Abstract
The maize (Zea mays) β-glucosidase Zm-p60.1 has been implicated in regulation of plant development by the targeted release of free cytokinins from cytokinin-O-glucosides, their inactive storage forms. The crystal structure of the wild-type enzyme was solved at 2.05-Å resolution, allowing molecular docking analysis to be conducted. This indicated that the enzyme specificity toward substrates with aryl aglycones is determined by aglycone aromatic system stacking with W373, and interactions with edges of F193, F200, and F461 located opposite W373 in a slot-like aglycone-binding site. These aglycone-active site interactions recently were hypothesized to determine substrate specificity in inactive enzyme substrate complexes of ZM-Glu1, an allozyme of Zm-p60.1. Here, we test this hypothesis by kinetic analysis of F193I/Y/W mutants. The decreased Km of all mutants confirmed the involvement of F193 in determining enzyme affinity toward substrates with an aromatic aglycone. It was unexpected that a 30-fold decrease in kcat was found in F193I mutant compared with the wild type. Kinetic analysis and computer modeling demonstrated that the F193-aglycone-W373 interaction not only contributes to aglycone recognition as hypothesized previously but also codetermines catalytic rate by fixing the glucosidic bond in an orientation favorable for attack by the catalytic pair, E186 and E401. The catalytic pair, assigned initially by their location in the structure, was confirmed by kinetic analysis of E186D/Q and E401D/Q mutants. It was unexpected that the E401D as well as C205S and C211S mutations dramatically impaired the assembly of a catalysis-competent homodimer, suggesting novel links between the active site structure and dimer formation.
β-Glucosidases (β-glucoside glucohydrolases, EC 3.2.1.21) are a widespread group of enzymes hydrolyzing a broad variety of aryl- and alkyl-β-d-glucosides as well as glucosides with only a carbohydrate moiety. Interest in β-glucosidase research reflects essential functions of β-glucosidases in a variety of basic biological processes ranging from developmental regulation to chemical defense against pathogen attack, and in a number of industrial applications such as biomass conversion. In plants, β-glucosidases have been implicated in regulating various aspects of development, e.g. phytohormone activation (Smith and van Staden, 1978; Brzobohatý et al., 1994), cell wall degradation in the endosperm during germination (Leah et al., 1995), and pathogen defense reactions (Poulton, 1990).
Plant β-glucosidases are classified as family 1 of retaining glycosyl hydrolases according to their primary structure (Henrissat and Bairoch, 1993; Henrissat et al., 1995). Hydrolysis of a glycosidic bond involves two essential carboxylates, one acting as a general acid/base catalyst and the other as a nucleophile (Sinnott, 1990). The hydrolysis is initiated by the nucleophilic attack at the anomeric carbon (C1) of the substrate that results in the formation of glycosyl enzyme intermediate followed by the release of the aglycone facilitated by protonation of the glycosidic oxygen by the acid catalyst (the glycosylation step). The deprotonated acid catalyst acting now as a base (anion) removes a proton from water, and the resulting hydroxyl anion attacks the covalent bond of the glycosyl enzyme intermediate. As a result of the attack, Glc is released and nucleophile regenerated (the deglycosylation step). For example, in the homodimeric Agrobacterium faecalis β-glucosidase (further referred to as A. faecalis β-glucosidase) E358 was identified as the nucleophile by means of mechanism-based inactivators and site-directed mutagenesis (Withers et al., 1990; Withers et al., 1992). E170 subsequently was demonstrated to serve as the general acid/base catalyst based on chemical rescue of inactive mutants in A. faecalis β-glucosidase (Wang et al., 1995). E358 and E170 are conserved in all members of family 1 of glycosyl hydrolases, and reside in the ITENG and TXNEX motifs (where X is a hydrophobic amino acid residue), respectively.
Three-dimensional structures of family 1 β-gly-cosidases from six divergent species have been solved recently (Barrett et al., 1995; Wiesmann et al., 1995; Aguilar et al., 1997; Burmeister et al., 1997; Sanz-Aparicio et al., 1998; Chi et al., 1999). Although levels of sequence identity vary between 17% and 44% in the β-glycosidases, their structures have proved to be highly similar. The overall fold in the enzymes is a single domain (β/α)8 barrel as predicted for family 1 glycosyl hydrolases. Glu residues residing in the conserved TXNEX and ITENG motifs are located at the C termini of strands 4 and 7, respectively, and are separated by approximately 5.5 Å, a feature typical for retaining glycosyl hydrolases. This is consistent with the classification of these β-glycosidases into the 4/7 superfamily (Henrissat et al., 1995; Jenkins et al., 1995). In myrosinases (β-thioglucosidases), the position of E acting as the acid/base catalyst is occupied by Q. In myrosinase substrates, glucosinolates, the aglycone is an excellent leaving group; thus, there is no need to provide protonation assistance for aglycone departure as reported for Sinapis alba myrosinase (SAMyr; Burmeister et al., 1997).
Thus, substantial progress has been achieved in understanding the mechanism of glucosidic bond cleavage and elucidating the roles of the two catalytic Glu residues within the active site that are involved in catalysis (the catalytic pair). However, until very recently, no experimental data on molecular determination of aglycone specificity in β-glucosidases were available. Yet, given the tremendous diversity of aglycone moieties in natural glucosides that reflects their varied biological functions, fine tuning of diverse biological processes in plants relies to a great extent on well-defined specificity in a number of β-glucosidases toward their respective aglycones. Elucidation of aglycone specificity in β-glucosidases is a key prerequisite toward uncovering their precise role(s) in biological processes that involve glucosylation and deglucosylation as regulatory elements. At the same time, the ability to modulate specificity in β-glucosidases holds considerable promise in terms of their biotechnological applications.
In maize (Zea mays), a β-glucosidase preferentially hydrolyzing cytokinin-O- and N3-glucosides in vitro and in vivo was identified, and a corresponding cDNA, Zm-p60.1, was cloned (Campos et al., 1992; Brzobohatý et al., 1993). Based on its enzyme activity and highly specific expression pattern, Zm-p60.1 has been suggested to be one of the key enzymes involved in the regulation of plant development by releasing active phytohomornes, cytokinins, from cytokinin-O-glucosides, their inactive storage and transport forms (Brzobohatý et al., 1993; Kristoffersen et al., 2000). Further characterization revealed that Zm-p60.1 dimer formation is an essential prerequisite for obtaining enzyme activity (Rotrekl et al., 1999). The enzyme has been localized to plastids/chloroplasts (Kristoffersen et al., 2000). As a first step toward addressing the mechanism of catalytic activity and substrate specificity the enzyme was purified, crystallized, and the crystals were subjected to preliminary x-ray analysis (Vévodová et al., 2001).
A cDNA containing an identical open reading frame has been cloned independently and sequenced by Esen and Shahid (1995; direct submission). Using this cDNA, ZM-Glu1, an allozyme of Zm-p60.1 processed five residues upstream of the N terminus identified in mature Zm-p60.1 (Brzobohatý et al., 1993), was produced in Escherichia coli. Substrate specificity analysis revealed that, apart from artificial chromogenic and fluorogenic glucosides designed specifically to monitor general β-glucosidase activity, ZM- Glu1 could hydrolyze 4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)-glucoside (DIMBOA- Glc; Cicek and Esen, 1999) in a manner similar to a β-glucosidase purified from maize seedlings (Babcock and Esen, 1994). Based on this finding, it has been suggested that ZM-Glu1 is involved in defense against pathogens by releasing the toxic aglycone (DIMBOA) from its storage form, DIMBOA-Glc. However, no direct experimental evidence confirming that ZM-Glu1 is actually involved in defense response in planta has been published. Crystal structure of the wild-type (WT) ZM-Glu1 at 2.5-Å resolution, and that of a complex of ZM-Glu1 with the non-hydrolyzable inhibitor p-nitrophenyl β-d-thioglucopyranoside, were solved recently (Czjzek et al., 2001). The cocrystal structure of an inactive mutant of ZM-Glu1 and DIMBOA-Glc complex subsequently was solved at 2.1-Å resolution. The data permitted visualization of the aglycone within ZM-Glu1 active site. The aglycone is sandwiched between W378 on one side and F198, F205, and F466 (equivalent to positions W373, F193, F200, and F461 in the sequence of Zm-p60.1) on the other side of the active center. The structure prompted the hypothesis that the specific conformation of these four hydrophobic amino acids and the shape of the aglycone-binding site they form determine aglycone recognition and substrate specificity in ZM-Glu1 (Czjzek et al., 2000).
