP2/EREBP family of DNA-binding proteins (Riechmann and Meyerowitz, 1998), recognize the cold- and dehydration-responsive DNA regulatory element designated the CRT (C-repeat; Baker et al., 1994)/DRE (dehydration-responsive element; Yamaguchi-Shinozaki and Shinozaki, 1994). CRT/DRE elements, which have a conserved 5-bp core sequence of CCGAC, are present in the promoter regions of many cold- and dehydration-responsive genes of Arabidopsis including those designated COR (cold-regulated; Thomashow, 1999). The CBF genes are induced within 15 min of plants being exposed to low nonfreezing temperatures followed at about 2 h by induction of cold-regulated genes that contain the CRT/DRE-regulatory element, i.e. the “CBF regulon” (Gilmour et al., 1998; Liu et al., 1998). Over the next few days at low temperature, the plants increase in freezing tolerance reaching a maximum level within 1 to 2 weeks.
A role for the CBF regulon in the enhancement of freezing tolerance is indicated by the results of CBF overexpression experiments. Constitutive expression of the CBF genes in transgenic Arabidopsis plants results in the induction of COR gene expression and an increase in freezing tolerance without a low temperature stimulus (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000). It is significant that multiple biochemical changes that are associated with cold acclimation and thought to contribute to increased freezing tolerance, including the accumulation of sugars and Pro, occur in nonacclimated transgenic Arabidopsis plants that constitutively express CBF3 (Gilmour et al., 2000). Thus, it has been proposed that the CBF genes act to integrate the activation of multiple components of the cold acclimation response (Gilmour et al., 2000).
The discovery of the Arabidopsis CBF cold-response pathway raises a number of fundamental questions about plant freezing tolerance. Do plants other than Arabidopsis have CBF genes that are cold induced? If so, do they activate expression of CBF regulons that increase freezing tolerance? Are cold-regulated orthologs of CBF genes limited to plants that cold acclimate? The results presented here begin to address these questions.
RESULTS
A CBF Cold-Response Pathway in Brassica napus
B. napus, like Arabidopsis, cold acclimates and is a member of the Cruciferae family. As a first step to determine whether B. napus has a cold-response pathway related to the CBF cold-response pathway of Arabidopsis, we asked whether B. napus encoded CBF-like proteins. The results indicated that it did. cDNA clones encoding two different CBF-like proteins (accession nos. AF370733 and AF370734) were identified by screening B. napus cDNA libraries using PCR-generated probes (see “Materials and Methods”). The B. napus CBF-like proteins were 92% identical in amino acid sequence to each other and approximately 76% identical in sequence to Arabidopsis CBF1. An alignment of the B. napus proteins with Arabidopsis CBF1 indicated that the sequence identity extended throughout the protein, but was greatest in the AP2/EREBP DNA-binding domain (Fig. 1 includes an alignment of one B. napus CBF protein against Arabidopsis CBF1). A sequence for a third B. napus CBF polypeptide has been deposited by others (accession no. AF084185; N. Zhou, G. Wu, Y.-P. Gao, R.W. Wilen, and L.V. Gusta).
Figure 1.
Alignment of CBF-like proteins. The amino acid sequence shown are for: At, Arabidopsis CBF1 (accession no. AAC49662); Bn, B. napus CBF (accession no. AF370733); Le, tomato (Lycopersicon esculentum CBF (accession no. AY034473); Sc, rye (Secale cereale) CBF (accession no. AF370730); and Ta, wheat CBF (accession no. AF376136). The AP2/EREBP domain is indicated by an over line and the signature sequences PKK/RPAGRxKFxETRHP and DSAWR are indicated by black circles and white boxes, respectively.
