Abstract
The expression of light-regulated genes in plants is controlled by different classes of photoreceptors that act through a variety of signaling molecules. During photomorphogenesis, the early light-induced protein (Elip) genes are among the first to be induced. To understand the light signal transduction pathways that regulate Elip expression, the two Elip genes, Elip1 and Elip2, in Arabidopsis were studied, taking advantage of the genetic tools available for studying light signaling in Arabidopsis. Using two independent quantitative reverse transcriptase-PCR techniques, we found that red, far-red, and blue lights positively regulate expression of the Elip genes. Phytochrome A and phytochrome B are involved in this signaling. The cryptochrome or phototropin photoreceptors are not required for blue-light induction of either Elip gene, suggesting the involvement of an additional, unidentified, blue-light receptor. Although the COP9 signalosome, a downstream regulator, is involved in dark repression of both Elips, Elip1 and Elip2 show different expression patterns in the dark. The transcription factor HY5 promotes the light induction of Elip1, but not Elip2. A defect in photosystem II activity in greening of hy5 seedlings may result from the loss of Elip1. Heat shock positively controlled Elip1 and Elip2 in a light-independent fashion. This induction is independent of HY5, indicating that heat shock and light activate transcription of the Elip genes through independent pathways.
Light has three main effects on plant development (for review, see Mustilli and Bowler, 1997; Batschauer, 1998). First, it is the source of energy that fuels growth through photosynthesis. Second, light is a developmental signal that modulates morphogenesis, such as de-etiolation and the transition to reproductive development. Third, light is also deleterious for plants because excess light, absorbed by the photosynthetic apparatus, promotes the formation of dangerous compounds such as active oxygen species. Because plants must quickly respond to changing and often extreme light conditions, sophisticated photosensory networks have evolved that enable plants to maximize photosynthesis while minimizing damage. One of the main mechanisms of this overall control is accomplished through regulation of gene expression.
Light is perceived in plants by a sophisticated system of photoreceptors that detect different light wavelengths. Five phytochromes mediate red and far-red light responses in Arabidopsis. Among these, phytochrome A (PhyA) is primarily responsible for the perception of constant far-red light, whereas PhyB is primarily responsible for the perception of constant red light. Three photoreceptors for blue light have been identified in Arabidopsis (for review, see Lin, 2000). Crytpochrome 1 (Cry1) is the principal blue/UV-A light receptor, modulating growth at medium and high-light intensities. Cry2 has major functions in responding to low intensities of blue light (Lin et al., 1996). Phototropin, encoded by the non-phototropic hypocotyl 1 (NPH1) gene, is the photoreceptor for phototropism (Liscum and Briggs, 1995). Various reports have discussed the possibility of other blue-light receptors, though their identities were enigmatic (Zeiger and Zhu, 1998; Briggs and Huala, 1999; Frechilla et al., 1999). NPL1 (NPH-like 1), a fourth blue-light receptor, recently was identified that is partly functionally redundant with NPH1, and has a major role in the chloroplast high-blue-light avoidance response (Jarillo et al., 2001; Kagawa et al., 2001; Sakai et al., 2001).
Downstream from the photoreceptors are a plethora of positive and negative regulators of light signaling (for review, see Nagy et al., 2000; Neff et al., 2000). Among these, the COP9 signalosome (CSN) is a multisubunit regulatory complex that functions through unknown mechanisms as a master repressor of photomorphogenesis in the dark (for review, see Karniol and Chamovitz, 2000). One of the targets of the CSN-mediated repression is HY5. HY5 is a basic Leu zipper transcription factor directly involved in the expression of light-inducible genes (Oyama et al., 1997; Chattopadhyay et al., 1998). No role for these receptors or signaling molecules has been reported for responses to light stress.
The effect of light on plant development is particularly evident in seedling development and the transition from growth under soil (dark) to growth above the ground (light). As photomorphogenesis is initiated, cellular and subcellular processes are initiated to allow the development of photosynthetic capable tissues. This includes chloroplast development, pigment synthesis, and assembly of the photosystems in the thylakoids. All of these processes are accomplished by and depend on the differential expression of a large number of genes. However, before a chloroplast is competent for performing photochemistry, it is saturated with photons that have no outlet, and thus form toxic compounds that can kill the developing cell. In response to this light-induced stress, plants produce photoprotective pigments such as carotenoids and xanthophylls, and protective proteins.