The work presented now provides a test of this hypothesis via a kinetic analysis of mutants at several active site residues, including F193 in the aglycone-binding pocket. An improved crystallographic structure at 2.05-Å resolution allowed computer modeling of the structural consequences of the F193 mutations. Together, these analyses confirm that F193 is involved in determining the affinity of Zm-p60.1 toward aromatic aglycones as reflected in reduced Km values in the mutants, but, unexpectedly, they clearly demonstrate a further role of F193 in determining the rate of glycosidic bond hydrolysis. Furthermore, a kinetic analysis of E186D/Q and E401D/Q confirmed that E186 and E401 constitute the catalytic pair, and, surprisingly, that the E401D mutation hinders the assembly of enzymatically active dimers suggesting a previously unrecognized link between the active site architecture and dimer interface.
RESULTS AND DISCUSSION
Structure of the Zm-p60.1 β-Glucosidase
The crystal structure of the Zm-p60.1 β-glucosidase was solved by molecular replacement at 2.05-Å resolution. Concomitantly with solving the Zm-p60.1 structure, structures of ZM-Glu1, an allozyme of Zm-p60.1, and ZM-Glu1 inactive mutant substrate complex have been published by an independent group (Czjzek et al., 2000, 2001). The Zm-p60.1 structure presented here is more appropriate for molecular docking and computer modeling because the ZM-Glu1 structure was solved at lower resolution (2.5 Å). The inactive mutant substrate structure was solved at a resolution (2.1 Å) comparable with our structure. However, employment of the complex structure for substrate docking is not optimal because the structure must be different to that one found in the wild type to prevent a sterical clash between the mutated amino acid residue and the ligand. Otherwise, the structures of the wild-type enzymes are, within an experimental error, identical. To prevent unnecessary duplications, here, we restrict description of the structure of the protein to presentation of basic overall features of Zm-p60.1 structure, and we focus mainly on those that are essential for interpretation of our functional analysis of the catalytic center.
The protein is a homodimer in solution and it crystallized as a homodimer in one asymmetric unit as well. However, due to the different packing environments, there are some differences between these two monomers. For example, clear electron density from residue A6 to A495 can be seen in monomer A, whereas B11 to B502 was shown for monomer B. Some of the residues from solvent-accessible areas (mainly Arg, Lys, glutamic, and aspartic acids) are disordered and have been refined with the partial occupancies. Crystallographic results are summarized in Table I.
Table I.
Summary of crystal data and refinement statistics
| Structure Refinement | Zm-p60.1 |
|---|---|
| Data collection | |
| Space group | P21 |
| a (Å) | 55.66 |
| b (Å) | 110.72 |
| c (Å) | 72.94 |
| β (°) | 92.10 |
| Resolution (Å) overall (last shell) | 68–2.05 (2.18–2.05) |
| Unique reflections | 52,651 |
| Redundancy overall (last shell) | 9.7 (8.8) |
| Completeness (%) overall (last shell) | 94.7 (85.7) |
| Rmergea (last shell) | 5.3 (17.6) |
| Refinement statistics | |
| R factorb | 0.169 |
| Rfreec | 0.230 |
| No. of protein atoms | 7,943 |
| No. of water molecules | 909 |
| Root-mean-square deviations from ideal geometry | |
| Bond length (Å) | 0.005 |
| Bond angles (°) | 1.3 |
| Average B factor (Å2) | 22.2 (monomer A) |
| 18.8 (monomer B) |
Rmerge = (∑ ‖I − <I>‖ / ∑ I ) where I is the observed intensity and <I> is the average intensity of symmetry-related reflections.
R factor = ∑h ‖ ‖Fobs‖ − ‖Fcalc‖ ‖ / ∑h‖Fobs‖, where Fcalc and Fobs are calculated and observed structure factors.
Rfree = R factor for a 6.1% subset of the reflections that were not included in the refinement.
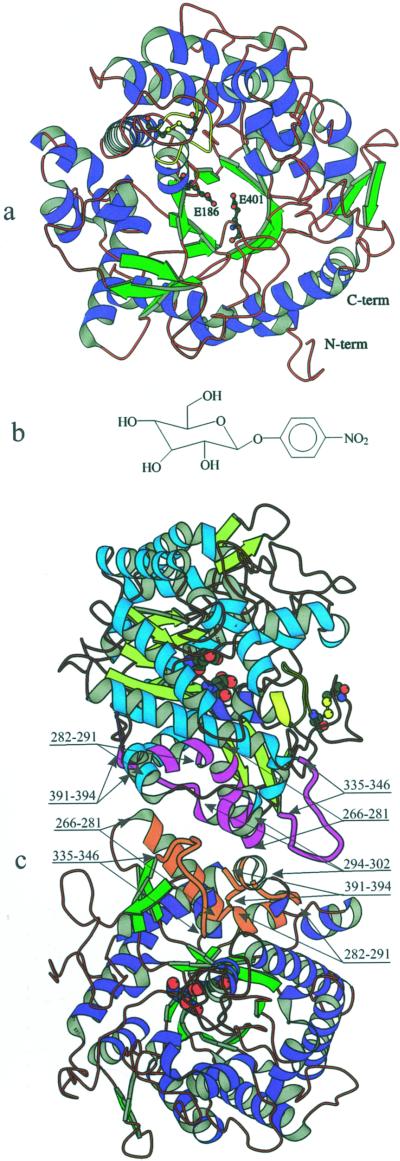
Zm-p60.1 monomer is comprised of a single (β/α)8 barrel domain (Figs. 1 and 2). Structure analysis confirmed that C205 and C211 form a disulfide bridge that has been identified previously (Rotrekl et al., 1999; Fig. 2).
Figure 1.
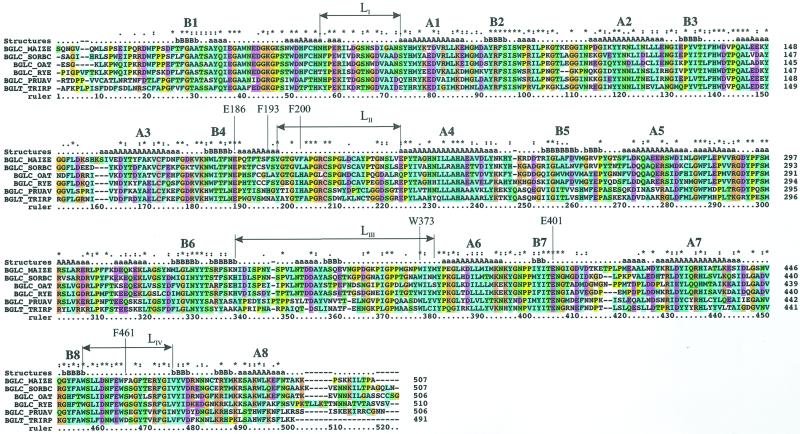
Multiple sequence alignment of proteins closely related to Zm-p60.1 β-glucosidase with the secondary structure labeled. Sequences were obtained from the Swall database using the on-line FASTA search page (http://www.ebi.ac.uk/fasta3). SORBC, Sorghum bicolor/Q41290; oat (Avena sativa)/Q9ZP27; and rye (Secale cereale). PRUAV, Prunus avium/Q43014. TRIRP, Trifolium repens/P26205. In the secondary structure labeling, letter a or A represents α-helices, and b or B represents β-strands. A1 through A8 and B1 through B8 represent the eight α-helices and β-strands that form the (β/α)8 barrel core. The asterisks on top of the alignment indicate identical residues, whereas a colon and a period are similar residues. The four enzymes in between Zm-p60.1 and T. repens cyanogenic β-glucosidase (TRCB-Glu) are related to Zm-p60.1 by sequence with identities to Zm-p60.1 of 71%, 62%, 63%, and 49%, respectively.