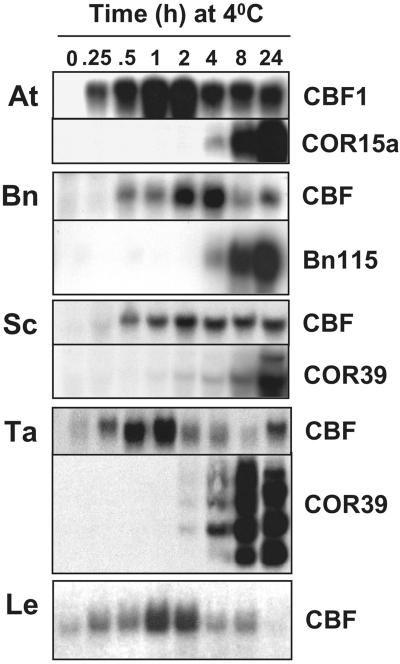
Transcripts encoding B. napus CBF-like proteins were found to accumulate rapidly (within 30 min) upon exposure of plants to low temperature (Fig. 2). This was closely followed by expression of Bn115 (Weretilnyk et al., 1993), a cold-regulated ortholog of Arabidopsis COR15a (Hajela et al., 1990). Arabidopsis COR15a is cold regulated, has CRT/DRE regulatory elements, and is induced in response to the CBF transcriptional activators (Gilmour et al., 1998; Jaglo-Ottosen et al., 1998). Cold-regulated expression of the B. napus Bn115 gene involves a DNA regulatory element, the low temperature responsive element, that contains the CRT/DRE core sequence CCGAC (Jiang et al., 1996). As with Arabidopsis CBF transcripts, B. napus CBF transcripts reached maximum levels within a few hours of plants being transferred to low temperature, after which time they decreased, but at 24 h remained elevated over the level found in nonacclimated plants.
Figure 2.
Accumulation of CBF and putative target gene transcripts in response to low temperature. Plants were grown at normal growth temperatures (20°C–22°C) and transferred to low temperature (4°C) for the indicated times. Total RNA was isolated from leaves and northern analyses performed using probes for CBF transcripts and putative CBF-targeted cold-regulated genes for B. napus (Bn115), wheat and rye (Wcs120/COR39), and Arabidopsis (COR15a) as described in “Materials and Methods.” At, Arabidopsis; Bn, B. napus; Sc, rye; Ta, wheat; Le, tomato.
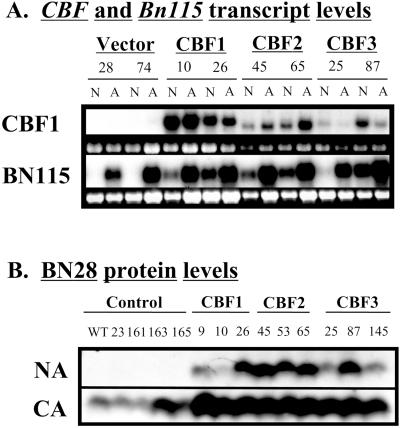
Constitutive expression of Arabidopsis CBF1, CBF2, or CBF3 in transgenic Arabidopsis plants activates expression of the target CRT/DRE-containing COR genes and increases freezing tolerance without a low temperature stimulus (Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Liu et al., 1998; S.J. Gilmour and M.F. Thomashow, unpublished data). We reasoned that if B. napus had a similar CBF-like cold-response pathway, then expression of the Arabidopsis CBF genes in transgenic B. napus might also activate expression of Bn115 and other cold-regulated genes containing the CRT/DRE-related regulatory elements and increase plant freezing tolerance. This was found to be the case. Constitutive expression of Arabidopsis CBF1, CBF2, and CBF3 in transgenic B. napus caused the accumulation of transcripts for Bn115 (Fig. 3A) and Bn28 (not shown) without a low temperature stimulus; Bn28 encodes an ortholog of the CRT/DRE-regulated cold-responsive gene COR6.6 (Hajela et al., 1990). Immunoblot analysis further indicated that the BN28 protein accumulated in nonacclimated plants that expressed CBF1, CBF2, or CBF3 (Fig. 3B). Finally, the levels of the BN28 protein were higher in cold-acclimated CBF-expressing plants than they were in control plants (Fig. 3B).
Figure 3.