An example of protective proteins are the early light-induced proteins (ELIPs), nuclear-encoded thylakoid membrane proteins that are transiently expressed immediately after light stress. Elip transcript and protein appear considerably faster than those of other light-induced genes during the early stage of de-etiolation, and disappear before chloroplast development is completed (Grimm and Kloppstech, 1987). In mature plants, ELIP accumulation under light stress conditions correlates with the photoinactivation of photosystem II (PSII), degradation of the D1 protein, and changes in the level of pigments (Adamska et al., 1992a, 1993). ELIPs bind chlorophyll a and lutein and have been proposed to function as transient pigment carriers or chlorophyll exchange proteins (Adamska et al., 1999).
The regulation of ELIP expression is modulated by light and other stress signals. Blue and red light induce Elip transcription in etiolated plumulas of pea (Pisum sativum) seedlings (Adamska, 1995), whereas blue and UV-A light induce ELIP in adult tissues (Adamska et al., 1992a, 1992b). ELIP homologs from various systems have been implicated in various stress responses. For example, one of the responses to extreme dehydration of the “resurrection” plant Craterostigma plantagineum is the expression of the ELIP homolog dsp-22 (Bartels et al., 1992).
In pea (Scharnhorst et al., 1985; Kolanus et al., 1987) and tobacco (Nicotiana tabacum; Blecken et al., 1994), ELIP is encoded by a single gene, whereas two ELIPs was reported to exist in barley (Hordeum vulgare; Grimm and Kloppstech, 1987) and in Arabidopsis (Moscovici-Kadouri and Chamovitz, 1997; Heddad and Adamska, 2000). The functions of the two genes are unclear, as is the genetic regulation of Elip transcription.
To understand genetic mechanisms regulating light and stress control of ELIP induction, we have initiated a study of ELIP in Arabidopsis. Previous studies on Elip in other plants were limited to characterizing the light quality and intensity that regulate Elip expression, and often in dismembered leaves. Arabidopsis provides a convenient system for studying Elip transcription due to the large collection of characterized light-signaling mutants available. We show here that multiple photoreceptors, including a cryptic blue-light receptor, regulate Elip transcription, and that the two ELIP genes have differing regulation patterns, which hint at different functions.
RESULTS
Development of Semiquantitative Reverse Transcriptase (RT)-PCR Experiment Conditions
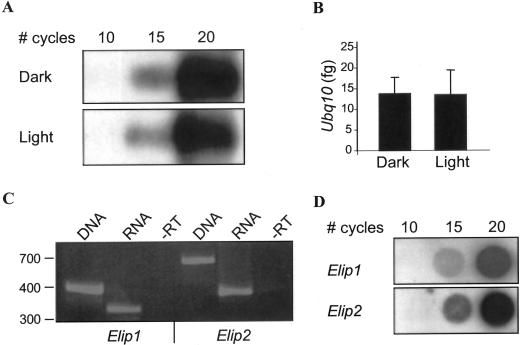
Previous studies showed that light regulation of Elip steady-state transcript levels is manifested at the level of transcription, allowing for a correlation between steady-state transcript levels and transcriptional control (Adamska, 1995). To study the regulation of Elips in multiple genetic backgrounds under different conditions, we developed a sensitive and fast RT-PCR method for analyzing Elip transcript levels. We used two PCR techniques: conventional RT-PCR followed by hybridization, and the LightCycler system. Ubiquitin10 (Ubq10) was used as an internal RT-PCR control because it was previously shown by RNA gel-blot analysis to be constitutively expressed in light and dark conditions (Sun and Callis, 1997). To confirm this result by RT-PCR, and to determine if the steady-state levels of Ubq10 are affected by the developmental stage of the plant, we checked Ubq10 levels in dark- and light-grown seedlings by both RT-PCR techniques. As seen in Figure 1, A and B, the steady-state levels of Ubq10 in dark-grown seedlings kept in the dark or exposed to 1 h light are equal as determined by conventional RT-PCR and LightCycler RT-PCR. This endogenous mRNA standard has the advantage of serving as a control for RNA recovery and integrity, as well as for sample-to-sample variations in RT and PCR.
Figure 1.