Figure 2.
Overview of the tertiary and quaternary structure of Zm-p60.1 β-glucosidase and structural formula of p-nitrophenyl β-d-glucopyranoside (PNPG) substrate (b). Positions of the acid/base (E186), the nucleophile (E401), the disulfide bridge between C205 and C211, and a portion (F193-C205) of a loop carrying F193 and F200 (yellow) are highlighted in the ribbon diagram presentation of a Zm-p60.1 monomer when viewed from the entrance into the interior of the active site (a). Mutual monomer orientation in the catalytically active dimer structure, and the positions of helices and loops forming the dimer interface (c). α-Helices and loops interacting at the dimer interface are indicated in magenta and violet in monomer A and B, respectively. The individual secondary structure elements located at the dimer interface are identified by their terminal amino acid residue numbers. The disulfide bridge and a loop carrying F193 and F200 (yellow) are shown only in monomer B to prevent possible confusions caused by symbol overlaps in the monomer A caused as a consequence of viewing the later one from a different angle. The panel was generated by BobScript (Esnouf, 1999).
Zm-p60.1 monomers do not possess any detectable catalytic activity (Rotrekl et al., 1999), and no experimental evidence on the mechanism by which homodimer assembly contributes to formation of catalysis-competent state of the enzyme is available yet. Description of the dimerisation interface is the first step toward understanding the role of homodimer formation in the assembly of a catalysis-competent structure in Zm-p60.1 and related β-glucosidases. In the crystal, two monomers per asymmetric unit related by a non-crystallographic symmetry were identified. The surface area buried on the dimer interface is 2,131 Å2 corresponding to 1,066 Å2 per monomer when the calculation is performed with the program Crystallography & NMR System (Brünger et al., 1998) and the probe diameter is 1.4 Å. The dimer interface is formed by residues of three α-helices (S266-L281, G282-G291, and P294-R302) and two loops (N335-L346 and K391-N394). Mutual orientation of the secondary structure elements within the dimer is indicated in Figure 2. Most of the monomer contacts in the dimer interface are represented by aromatic stacking and hydrogen and ionic bonds. No intermolecular disulfide bridge connects the monomer subunits in the dimer. An intramolecular disulfide bridge was identified in each monomer (Fig. 2), and, recently, we have shown that its formation is an essential prerequisite for gaining monomer competence to assemble into an active dimer (Rotrekl et al., 1999).
Active Site
The active site is a slot-like structure intruding from the surface to the barrel core of the protein. The third layer of residues constituting the barrel interior represents the base of the slot. Slot walls are formed mainly by four extended loops (I–IV in Fig. 1). The loops consist of residues N54 through S71 (loop I), Y195 through E221 (loop II), N335 through M376 (loop III), and W452 through V471 (loop IV). The slot-forming loops represent the sites of the highest variability in the β-glucosidase family as expected for the sites participating in the determination of substrate specificity. At the entrance, the slot is approximately 22 Å long and 8 Å wide. The upper part of the active site represents a putative aglycone-binding site, whereas the glycone-binding site is located in the bottom one-half of the active site slot.
The glycone-binding site is formed by a number of polar and aromatic residues that are commonly found within carbohydrate recognition sites. The hydrophobic site of the Glc ring can be stacked onto W452 that is highly conserved in family 1 of β-glycosidases and represents the last amino acid residue on the C-terminal end of β-strand 8. For hydrogen-bonding interactions with the Glc moiety, several conserved amino acid residues are available in appropriate orientations including Q33 (Nε2 and Oε1 H bond to O4 and O3, respectively), H137 (Nε2 H bonds to O3), N185 (Nδ2 H bonds to O2), and E459 (Oε1 and Oε2 H bond to O4 and 06, respectively). It is interesting that an unknown electron density was observed in the glycone-binding pocket. To interpret the electron density, enzyme kinetic analysis was employed to investigate the affinity of the enzyme active site to glycerol, a cryoprotectant used in crystal diffraction analysis. Glycerol proved to be a weak competitive inhibitor of Zm-p60.1 (dissociation constant of an enzyme-inhibitor complex [Ki] = 215 mm). Modeling experiments indicated that the electron density can be interpreted as two glycerol molecules. Electron densities in substrate-unoccupied active sites were assigned as glycerol molecules in SAMyr (Burmeister et al., 1997) and a ZM-Glu1 mutant (Czjzek et al., 2000), however, without providing independent experimental evidence to support the assignment.
E186 and E401 Constitute the Catalytic Pair in Zm-p60.1
Multiple alignment using ClustalW program positioned E186 and E401 into the highly conserved TXNEX and ITENG motifs, respectively (not shown). In Zm-p60.1 structure, the conserved E186 and E401 were found in the loop regions close to the carboxy-terminal ends of β-strands 4 and 7, respectively. Furthermore, E186 and 401 are in close proximity—the distance between their carbonyl carbons Cδ is 4.98 Å and between E186 Oε1 and E401 Oε2 it is 3.52 Å (Fig. 2). Two conserved Glu residues located close to the C termini of β-strands 4 and 7 and separated by a distance of about 5 Å were proposed to function as the acid/base and nucleophile, respectively, in the 4/7 superfamily of glycohydrolases (Henrissat et al., 1995; Jenkins et al., 1995). Because Zm-p60.1 belongs to the 4/7 superfamily based on sequence similarities and crystal structure analysis, E186 and E401 can constitute the catalytic pair in Zm-p60.1.
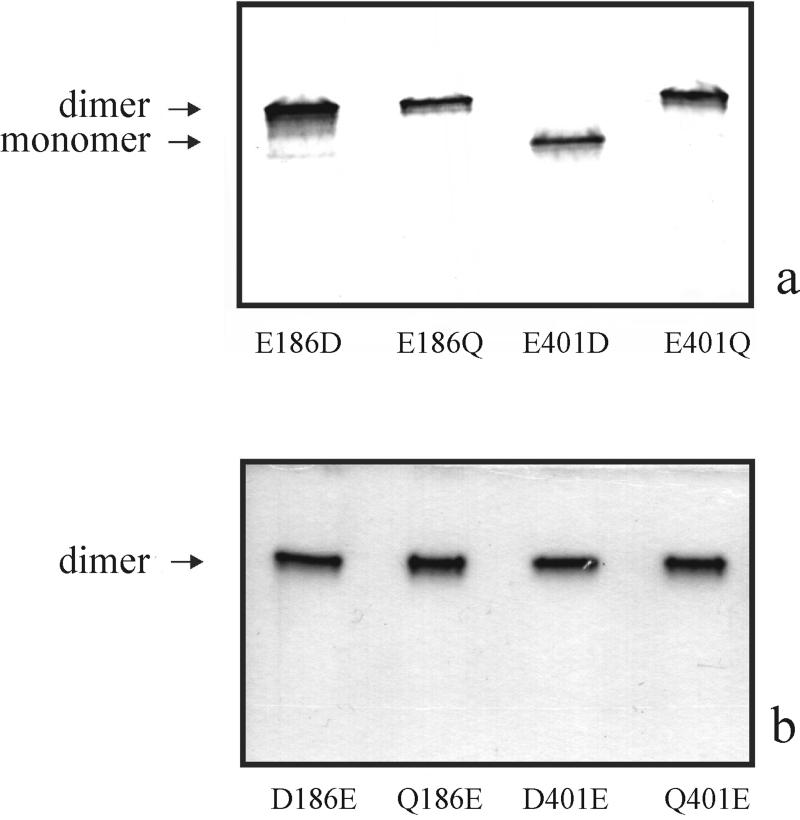
The contribution of E186 and E401 to Zm-p60.1 enzyme activity was investigated using site-directed mutagenesis followed by enzyme kinetic analysis of the mutants. In (His)6Zm-p60.r, a recombinant derivative of Zm-p60.1, the Glu residues were changed individually, in independent experiments, to Asp and Gln residues. The resulting mutants were produced in E. coli. To exclude effects of any gross alteration in quaternary structure on enzyme kinetic analysis, assembly of the mutants into catalysis-competent dimer structure (Rotrekl et al., 1999) was analyzed in soluble bacterial protein extracts using native PAGE followed by β-glucosidase activity “in-gel” staining, and immunodetection of the mutants on corresponding western blots. E186D, E186Q, and E401Q were found in the form of dimers indistinguishable from the (His)6Zm-p60.r dimer. It was surprising that the major fraction of E401D migrated at (His)6Zm-p60.r monomer position (Fig. 3a). Highly sensitive activity “in-gel” staining based on a fluorogenic substrate 4-methylumbelliferyl β-d-glucopyranoside (MUG) staining demonstrated a small but detectable enzymatic activity associated with the dimer form in E186D/Q and E401Q mutants. Detectable staining associated with expected dimer position in E401D indicated that a small fraction (below immunostaining detection limit—about 10 ng of protein per band in our assay conditions) of the mutant is present in the dimeric form and retained substantial enzyme activity. As expected from our previous experiments (Rotrekl et al., 1999), there was no enzyme activity associated with the monomeric form of the mutant (not shown). In addition to DNA sequencing, the sample homogeneity of the mutants was demonstrated by operational reversion. The revertants, Q186E, D186E, Q401E, and D401E, were detected in dimeric form on western blot (not shown) and were stained “in gel” using a chromogenic substrate 6-bromo-2-naphtyl β-d-glucopyranoside to an extent undistinguishable from the (His)6Zm-p60.r dimer (Fig. 3b).