Effect of overexpressing Arabidopsis CBF genes in transgenic B. napus plants on expression of endogenous cold-regulated genes Bn115 and Bn28. A, Transcript levels of the Arabidopsis CBF transgenes and the endogenous B. napus Bn115 gene in control (vector) and CBF-expressing (CBF1, CBF2, and CBF3) B. napus transgenic plants that were either nonacclimated (N) or cold acclimated (A) for 3 weeks. Total RNA was isolated from pooled plants of the indicated transgenic lines and subjected to northern analysis using probes prepared from cDNAs for either the Arabidopsis CBF1 gene or B. napus Bn115 gene. Numbers above the samples refer to the specific transgenic lines tested. Loading controls show the 18S ribosomal RNA band from the corresponding ethidium bromide-stained agarose gel used for the northern analysis. B, Levels of the B. napus BN28 protein in nonacclimated (NA) and cold-acclimated (CA) control and CBF-expressing transgenic B. napus plants. Total soluble protein (100 μg) prepared from nonacclimated and 3-week cold-acclimated plants was subjected to immunoblot analysis using antiserum raised to the BN28 polypeptide (Boothe et al., 1997). Numbers above each sample refer to the specific transgenic line tested. The sample designated WT was from plants that had not been transformed. Protein transfer for line 10 was inefficient in this experiment due to a bubble in the gel.
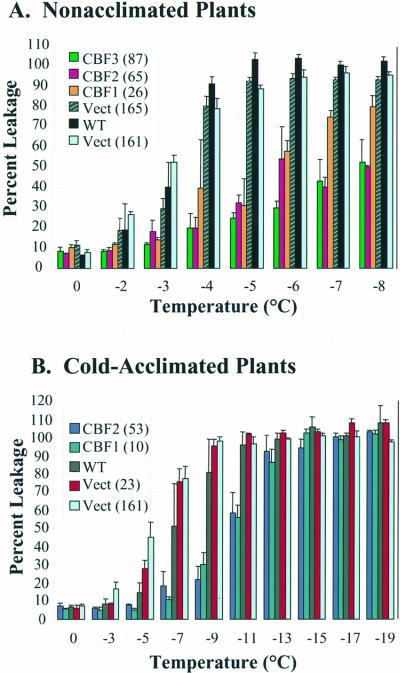
Electrolyte leakage experiments indicated that expression of the Arabidopsis CBF genes in B. napus resulted in an increase in freezing tolerance. In the experiment shown in Figure 4A, leaf tissue from nonacclimated control B. napus plants had EL50 values (the freezing temperature that causes leakage of 50% of total electrolytes) between −3°C and −4°C, whereas the leaf tissue of plants expressing CBF1, CBF2, or CBF3 had EL50 values of about −6°C. Combined results from multiple electrolyte leakage experiments indicated that leaf tissue from nonacclimated control B. napus plants had an EL50 value of about −2.1°C, whereas leaf tissue from nonacclimated CBF-expressing plants had an EL50 value of about −4.7°C (Table I). CBF expression was also found to cause an increase in the freezing tolerance of cold-acclimated plants. In the experiment shown in Figure 4B, leaf tissue from cold-acclimated control B. napus plants had EL50 values of about −6°C, whereas the leaf tissue of plants expressing either CBF1 or CBF2 had EL50 values of about −11°C. Combined results from multiple experiments indicated that leaf tissue from cold-acclimated control B. napus plants had an EL50 value of about −8.1°C, whereas leaf tissue from cold-acclimated CBF-expressing plants had an EL50 value of −12.7°C (Table I).
Figure 4.
Freezing tolerance of leaf tissue from nonacclimated (A) or cold-acclimated (B) control and CBF-expressing B. napus plants. Leaves from nonacclimated and cold-acclimated seedlings were frozen to the temperatures indicated and cellular damage assessed by measuring electrolyte leakage as described in “Materials and Methods.” Numbers in parentheses indicate the specific transgenic lines tested. Error bars indicate the sds of the three replicates of each data point.
Table I.
Freezing tolerance (EL50 values in °C) for nonacclimated and cold-acclimated control and CBF-expressing transgenic B. napus plantsa
| Plants | Nonacclimated | Cold Acclimated |
|---|---|---|
| Control | −2.1 ± 0.34 (10) | −8.1 ± 0.42 (8) |
| CBF expressing | −4.7 ± 0.40 (23) | −12.7 ± 0.52 (12) |
EL50 values were calculated using combined data from individual nonacclimated or cold-acclimated control and CBF-expressing plants (no. of plants used are indicated in parentheses). All values were significantly different from each other (P < 0.001) as determined by ANOVA. Nonacclimated control plants used were: wild type (2), vector-23 (2), vector-161 (4), vector-163 (1), and vector-165 (1). Nonacclimated CBF-expressing plants used were: CBF1-9 (1), CBF1-10 (3), CBF1-26 (3), CBF2-45 (1), CBF2-53 (2), CBF2-65 (3), CBF3-25 (2), CBF3-87 (2), CBF3-108 (1), CBF3-129 (1), and CBF3-145 (3). Cold-acclimated control plants used were: wild type (2), vector-23 (1), vector-161 (3), vector-163 (1), and vector-165 (1). Cold-acclimated CBF-expressing plants used were: CBF1-9 (1), CBF1-10 (2), CBF1-26 (2), CBF2-45 (1); CBF2-53 (1); CBF2-65 (1); CBF3-25 (1); CBF3-87 (1); CBF3-145 (2).