Figure 3.
E401D mutant accumulates predominantly as a monomer when expressed in E. coli. Soluble protein extracts (100 μg of protein per lane) of E. coli cells expressing E186D/Q and E401D/Q mutants were separated by 10% (w/v) PAGE under native conditions followed by western blotting and immunodetection of mutant monomer and dimer forms (indicated by arrows, a). Operational reversion was employed to confirm that ability to form dimer was lost solely as a result of E401D mutation. Soluble protein extracts of E. coli cells expressing revertants D/Q186E and D/Q401E were analyzed by native PAGE followed by activity staining (indicated by an arrow, b).
Enzyme kinetic analysis was performed on (His)6Zm-p60.r and individual mutants purified in a single step on metal chelate affinity chromatography (MCAC) columns. The kinetic parameters kcat(WT)/kcat(mut) and Km of the mutants obtained for the fluorogenic substrate MUG are presented in Table II. Km values are fairly similar to those of the wild-type enzyme for all mutants, suggesting that the Glu residues are not significantly involved in substrate binding as expected for the catalytic pair. The dramatic decrease of kcat observed in the mutant enzymes E186D/Q and E401Q strongly suggests that these are the catalytic amino acid residues. The conservative substitution E401D resulted in a kcat decrease similar to E401Q when judged on a total enzyme quantity in an assay mixture. However, native PAGE electrophoresis followed by MUG staining demonstrated that the enzyme activity was associated solely with the dimer form. Western blot followed by immunostaining and densitometry analysis of the purified mutant E401D revealed that only approximately 0.5% of the protein was present as the active dimer in the preparation subjected to the enzyme kinetics analysis. After including a correction for the active component of the protein preparation, only a moderate decrease of kcat in E401D was found [kcat(WT)/kcat(E401D) = 50 with MUG as a substrate] that is close to the value found in an analogous A. faecalis β-glucosidase nucleophile mutant [kcat(WT)/kcat(E358D) = 122 with 2′,4′-dinitrophenyl β-d-glucopyranoside as a substrate; Withers et al., 1992]. Thus, Asp residue can substitute for the Glu residue more efficiently as a nucleophile, rather than as an acid/base catalyst. In the extensively characterized A. faecalis β-glucosidase, replacement of the acid/base Glu residue by a Gly residue (E170G) resulted in a dramatic reduction of rate of glycosyl enzyme formation for substrates needing acid catalysis, whereas the rate remained almost unchanged for substrates not requiring protonic assistance (Wang et al., 1995). In Sulfolobus solfataricus β-glycosidase, substitution of a putative acid/base, E206 identified in an NEP motif, by Q resulted in a 60-fold decrease in catalytic activity on PNPG (Moracci et al., 1996). The more pronounced effect on kcat observed in the Zm-p60.1 mutant enzyme E186Q [kcat(WT)/kcat(E186Q) = 1.5 × 103] might, at least in part, be account for by methylumbelliferone being a less efficient leaving group compared with p-nitrophenol.
Table II.
Kinetic parameters of mutants in the proposed catalytic pair of Zm-p60.1 with MUG as substrate
| Enzyme | Km | kcat(WT)/kcat(mut) |
|---|---|---|
| mm | ||
| WT | 0.14 ± 0.04 | – |
| E186D | 0.17 ± 0.03 | 8 × 103 |
| E186Q | 0.19 ± 0.03 | 1.5 × 103 |
| E401D | 0.16 ± 0.04 | 50a (1 × 104)b |
| E401Q | 0.17 ± 0.04 | 4 × 104 |
kcat(WT) was 140.5 ± 3.7 s−1 · kcat(mut) were estimated with accuracy comparable to kcat(WT).
kcat(E401D) related to dimer form.
kcat(E401D) related to mixture of dimer and monomer forms.
The microenvironments of E186 and E401 are consistent with their proposed roles in the catalytic step. As a proton donor, E186 is expected to be protonated in the basic state of the enzyme. Hydrophobic nature of W138 and T189 located close to Oε1 might contribute to the increased pKa necessary for catalysis at pH 5.5 to 6.0 (the optimum reaction pH range). Oε2 of E186 forms a hydrogen bond with Nδ2 of N326. In contrast, E401 is located close to mostly polar residues (R91, N185, N326, and Y328). Moreover, Oε2 of E401 forms a salt bridge with R91, and Oε1 forms two hydrogen bonds with hydroxyl group of the Y328 and a water molecule (Fig. 4b). Thus, E401 is deprotonated and therefore available for nucleophilic attack. A similar microenvironment was reported for the proton donor and the nucleophile in TRCB-Glu (Barrett et al., 1995), and for the nucleophile in SAMyr (Burmeister et al., 1997).
Figure 4.
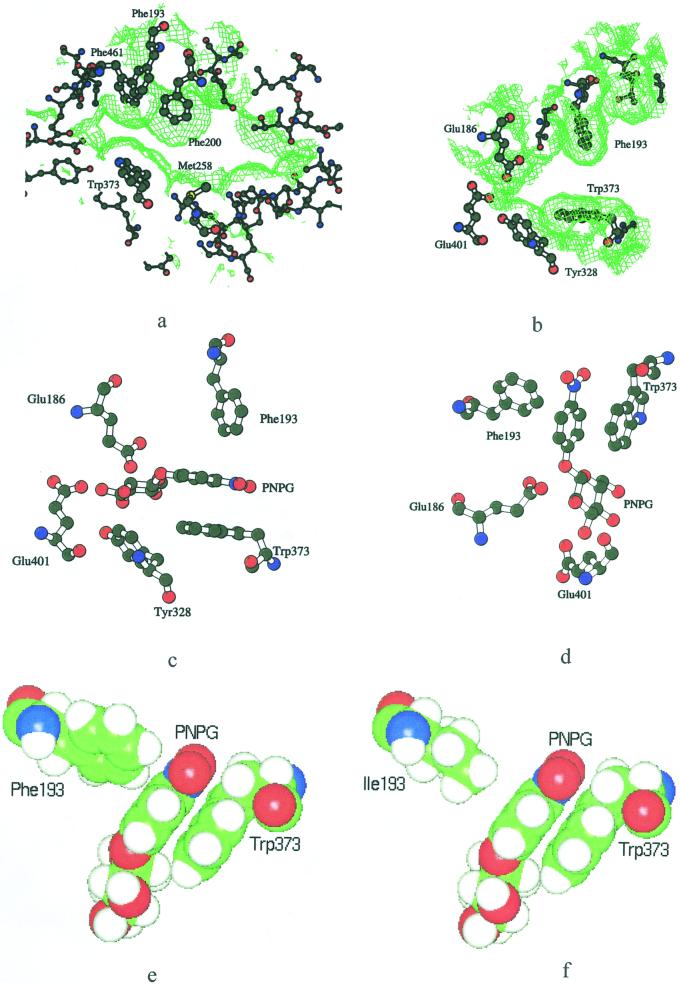
The aglycone-binding site of Zm-p60.1 β-glucosidase. Mutual orientation of the cluster F193-F200-F461-W373 determining specificity toward substrates with aromatic aglycone and the catalytic pair (E186 and E401, a), a close-up view of the “bottleneck” region of the aglycone-binding site formed by F193 and W373 (b), model of PNPG docked into the aglycone-binding site of the wild-type enzyme (c–e), and F193I mutant (f). The surfaces were generated with a probe radius of 1.1 Å using VOIDOO (Kleywegt and Jones, 1994). a through d, Generated by BobScript; e and f, generated by InsightII.
Taken together, the sequence alignments, structure, and site-directed mutagenesis followed by enzyme kinetics prove that E186 and E401 are the acid/base and the nucleophile, respectively, in Zm-p60.1.
E401D Mutation Hinders Dimer Assembly
As mentioned above, native PAGE analysis revealed a remarkable effect of E401D substitution on the enzyme structure. The electrophoretic mobility of the mutant on native gels is indistinguishable from that of (His)6Zm-p60.r monomer, confirming that the overall fold is retained in the mutant. However, the dramatically decreased ability of mutant monomer to assemble into a homodimer suggests that it adopts a distinct conformational state, an interpretation that is consistent with the observation that E401D exhibits altered chromatographic behavior observed during purification. Although (His)6Zm-p60.r, E186D/Q and E401Q bind readily to a zinc-charged iminodiacetate-POROS-column (IDA-Zn2+) used routinely for the single-step purification of the wild type and mutants, the E401D mutant is not retained by the column to any detectable extent. However, no difference in chromatographic behavior was observed between E401D and (His)6Zm-p60.r on a nickel-charged nitrilotriacetate-Sepharose-column (NTA-Ni2+) allowing single-step purification of E401D (not shown). Lack of E401D binding to IDA-Zn2+ might be explained if the proposed change in conformational state causes the His tag on E401D surface to become less accessible. Because in general the His tag binds more tightly to NTA-Ni2+ compared with IDA-Zn2+, the partly exposed His tag could still efficiently interact with a stronger coordination partner, NTA-Ni2+. The His tag and E401 are located on the opposite sides of the (β/α)8 barrel and are not connected by a common β-strand or α-helix. Thus, the conformational change extends beyond the β-7 strand and the adjacent loop carrying the ITENG motif, and is likely to be more complex than a conformational change in a single element of the secondary structure. Substitution of the Glu residue in the nucleophile position by an Asp residue in previously studied glycosidases in the 4/7 superfamily (including, for example, A. faecalis β-glucosidase, Withers et al., 1992; E. coli β-galactosidase, Yuan et al., 1994; and Bacillus circulans xylanase, Wakarchuk et al., 1994) has not resulted in gross conformational changes detectable as altered CD spectra, and native PAGE mobility. However, although the E to Q/N mutants in A. faecalis β-glucosidase, and E to Q/V mutants in E. coli β-galactosidase exhibited slightly higher or very similar thermal stability compared with the corresponding wild-type enzymes, all examined E to D mutations exerted a clear decrease in thermal stability. Inspection of E401 microenvironment clearly indicates also that the E401D mutation in Zm-p60.1 results in a loss of stabilizing interactions (a hydrogen bond between E401 and Y328, and a salt bridge between E401 and R91). Earlier studies of peptide models suggested that the process of folding is guided by interactions that also stabilize the final native structure. Thus, the distinct conformational state in E401D might be a consequence of perturbation of interactions that guide folding. It is interesting that we have recently identified an independent mutation, I404D, in the ITENG loop that also hinders dimer formation in the same experimental system (J. Zouhar and B. Brzobohatý, unpublished data).
Though the Zm-p60.1 crystal structure proves that all residues forming the active center are provided by a single monomer, dimer formation is an essential prerequisite for activity of the enzyme (Rotrekl et al., 1999). Because the dimerization interface and active site are not in close proximity with each other (Fig. 2c), the reason for the necessity of dimerization for enzyme activity is not obvious. However, the dramatic influence of E401D and I404D mutations on the assembly of catalysis-competent homodimer might represent the first experimental indication of interactions between the ITENG loop and loops forming the dimerization area. Independent evidence supporting mutual dependence of the active site architecture and adoption of dimerization competent conformation of loops forming the dimerization area comes from our earlier analysis of C205A/S/R/D and C211A/S/R/D mutants. The mutations resulted in a dramatic reduction of monomer competence to assemble into dimers, and reduced catalytic efficiency of the enzyme (Rotrekl et al., 1999), though the disulfide bridge is neither a part of the catalytic center nor dimerization area (Fig. 2). However, the enzyme structure analysis revealed that formation of the disulfide bridge is involved in precise positioning of F193 and F200, two of four aglycone-binding site key residues involved in enzyme-aglycone interactions (Figs. 2 and 4). Thus, conformational changes within the active site induced by diverse mutations seems to uncover a link between fine tuning of formation of catalysis competent structure of the active site and dimerization-competent architecture of the monomer-monomer interface in the course of dimer assembly. Structure analysis of E401D and I404D mutants could represent an important step in uncovering structural principles underlying mutual dependence of dimer formation and recovery of enzyme activity in Zm-p60.1 and related β-glucosidases. The experiments addressing this issue are in progress.
F193 Determines Both Substrate Affinity and Catalytic Rate
To obtain preliminary information on the nature of the molecular determination of Zm-p60.1 specificity toward substrate aglycone moiety, a preliminary attempt to locate putative amino acid residues forming the aglycone binding site was performed on a refined homology-based model of the three-dimensional structure of Zm-p60.1 constructed earlier (Rotrekl et al., 1999). Inspection of the active site slot showed that its upper part, the proposed aglycone-binding site, is formed mainly by hydrophobic residues. The limited accuracy of the model did not justify any detailed molecular docking that could identify residues involved in enzyme-substrate interactions. However, F193 appeared as a key determinant of the width of the slot-like aglycone-binding site, and therefore a potential determinant of aglycone specificity. A similar, though less convincing prediction could be made for M258, which appeared as a width codeterminant located opposite F193. Later, solving of the crystal structure led to confirmation of the proposed role for F193, and facilitated reliable interpretations of kinetic data obtained in F193 mutants. Though the prediction made for M258 turned out not to be precise, M258 is just adjacent to the residues determining the narrowest region of the aglycone-binding site. Therefore, kinetic analysis of M258 mutants brings a support for highly localized character of the “bottleneck” region of the slot-like aglycone-binding site deduced from the crystal structure (Fig. 4).
The functional contribution of F193 and M258 to catalytic activity was investigated by site-directed mutagenesis followed by enzyme kinetic analysis. In (His)6Zm-p60.r, F193 was changed in separate experiments to I, W, and Y. In a similar manner, M258 was replaced by I, F, and V. The resulting mutants were produced in E. coli. Native PAGE analysis of individual soluble bacterial protein extracts confirmed formation of a catalysis-competent dimer structure in all investigated mutants (not shown). For enzyme kinetic analysis, the mutants were purified by single step MCAC chromatography. The kinetic parameters Km and kcat obtained for the chromogenic substrate PNPG are summarized in Table III. Although systematic increase in Km in all the F193 mutants supported the proposed role of F193 in determination of the enzyme substrate affinity, distinct decreases in kcat in the individual F193 mutants indicated a yet unrecognized function of F193. Kinetic parameters of M258 mutants displayed no or only moderate changes compared with the wild type.