Cold-Responsive CBF-Like Genes in Wheat and Rye
The results presented above indicated that B. napus encodes a CBF cold-response pathway related to that found in Arabidopsis. We next asked whether more distantly related plants that cold acclimate have CBF-like genes that are rapidly induced in response to low temperature. cDNA libraries of rye and wheat were screened for clones encoding CBF-like proteins using probes generated by PCR (see “Materials and Methods”). This resulted in the identification of cDNA inserts encoding one wheat (accession no. AF376136) and three rye (accession nos. AF370728, AF370729, and AF370730) CBF-like polypeptides. The rye and wheat polypeptides shared 30% to 34% sequence identity with Arabidopsis CBF1, most of which was due to a high degree of identity between the AP2/EREBP DNA-binding domains (Fig. 1 includes an alignment of the wheat and a rye CBF protein with Arabidopsis CBF1). However, a striking feature of the wheat and rye proteins was that they had in common with the Arabidopsis and B. napus CBF proteins short polypeptide sequences that flanked the AP2/EREBP sequence; PKK/RPAGRxKFxETRHP immediately upstream of the AP2/EREBP domain and the sequence DSAWR just downstream from it (see Fig. 1). It is significant that of the more than 140 AP2/EREBP domain proteins predicted to be encoded by Arabidopsis (Riechmann et al., 2000), only CBF1, CBF2, and CBF3 were found to have the PKK/RPAGRxKFxETRHP and DSAWR “signature sequences” surrounding the AP2/EREBP domain. The AP2/EREBP domains of three additional Arabidopsis AP2/EREBP proteins (accession nos. 3241926, AC025417, and AC010795) were also found bracketed by the nearly identical sequences PKK/RRAGRxxFxETRHP and DSAWR.
As in Arabidopsis and B. napus, CBF-like transcripts accumulated rapidly (within 15–30 min) in response to low temperature in both wheat and rye (Fig. 2). This was followed at about 2 h by accumulation of transcripts for the cold-responsive Wcs120/COR39 gene family (Guo et al., 1992; Houde et al., 1992; Fig. 2). Wcs120/COR39, which is an ortholog of the CBF-targeted cold-regulated COR47 gene of Arabidopsis (Gilmour et al., 1992), is a potential CBF target because its promoter is activated in response to low temperature and has multiple copies of the CRT/DRE core sequence CCGAC (Ouellet et al., 1998).
Cold-Responsive CBF-Like Genes in Tomato
The results presented above supported the hypothesis that a common feature of cold acclimation is rapid cold induction of genes encoding CBF-like transcriptional activators. A fundamental question raised was whether plants that do not cold acclimate encode CBF-like proteins and whether transcripts encoding them accumulate rapidly in response to low temperature. A search of the public databases indicated that tomato encoded multiple AP2/EREBP proteins that share significant sequence identity with Arabidopsis CBF1. A clone for one expressed sequence tag (EST; accession no. AI89824) was obtained and the complete DNA sequence of the insert was determined (accession no. AY034473). The deduced polypeptide was found to share 53% amino acid sequence identity with Arabidopsis CBF1 and contain the PKK/RPAGRxKFxETRHP and DSAWR signature sequences (Fig. 1.). Moreover, CBF-like transcripts were found to accumulate rapidly upon exposure of tomato plants to low temperature (Fig. 2). The results shown are from an experiment using tomato var. Castle Mart, but similar results were obtained with Bonny Best, Micro-Tom, and D Huang (not shown). Unlike in Arabidopsis, B. napus, rye, and wheat, however, the transcript levels of the tomato CBF transcripts in Castle Mart (Fig. 2) and the other varieties (not shown) appeared to return to those found in warm-grown plants after 24 h of exposure to low temperature and remained at low levels after 1 week of cold treatment (not shown). We were unable to test for the expression of tomato cold-regulated genes containing active CRT/DRE-like elements because to our knowledge, such genes have not yet been identified.