Table III.
Kinetic parameters of mutants in F193 and M258 of Zm-p60.1 with PNPG as substrate
| Enzyme | Km (mM) | kcat(mut)/kcat(WT) | [kcat/Km(mut)]/[kcat/Km(WT)] |
|---|---|---|---|
| WT | 0.64 ± 0.08 | – | – |
| F193I | 1.76 ± 0.06 | 0.03 | 0.01 |
| F193Y | 1.29 ± 0.01 | 0.62 | 0.31 |
| F193W | 1.61 ± 0.17 | 1.12 | 0.45 |
| M258I | 1.18 ± 0.36 | 0.76 | 0.41 |
| M258V | 0.59 ± 0.08 | 0.34 | 0.37 |
| M258F | 0.59 ± 0.08 | 0.30 | 0.33 |
kcat(WT) was 28.0 ± 0.8 s−1 · kcat(mut) were estimated with accuracy comparable to kcat(WT).
Prompted by the dramatic effect of F193I on the Zm-p60.1 specificity constant (kcat/Km) with PNPG, we focused on a detailed analysis of the F193 microenvironment in the Zm-p60.1 crystal structure. Detailed inspection of the structure followed by molecular docking revealed that the aglycone interaction with the slot-like aglycone-binding site is largely determined by W373 stacking interactions with the aglycone aromatic system, and van der Waals interactions with the edges of the phenyl rings provided by F193, F200, and F461 (Fig. 4). The W373 orientation favorable for stacking interactions is stabilized by a hydrogen bond between Nε1 of W373 and water that is firmly positioned by additional hydrogen bonds with E466 and Y468. Therefore, E466 and Y468 can contribute indirectly to substrate specificity and catalytic efficiency. Proper positioning of F193, F200, and F461 appears to be stabilized by a large hydrophobic cluster formed by F51, W48, W138, F190, and W460 (not shown). In addition, the disulfide bridge between C205 and C211 stabilizes the loop containing F193 and F200 (Fig. 2). It is interesting that we have found a dramatic drop in enzyme activity in mutants C205S and C211A/S (Rotrekl et al., 1999). Though the drop might be explained in part by the observed decreased ability of these mutants to form the catalysis-competent dimer (Rotrekl et al., 1999), the structural analysis presented here suggests that the decrease in catalytic efficiency may partly be due to loss of F193 and F200 position stabilization.
The molecular docking together with increase in Km in F193/I/W/Y mutants presented above support a recent assignment of F193 as a determinant of enzyme affinity toward glucosides with an aromatic aglycone that was based on structural analysis of inactive mutant enzyme-substrate complex (Czjzek et al., 2000). It was unexpected that our kinetic results clearly demonstrate that the dramatic drop in the F193I specificity constant is caused mainly by drop in kcat. Based on this novel finding, structure inspection allowed us to propose that the F193-aglycone-W373 interaction represents a major contribution to the positioning of the glucosidic bond in an orientation favorable for attack by E186 and E401. Therefore, the interaction is expected to codetermine catalytic rate of the enzyme. In contrast, glycone-enzyme interactions do not seem to contribute to the appropriate positioning of the glucosidic bond to large extent. Previous enzyme kinetic analysis indicated that the ground state of the glycone-binding pocket is complementary to Glc in the half chair conformation rather than the chair conformation. Glc was a much weaker competitive inhibitor compared with d-glucono-1,5-lactone, which has a half-chair conformation and inhibits the enzyme by acting as a transition state analog (Babcock and Esen, 1994; J. Zouhar, J. Vévodová, J. Marek, J. Damborský, X.-D. Su, and B. Brzobohatý, unpublished data). Thus, the narrow slot of the aglycone-binding site formed by F193, F200, F461, and W373 appears to be a stereochemical determinant of the interaction strength (Fig. 4). Although W373 is highly conserved among family 1 β-glucosidases, F193 is highly variable (Fig. 1), indicating that a residue at this position is likely to be involved in fine-tuning substrate specificity and reaction rate. This is consistent with the fact that the F193I mutation caused approximately 100-fold decrease in the specificity constant (kcat/Km) with PNPG (Table III).
The analysis of the topological consequences of F193I substitution in Zm-p60.1 crystal structure correlates well with the dramatic effect on kcat. When F193I substitution is modeled, the width of the slot increases from 7.3 Å in WT to 8.5 Å. An even more dramatic increase is found for mouth opening (Table IV), which determines the cross section of the slot at the respective position more precisely (Lee and Richards, 1971; Connolly, 1983). One can hypothesize intuitively that the increased cross section of the slot might allow higher freedom of movement of the aglycone moiety; that in turn would result in decreased average time for which the glucosidic bond is located in the orientation favorable for attack by the catalytic pair. As a consequence, the rate of glucosidic bond cleavage will decrease, and reaction rate represented by kcat will drop. Consistent with this hypothesis, in F193W, the decrease in width and mouth opening that can be calculated for the mutant correlate with a slight increase in reaction rate. Increased Km found in F193W might reflect steric constraints connected with substrate penetration into, and accommodation within, the active center due to narrowing of the slot. Although the topological parameters in F193Y reach values between WT and F193W (Table IV), kcat decreased to 62% of WT (Tables III). The kcat decrease might reflect the different chemical nature of the atoms forming contacts with the aglycone and the residue at position 193. Although in WT and F193W an H atom of the respective aromatic rings form the van der Waals interactions with the aglycone, the more polar H of a hydroxyl group is involved in the interaction in F193Y.
Table IV.
Mouth opening calculated for Zm-p60.1 mutants in F193 and M258
| Enzyme | Solvent-Accessible Surface Area | Molecular Surface Area |
|---|---|---|
| Å2 | ||
| WT | 14 | 94 |
| F193I | 23 | 113 |
| F193Y | 10 | 78 |
| F193W | 11 | 88 |
| M258I | 16 | 94 |
| M258V | 40 | 135 |
| M258F | n.a.a | n.a.a |
Substitutions were introduced in the structure using INSIGHTII v95 (Biosym/MSI Accelrys). Solvent-accessible (Lee and Richards, 1971) and molecular (Connolly, 1983) surfaces of the mouth openings were calculated using CASTP v1.1 (Liang et al., 1998) with a solvent probe diameter of 1.4 Å.
Opposite trends in change of active site size were obtained for two distinct rotamers in M258F; thus, unambiguous values are not available (n.a.) for the mutant.
The crucial role of a precise stereochemical architecture of the aglycone binding site in the region formed by F193, F200, F461, and W373 for enzyme activity becomes even more evident when modeled topological alterations and kinetic parameters are compared for mutations in a position close but clearly outside this region. As mentioned above, analysis of the homology-based model of the three-dimensional structure of Zm-p60.1 suggested that the width of the slot in the aglycone-binding site might be determined by F193 and M258 located opposite each other along the sides of the slot. Therefore, three mutations in position 258 were constructed in addition to mutations in position 193. In all the mutants analyzed, the most dramatic increase in mouth opening was calculated for M258V mutant (Table IV). However, kinetic analysis of the mutant revealed only a moderate decrease in kcat (Table III). On the other hand, only minor changes in mouth opening calculated for M258I are accompanied by a clearly decreased specificity constant. No unambiguous solution for mouth opening calculation can be obtained for M258F precluding any correlation with kinetic data obtained for M258F. The available data are consistent with the location of M258 at the edge of the aglycone-binding site.
A systematic assessment by site-directed mutagenesis, enzyme kinetic analysis, and computer modeling of the relative contributions of the individual residues identified in the active center to substrate specificity and catalytic rate remains a challenge for subsequent studies.
MATERIALS AND METHODS
Site-Directed Mutagenesis
The GeneEditor in vitro site-directed mutagenesis system (Promega, Madison, WI) was used to introduce the desired mutations into (His)6Zm-p60.r, a recombinant Zm-p60.1 derivative lacking the plastid targeting sequence [the N-terminal sequence of (His)6Zm-p60.r is Mr(H)6GMAS, the last residue of which corresponds to the Ser determined at the N terminus of Zm-p60.1 isolated from maize (Zea mays) coleoptiles; Brzohohatý et al., 1993], in pRSET::Zm-p60.r as described earlier (Rotrekl et al., 1999; Zouhar et al., 1999). The resulting mutants are designated pRSET::Zm-p60.rm. The mutagenic oligonucleotides were as follows: E186D, 5′-GTCTGGGGGTCATTAAAGG-3′; E186Q, 5′-TCTGGGGCTGATTAAAGGT-3′; E401D, 5′-GATTCCGTTGTCCGTGATG-3′; E401Q, 5′-TTCCGTTCTGCGTGATGTAG-3′; F193I, 5′-TCCGTAGGAAATGGAAGTAAATG-3′; F193Y, 5′-TCCGTAGGAATAGGAAGTAA-3′; F193W, 5′-TCCGTAGGACCAGGAAGTAAATG-3′; M258I, 5′-GCACACGACCTATTACGTCAAAC-3′; M258F, 5′-GCACACGACCAAATACGTCAAAC-3′; and M258V, 5′-GCACACGACCCACTACGTCAAAC-3′ (the changed nucleotides are underlined). Mutations were confirmed by DNA sequencing. The site-directed mutagenesis resulted in pRSET(AmpRM)::Zm-p60.rm, where AmpRM is modified/enhanced ampicillin resistance.
D/Q186E and D/Q401E revertants were generated in an analogous way using individual pRSET::Zm-p60.rm as templates. The codon GAA (Glu) was introduced into positions 186 and 401 by oligonucleotides (5′-AAATGTCTGGGGTT-CATTAAAGGTCAA-3′ and 5′-CCGATTCCGTTTTCCGT-GATGTAGATAG-3′, respectively). The presence of GAA enabled unequivocal revertant identification because E186 and E401 are encoded by GAG in (His)6Zm-p60.r.
Expression and Purification of (His)6Zm-p60.rm
The mutants were expressed in the Escherichia coli strain BL21(DE3) pLysS as described earlier (Kuderová et al., 1999; Zouhar et al., 1999). Single-step purification on MCAC columns (POROS MC/M peak column 4.6 × 100 mm, BioCAD workstation, PerSeptive, Framingham, MA) was facilitated by His tag engineered at the N termini of (His)6Zm-p60.r and the mutants (Zouhar et al., 1999). Elution was triggered by EDTA, the purified protein preparation was diafiltered against water, and used directly for kinetic and electrophoretic analysis. Purity of the mutants was higher than 95% as determined by SDS-PAGE followed by Coomassie Blue staining and densitometry.
Electrophoresis and Western Blotting
Native PAGE was performed in 10% (w/v) gels (Laemmli, 1970) followed by either activity “in-gel” staining (zymograms) or semidry western blotting. Zymograms were developed with 6-bromo-2-naphtyl β-d-glucopyranoside as a substrate and Fast Blue BB as a coupling dye (Esen and Cokmus, 1990). A fluorogenic substrate MUG was employed when increased zymogram sensitivity was desirable (Jefferson et al., 1987). Protein transfer on polyvinylidene difluoride membrane (Immobillon P, Millipore, Bedford, IN) was performed according to Towbin (1979). Positions of dimer and/or monomer forms of (His)6Zm-p60.rm were visualized by an alkaline phosphatase-mediated immunostaining procedure (Blake et al., 1984). Anti-Zm-p60 polyclonal antibodies were raised in rabbits against recombinant (His)6Zm-p60.r produced in E. coli. Anti-rabbit-IgG antibody, conjugated to alkaline phosphatase, was from Sigma (Deisenhofen, Germany).
Enzyme and Protein Assays
Enzyme activity was assayed using MUG and PNPG as the fluorogenic and chromogenic substrates, respectively (Babcock and Esen, 1994; Rotrekl et al., 1999). Protein concentration was determined according to Bradford (1976) using protein assay (Bio-Rad Laboratories, Hercules, CA) and bovine serum albumin as a standard.
X-Ray Crystallography
Diffraction data with single crystal of (His)6Zm-p60.r was collected as reported previously (Vévodová et al., 2001). In brief, the crystals of recombinant protein grew from 20% to 25% (w/v) PEG 4000, 0.1 m citrate buffer, pH 5.3 to 5.9, and 0.2 m ammonium acetate at room temperature and the data were collected at 100.0 K with cryoprotectant 20% (w/v) PEG 4000 and 5% (v/v) glycerol and wavelength λ = 0.9420 Å at the crystallographic beamline BL711 at the MAX-II synchrotron in Lund (Sweden). Data were processed by DENZO and SCALEPACK packages (Otwinowski and Minor, 1997). The Zm-p60.1 structure was solved by molecular replacement by AMoRe (Navaza, 1994) with the coordinates of the TRCB-Glu (Protein Data Bank entry 1CBG, Barrett et al., 1995) as a model. The model was refined first by rigid body refinement, by several steps of the crystallographic refinement including the simulated annealing, and finally by simulated annealing and restrained maximum-likelihood methods. All calculations were performed with Crystallography & NMR System (Brünger et al., 1998). Model inspection and rebuilding was carried out using program O (Jones et al., 1991). The final model contained 7,943 non-hydrogen protein atoms and 909 water molecules and converged at R and Rfree (Brünger, 1992) 16.9% and 23.0%. No ς cut-off was used during the refinement. The main-chain dihedral angles of all residues are within energetically allowed regions of the Ramachandran plot—85.6% of all residues lie in the most favored regions and the rest are in additional allowed regions. Data statistics are summarized in Table I.
Coordinates
The atomic coordinates of the refined model have been deposited at the Protein Data Bank (Rutgers, NJ) with reference code 1HXJ.
Molecular Modeling
The enzyme-substrate complexes of Zm-p60 with MUG and PNPG were prepared using the molecular modeling package InsightII (Biosym/MSI Accelrys; www.accelrys.com). The substrate molecules were built in InsightII and optimized using AM1 semi-empirical quantum mechanical calculations. Polar hydrogens were added to the protein structure using the WHATIF 5.0 program package (Vriend, 1990). Starting models of enzyme-substrate complexes were constructed manually according to experimental structures of enzyme-substrate and enzyme-inhibitor complexes (Czjzek et al., 2000) and refined by energy minimization using 100 steps of steepest descent and 500 steps of conjugate gradient with consistent valence force field of Discover95.0/3.0 (Biosym/MSI Accelrys).
ACKNOWLEDGMENTS
We wish to thank Dr. Jana Klánová (Department of Functional Genomics and Proteomics, Masaryk University, Brno, Czech Republic) for DNA sequencing and Dr. Hana Konečná (Department of Functional Genomics and Proteomics, Masaryk University) for oligonucleotide synthesis. Anti-(His)6Zm-p60.r antibodies were prepared in cooperation with the Veterinary Research Institute (Brno, Czech Republic). We wish to thank Dr. Ian Moore (Department of Plant Sciences, University of Oxford) and Kiran Nagavalli Subbana, MSc (Department of Functional Genomics and Proteomics, Masaryk University), for critically reading the manuscript.
Footnotes
This work was supported by the Ministry of Education of the Czech Republic (grant nos. VS96096 and MSM143100008), by the INCO-Copernicus Program (grant no. ERB3512–PL966135), by the National Science Foundation, U.S. (grant no. INT–9600462), by the Socrates Erasmus Free Movers and Swedish Institute (grants to J.V.), and by the Swedish Foundation for Strategic Research and Structural Biology Network (support to X.-D.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010712.