DISCUSSION
Cold acclimation in Arabidopsis involves action of the CBF cold-response pathway (Thomashow, 2001). The hallmark characteristics of this pathway are rapid induction of the CBF genes in response to low temperature followed by expression of the CBF regulon, which includes genes that increase plant freezing tolerance. Here, we report that B. napus encodes CBF-like proteins, that transcripts encoding these proteins accumulate rapidly in response to low temperature, and that this is closely followed by induction of Bn115, an ortholog of the CBF-targeted Arabidopsis gene COR15a. Moreover, we demonstrate that overexpression of Arabidopsis CBF genes in B. napus induces expression of Bn115 and Bn28, an ortholog of the CBF-targeted Arabidopsis gene COR6.6, and increases freezing tolerance in both nonacclimated and cold-acclimated plants. From these results we conclude that B. napus, a close relative of Arabidopsis that cold acclimates, encodes a CBF cold-response pathway related to that found in Arabidopsis. In addition, we conclude that components of the CBF cold-response pathway are conserved in wheat and rye, more distant relatives of Arabidopsis that also cold acclimate. In particular, we show that these cereals encode CBF-like proteins, that transcripts for these proteins accumulate rapidly in response to low temperature and that this is quickly followed by induction of Wcs120/COR39, a gene with a cold-inducible promoter that has multiple copies of the CRT/DRE core sequence, CCGAC (Ouellet et al., 1998).
It is significant that the results presented also indicate that cold-regulated CBF-like genes are not limited to plants that cold acclimate. To be specific, we show that transcripts encoding a CBF-like protein(s) rapidly accumulate in response to low temperature in tomato, a chilling-sensitive plant that does not cold acclimate. Thus, tomato appears to have components of a CBF cold-response pathway. Thus, a fundamental question raised is why doesn't tomato cold acclimate? One possibility is that tomato has a completely functional CBF cold-response pathway, but that some other component(s) of the cold acclimation response is limiting. In an alternate manner, tomato might not have a fully functional CBF cold-response pathway. There might, for instance, be differences in the activities of the CBF-like proteins, though we have found that overexpression of the tomato CBF coding sequence (accession no. AY034473) in transgenic Arabidopsis plants activates expression of COR15a and COR6.6 without a low temperature stimulus (X. Zhang and M.F. Thomashow, unpublished data). Other possibilities would include differences in the composition of the CBF regulons and differences in regulation of the CBF genes. The results presented indicate that the levels of the tomato CBF transcripts do not remain elevated at low temperature as Arabidopsis CBF transcripts do (Fig. 2). If true, it may be that an inability of tomato to sustain CBF expression results in only transient expression of CBF-targeted genes, which in turn may not allow the development of freezing (and possibly chilling) tolerance.
The AP2/EREBP protein family is characterized by a DNA-binding motif that is unique to plants, the AP2/EREBP domain (Riechmann and Meyerowitz, 1998). The domain consists of an α-helix and a three-stranded antiparallel β-sheet that interacts with base pairs within the DNA major groove (Allen et al., 1998). The AP2/EREBP domain is found in a large number of plant proteins including more than 140 proteins in Arabidopsis (Riechmann et al., 2000). The results presented here indicate that the Arabidopsis CBF1, CBF2, and CBF3 proteins form a subset of the AP2/EREBP proteins that is characterized by two additional sequences that immediately surround the AP2/EREBP domain, PKK/RPAGRxKFxETRHP upstream of the domain and DSAWR downstream of it (Fig. 1). These “signature sequences” are present in CBF-like proteins from B. napus, wheat, rye, and tomato (Fig. 1). Conservation of these sequences across evolutionarily diverse plant species suggests that they have an important functional role. The resemblance of the PKK/RPAGRxKFxETRHP sequence to nuclear transport signals (Smith and Raikhel, 1999) indicates that it might be involved in protein trafficking as previously suggested (Stockinger et al., 1997). The signature sequences would not appear to be involved in recognition of the CRT/DRE regulatory element because they (or closely related sequences) are not present in the Arabidopsis AP2/EREBP protein DREB2a (Liu et al., 1998). This protein has been demonstrated to bind to the CRT/DRE element and activate gene expression in Arabidopsis in a transient assay (though interestingly not in stable Arabidopsis transformants; Liu et al., 1998). The DREB2a gene is not induced by low temperature, but instead is induced in response to dehydration stress (Liu et al., 1998). Expression of the DREB2a protein in drought-stressed plants is proposed to account, at least in part, for the dehydration responsiveness of the CRT/DRE element (Liu et al., 1998).
Understanding the mechanisms that plants have evolved to tolerate environmental stresses has the potential to provide new tools and strategies to improve the environmental stress tolerance of plants. The discovery of the Arabidopsis CBF cold-response pathway has possibilities in this regard. Previous studies demonstrated that increased expression of the CBF genes in Arabidopsis results in an increase in both freezing and drought tolerance (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000). Here, we extend these findings to an important agronomic crop plant, Brassica oilseed rape (canola). We show that the freezing tolerance of B. napus can be enhanced through CBF-mediated “regulon engineering.” It is important to bear in mind, however, that constitutive high-level overexpression of the CBF genes can result in undesirable agronomic traits. In Arabidopsis, high-level CBF overexpression can cause a “stunted” growth phenotype, a decrease in seed yield and a delay in flowering (Liu et al., 1998; Gilmour et al., 2000). The CBF-expressing B. napus plants used in the experiments described here were grown in environmental chambers under constant light and did not exhibit overt adverse effects in growth and development, but when grown under greenhouse conditions, display stunted growth and delayed flowering phenotypes (V. Haake and J. Zhang, unpublished data). Whether strategies such as using stress-inducible promoters to drive CBF expression (Kasuga et al., 1999) can be developed to attain the potential positive effects of CBF regulon engineering without incurring undesirable negative traits remains to be determined.
MATERIALS AND METHODS
Plant Material
Brassica napus cv Westar (a spring-type canola), winter wheat (Triticum aestivum L. cv Norstar), winter rye (Secale cereale L. cv Puma), and tomato (Lycopersicon esculentum var. Bonny Best, Castle Mart, Micro-Tom, and D Huang) were grown in pots containing Baccto Planting Mix (Michigan Peat, Houston) in controlled environment chambers at 20°C to 22°C under continuous cool-white fluorescent illumination of 100 to 150 μmol m−2 s−1 light intensity as described by Gilmour et al. (1988). For cold acclimation, plants were incubated at 4°C under continuous cool-white fluorescent illumination at approximately 50 μmol m−2 s−1 light intensity.
Isolation of cDNAs Encoding CBF-Like Proteins
A B. napus genomic DNA fragment encoding a CBF-like polypeptide was isolated by PCR (Innis et al., 1990) using degenerate primers O368 (CAYCCNATHTAYMGNGGNGT) and O378 (GGNARNARCATNCCYTCNGCC) based on conserved regions of the Arabidopsis CBF proteins at the beginning of the AP2/EREBP domain and putative activation domain, respectively. Full-length cDNAs were isolated based on the partial gene sequence using 5′ and 3′ RACE (MarathonTM cDNA amplification kit, CLONTECH, Palo Alto, CA). The isolation of cDNAs for rye and wheat CBF-like proteins was based on the sequence for a putative rice CBF1 homolog present in the GenBank EST database (accession no. AB023482). The rice gene was isolated from genomic DNA by PCR using primers O18016 (acgcgtcgacCCATCATCACCGAGATCGACTCGAC) and O18017 (ataagaatgcggccgcTCATTGTTCGCTCACTGGGAG). Based on the rice sequence, primers O18065 (GGCCGGCGGGGCGAACCAAGTTCC) and O18066 (AGGCAGAGTCGGCGAAGTTGAGGC) were synthesized and PCR used to isolate CBF gene fragments from rye cDNA libraries of RNA prepared from cold-acclimated plants (J. Zhang and V. Haake, unpublished data). cDNAs encoding full-length rye CBF-like proteins were isolated by screening cDNA libraries using the cloned partial genes as probes. The wheat cDNA was isolated by screening a cDNA library (Guo et al., 1992) with one of the rye cDNAs (accession no. AF370730). A tomato EST encoding a CBF-like protein (accession no. AI484513) was obtained from the Clemson University Genomics Institute (Clemson, SC). The sequence for the entire cDNA insert was determined and deposited (accession no. AY034473).