LITERATURE CITED
- Aguilar CF, Sanderson I, Moracci M, Ciaramella M, Nucci R, Rossi M, Pearl LH. Crystal structure of the beta-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J Mol Biol. 1997;271:789–802. doi: 10.1006/jmbi.1997.1215. [DOI] [PubMed] [Google Scholar]
- Babcock GD, Esen A. Substrate specificity of maize beta-glucosidase. Plant Sci. 1994;101:31–39. [Google Scholar]
- Barrett T, Suresh CG, Tolley SP, Dodson EJ, Hughes MA. The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1glycosyl-hydrolase. Structure. 1995;3:951–960. doi: 10.1016/s0969-2126(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EE, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blake MS, Johnston KH, Russel-Jones GJ, Gotschlich EC. A rapid, sensitive method for detection of alkaline-phosphatase conjugated anti-antibody on western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brünger AT. Free-R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Kristoffersen P, Bakó L, Campos N, Schell J, Palme K. Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Palme K. Cytokinin metabolism: implications for regulation of plant growth and development. Plant Mol Biol. 1994;26:1483–1497. doi: 10.1007/BF00016486. [DOI] [PubMed] [Google Scholar]
- Burmeister WP, Cottaz S, Driguez H, Iori R, Palmieri S, Henrissat B. The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidases. Structure. 1997;5:663–675. doi: 10.1016/s0969-2126(97)00221-9. [DOI] [PubMed] [Google Scholar]
- Campos N, Bakó L, Feldwisch J, Schell J, Palme K. A protein from maize labeled with azido-IAA has novel beta-glucosidase activity. Plant J. 1992;2:675–684. [Google Scholar]
- Chi Y-I, Martinez-Cruz LA, Jancarik J, Swanson RV, Robertson DE, Kim SH. Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Lett. 1999;445:375–383. doi: 10.1016/s0014-5793(99)00090-3. [DOI] [PubMed] [Google Scholar]
- Cicek M, Esen A. Expression of soluble and catalytically active plant (monocot) beta-glucosidases in E. coli. Biotechnol Bioeng. 1999;63:392–400. doi: 10.1002/(sici)1097-0290(19990520)63:4<392::aid-bit2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Connolly ML. Analytical molecular surface calculation. J Appl Cryst. 1983;16:548–558. [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Bevan RD, Henrissat B, Esen A. The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes. Proc Natl Acad Sci USA. 2000;97:13555–13560. doi: 10.1073/pnas.97.25.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czjzek M, Cicek M, Zamboni V, Burmeister WP, Bevan RD, Henrissat B, Esen A. Crystal structure of a monocotyledon (maize ZMGlu1) beta-glucosidase and a model of its complex with p-nitrophenyl beta-D-thioglucoside. Biochem J. 2001;354:37–46. doi: 10.1042/0264-6021:3540037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A, Cokmus C. Maize genotypes classified as null at the Glu locus have beta-glucosidase activity and immunoreactive protein. Biochem Genet. 1990;28:319–336. doi: 10.1007/BF02401422. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. Further additions to MolScript version 1.4 including reading and contouring of electron-density maps. Acta Cryst D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon J-P, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavangh TA, Bevan MW. GUS fusions-beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J, Leggio LL, Harris G, Pickersgill R. Beta-glucosidase, beta-galactosidase, family A cellulases, family F xylanases and 2 barley glycanases form a superfamily of enzymes with 8-carboxy-terminal ends of beta-strand-4 and beta-strand-7. FEBS Lett. 1995;362:281–285. doi: 10.1016/0014-5793(95)00252-5. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Cryst A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Cryst D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Kristoffersen P, Brzobohatý B, Höhfeld I, Bako L, Melkonian M, Palme K. Developmental regulation of the maize Zm-p60.1 gene encoding a beta-glucosidase located to plastids. Planta. 2000;210:407–415. doi: 10.1007/pl00008149. [DOI] [PubMed] [Google Scholar]
- Kuderová A, Nanak E, Truksa M, Brzobohatý B. Use of rifampicin in T7 RNA polymerase-driven expression of a plant enzyme: rifampicin improves yield and assembly. Protein Exp Purif. 1999;16:405–409. doi: 10.1006/prep.1999.1079. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leah R, Kigel J, Svendsen I, Mundy J. Biochemical and molecular characterization of a barley seed beta-glucosicase. J Biol Chem. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moracci M, Capalbo L, Ciaramella M, Rossi M. Identification of two glutamic acid residues essential for catalysis in the beta-glycosidase from the thermoacidophilic archaeon Sulfolobus solfataricus. Protein Eng. 1996;9:1191–1195. doi: 10.1093/protein/9.12.1191. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe-an automated package for molecular replacement. Acta Cryst A. 1994;50:157–163. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Poulton JE. Cyanogenesis in plants. Plant Physiol. 1990;94:401–405. doi: 10.1104/pp.94.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotrekl V, Nejedlá E, Kučera I, Abdallah F, Palme K, Brzobohatý B. Expression, single-step purification, and matrix-assisted refolding of a maize cytokinin glucoside-specific beta-glucosidase. Eur J Biochem. 1999;266:1056–1065. [Google Scholar]
- Sanz-Aparicio J, Hermoso JA, Martinez-Ripoll M, Lequerica JL, Polaina J. Crystal structure of beta-glucosidase A from Bacillus polymyxa: insight into thecatalytic activity in family 1 glycosyl hydrolases. J Mol Biol. 1998;275:491–502. doi: 10.1006/jmbi.1997.1467. [DOI] [PubMed] [Google Scholar]
- Sinnott M. Catalytic mechanism of enzymatic glycosyl transfer. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- Smith AR, van Staden J. Changes in endogenous cytokinin levels in kernels of Zea mays L. during imbibition and germination. J Exp Bot. 1978;29:1067–1073. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vévodová J, Marek J, Zouhar J, Brzobohatý B, Su X-D. Purification, crystallization and preliminary X-ray analysis of a maize cytokinin glucoside specific beta-glucosidase. Acta Cryst D. 2001;57:140–142. doi: 10.1107/s0907444900014001. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wakarchuk WW, Campbell RL, Sung WL, Davoodi J, Yaguchi M. Mutational and crystallographic analyses of the active-site residues of the Bacillus circulans xylanase. Protein Sci. 1994;3:467–475. doi: 10.1002/pro.5560030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Trimbur D, Graham R, Warren RAJ, Withers SG. Identification of the acid/base catalyst in Agrobacterium faecalis beta-glucosidase by kinetic-analysis of mutants. Biochemistry. 1995;34:14554–14562. doi: 10.1021/bi00044a034. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Beste G, Hengstenberg W, Schulz GE. The 3-dimensional structure of 6-phospho-beta-galactosidase from Lactococcus lactis. Structure. 1995;3:961–968. doi: 10.1016/s0969-2126(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Withers SG, Rupitz K, Trimbur D, Warren RAJ. Mechanistic consequences of mutation of the active-site nucleophile Glu-358 in Agrobacterium beta-glucosidase. Biochemistry. 1992;31:9979–9985. doi: 10.1021/bi00156a017. [DOI] [PubMed] [Google Scholar]
- Withers SG, Warren RAJ, Street IP, Rupitz K, Kempton JB, Aebersold R. Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. J Am Chem Soc. 1990;112:5887–5889. [Google Scholar]
- Yuan J, Martinez-Bilbao M, Huber RE. Substitutions for Glu-537 of beta-galactosidase from Escherichia coli cause large decreases in catalytic activity. Biochem J. 1994;299:527–531. doi: 10.1042/bj2990527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Nanak E, Brzobohatý B. Expression, single-step purification, and matrix-assisted refolding of a maize cytokinin glucoside-specific beta-glucosidase. Protein Exp Purif. 1999;17:153–162. doi: 10.1006/prep.1999.1108. [DOI] [PubMed] [Google Scholar]