Transformation of B. napus
The coding sequences for Arabidopsis CBF1, CBF2, and CBF3 were placed under control of the strong constitutive cauliflower mosaic virus 35S promoter in the plant expression vector pGA643 (An, 1995) which includes the NPTII gene to select for kanamycin resistance. The vector, with and without inserts, was introduced into Agrobacterium tumefaciens strain GV3101 and used to transform B. napus cotyledonary petioles selecting for kanamycin resistance (Moloney et al., 1989). Regenerated plants were tested for T-DNA inserts using an NPTII ELISA kit (5 Prime-3 Prime, Inc., Boulder, CO). Positive T0 plants were self-pollinated and T1 seeds collected. Because T1 populations were not homozygous for T-DNA inserts, individual plants were tested either for expression of the NPTII gene using the NPTII ELISA assay or for the presence of the NPTII gene using the PCR (primers were 5′: TGGAGAGGCTATTCGGCTA and 3′: CACCATGATATTCGGCAAG) before being used in experiments.
RNA Hybridization
Total RNA was isolated from B. napus using TRIZOL reagent (GibcoBRL, Grand Island, NY), from wheat and rye plants using a Plant RNA Isolation Kit (Qiagen Inc., Valencia, CA), and from tomato (Howe et al., 1996) and Arabidopsis (Gilmour et al., 2000) as described. Northern transfers (5–20 μg total RNA) were prepared, hybridized, and washed as described (Stockinger et al., 1997). The probe for Arabidopsis CBF1 was prepared from a full-length cDNA of CBF1 (Stockinger et al., 1997; Gilmour et al., 2000). The probe for B. napus CBF transcripts was made by PCR amplification of genomic DNA using 5′ and 3′ primers, GGTTACGTTAGGCGGAGAGT and GGACGGCGGCGGCAAAAG, respectively, based on sequence AF084185. The probe for rye and wheat CBF transcripts was the entire insert from one of the cloned rye cDNAs (accession no. AF370730). The probe for tomato CBF transcripts was the entire cDNA insert from EST AI484513. Hybridization probes for BN28 (Orr et al., 1992) and BN115 (Weretilnyk et al., 1993) were the entire cDNA inserts in plasmids pBN28 and pBN115, respectively, kindly provided by Jas Singh (Agriculture Canada, Ottawa). The probe for wheat COR39 was the entire cDNA insert from pWG1 (Guo et al., 1992). DNA fragments were 32P radiolabeled (Stockinger et al., 1997; Gilmour et al., 2000) and gel purified (Sambrook et al., 1989) as described.
Immunoblot Analysis
Total protein was extracted by grinding frozen tissue (approximately 300 mg) in extraction buffer (approximately 300 μL) containing 50 mm Tris-HCl (pH 8.0), 5% (w/v) glycerol, 100 mm KCl, and 1.5% (w/v) polyvinyl-polypyrrolidone. Insoluble material was removed by centrifugation at 13,000g for 20 min at 4°C. Protein concentrations of supernatants were determined using the Bradford dye-binding assay (Bio-Rad, Hercules, CA). Total soluble protein (100 μg) was fractionated by 10% (w/v) acrylamide tricine SDS/PAGE (Schägger and von Jagow, 1987) and transferred to 0.1-μm nitrocellulose membranes by electroblotting (Towbin et al., 1979) as described (Artus et al., 1996). BN28 protein was detected using antiserum kindly provided by Anne Johnson (Boothe et al., 1997) and visualized using the enhanced chemiluminescence system (Amersham, Buckinghamshire, UK).
Freezing Tolerance Assays
B. napus T1 seedlings (approximately 2 weeks old) were screened for the presence of the transgene and thinned to one plant per pot. At 4 to 6 weeks, plants were either tested directly for freezing tolerance (nonacclimated plants) or were placed at 4°C under continuous fluorescent illumination of approximately 50 μmol m−2 s−1 for 3 weeks. Freezing tolerance was determined using the electrolyte leakage test as previously described (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000). Tissue from the smallest two leaves was obtained using a 6-mm paper punch. Three or four punches were used in each of three replicate samples for each temperature point tested. The EL50 values (temperature that caused leakage of 50% of the electrolytes) were determined by fitting model curves of up to third-order linear polynomials for each electrolyte leakage test. To ensure unbiased predictions of electrolyte leakage, trends significantly improving the model fit at the 0.2 probability level were retained. An unbalanced one-way analysis of variance (ANOVA), adjusted for the different number of EL50 values for each tissue type was determined using SAS PROC GLM (SAS Institute, 1989).
ACKNOWLEDGMENTS
We are grateful to Wilf Keller for hosting one of us (S.K.) in his laboratory to learn how to transform canola; Maurice Moloney for advice regarding canola transformation; Jas Singh for cDNAs encoding BN28 and BN115; Anne Johnson for the antibody to the BN28 protein; Trevor Wagner for conducting initial alignments of CBF proteins; Cai-Zhong Jiang, Mark Leibman, and Sanjeev Pillai for their help in isolating CBF homologs; and Steve Triezenberg and Sarah Gilmour for critical reading of the manuscript.
Footnotes
This research was supported by a subcontract (no. 593–0219–06) under the U.S. Department of Agriculture/Cooperative State Research, Education, and Extension Service Cooperative Agreement North Central Biotechnology Initiative (no. 96–34340-2711), by Mendel Biotechnology, Inc., and by the Michigan Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010548.
LITERATURE CITED
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. Binary Ti plasmid vectors. Methods Mol Biol. 1995;44:47–58. doi: 10.1385/0-89603-302-3:47. [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin CT, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15agene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Boothe JG, Sonnichsen FD, de Beus MD, Johnson-Flanagan AM. Purification, characterization, and structural analysis of a plant low-temperature-induced protein. Plant Physiol. 1997;113:367–376. doi: 10.1104/pp.113.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol. 1992;18:13–21. doi: 10.1007/BF00018452. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Hajela RK, Thomashow MF. Cold acclimation in Arabidopsis thaliana. Plant Physiol. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced CORgene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Guo W, Ward RW, Thomashow MF. Characterization of a cold-regulated wheat gene related to Arabidopsis cor47. Plant Physiol. 1992;100:915–922. doi: 10.1104/pp.100.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF. Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Danyluk J, Laliberte JF, Rassart E, Dhindsa RS, Sarhan F. Cloning, characterization, and expression of a cDNA encoding a 50-kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol. 1992;99:1381–1387. doi: 10.1104/pp.99.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;47:291–305. [Google Scholar]
- Innis MA, Gelfand GD, Sninsky JJ, White TJ. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press Inc.; 1990. [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces CORgenes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jiang C, Iu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney M, Walker JM, Sharma KK. High-efficiency transformation of Brassica-napus using Agrobacteriumvectors. Plant Cell Rep. 1989;8:238–242. doi: 10.1007/BF00778542. [DOI] [PubMed] [Google Scholar]
- Orr W, Iu B, White TC, Robert LS, Singh J. Complementary DNA sequence of a low temperature-induced Brassica napus gene with homology to the Arabidopsis thaliana kin1gene. Plant Physiol. 1992;98:1532–1534. doi: 10.1104/pp.98.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Vazquez-Tello A, Sarhan F. The wheat wcs120promoter is cold-inducible in both monocotyledonous and dicotyledonous species. FEBS Lett. 1998;423:324–328. doi: 10.1016/s0014-5793(98)00116-1. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR. Arabidopsistranscription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Sakai A, Larcher W. Frost Survival of Plants: Responses and Adaptation to Freezing Stress. Berlin: Springer-Verlag; 1987. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- SAS Institute. SAS/STAT User's Guide, version 6. Cary, NC: SAS Institute; 1989. [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Smith HM, Raikhel NV. Protein targeting to the nuclear pore: what can we learn from plants? Plant Physiol. 1999;119:1157–1164. doi: 10.1104/pp.119.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehlelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets; procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weretilnyk E, Orr W, White TC, Iu B, Singh J. Characterization of three related low-temperature-regulated cDNAs from winter Brassica napus. Plant Physiol. 1993;101:171–177. doi: 10.1104/pp.101.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsisgene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]