Development of RT-PCR experimental conditions. A and B, Ubq10 is a constitutive control. Four-day-old Arabidopsis dark-grown wild-type seedlings were kept in the dark (Dark) or exposed to 1 h light (Light), RNA was extracted, and amounts of Ubq10 were determined by conventional RT-PCR followed by Southern blot (A) or by the LightCycler (B). The amount of Ubq10 PCR products as a function of polymerization cycles is shown in A. The calculated initial amount of Ubq10 transcript is shown in B. C and D, RT-PCR controls for Elip1 and Elip2. C, Elip1 and Elip2 primer pairs were used for PCR on genomic DNA and cDNA (from “Light” above) templates, and in the absence of RT (−RT). D, “Light” sample from above was used as template to determine the kinetics of PC reaction as a function of number of PCR cycles, as detected by dot blot.
To avoid artifacts due to genomic DNA contamination in an RNA preparation, PCR primer pairs of Elip1 and Elip2 were designed around introns (see “Materials and Methods”). In addition, to avoid artifacts due to the high identity between the two Elip transcripts, the forward primers of Elip1 and Elip2 anneal to the divergent 5′ end of the genes. As shown in Figure 1C, PCR on genomic DNA and cDNA yield product sizes for Elip1 of 414 and 330 bp, respectively, and for Elip2, 705 and 400 bp, respectively. In addition, RNA was treated with DNase (during total RNA preparation) prior to cDNA synthesis and the control reaction was performed in which RT is omitted (−RT; Fig. 1C). Conventional PCR was carried out for 15 cycles where the kinetics of the PCR reactions allowed quantitative analysis for all three genes, Elip1, Elip2, and Ubq10 (Fig. 1, A and D).
Expression of Elip Is Mediated by Red, Far-Red, and Blue Light and by Heat Shock
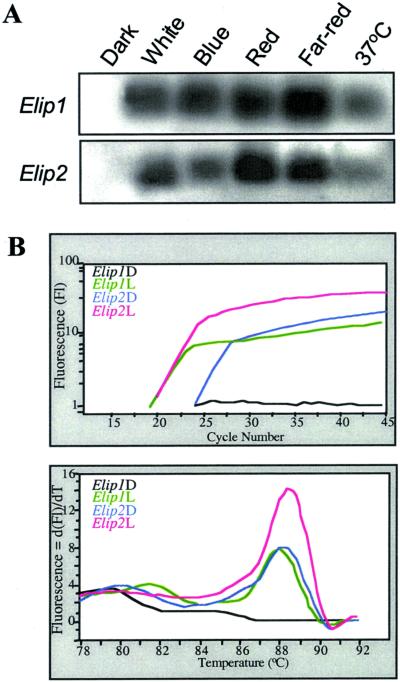
To examine the regulation of Elip expression during photomorphgenesis in wild-type Arabidopsis seedlings, the mRNA levels of Elip1 and Elip2 in seedlings grown under different light qualities were determined by RT-PCR. As seen in Figure 2A, both Elips are apparently not expressed, or expressed at levels below detection in 4-d-old dark-grown seedlings. Exposure of these seedlings to 1 h red, far-red, and blue light resulted in a detectable increase in transcript levels of both Elips. Elip expression is also induced in dark-grown seedlings following heat shock at 37°C for 1 h. This indicates that Elip expression is induced by high irradiance response (HIR) light in etiolated seedlings exposed to 1 h light during the etioplast/chloroplast conversion, and raises the question of which photoreceptors are involved in the positive regulation of Elip steady-state mRNA levels.
Figure 2.
Effect of light on expression of Elips. A, RT-PCR analysis of 4-d-old Arabidopsis dark-grown wild-type seedlings that were kept in the dark or exposed to 1 h of white, blue, red, or far-red lights, or to heat shock in the dark at 37°C. One-fifth of the PCR products were resolved in 1.7% (w/v) agarose gel and blotted onto Hybond N+. The membrane was hybridized with DIG labeled Elip1 and Elip2 probes. B, RT-PCR LightCycler analysis reveals the differences between Elip1 and Elip2 transcript levels. Four-day-old Arabidopsis dark-grown wild-type seedlings were kept in the dark (D) or exposed to 1 h of white light (L) before total RNA extraction. One-tenth of the cDNA was amplified by the LightCycler. Top, Amplification curves of Elip1 and Elip2. The diagram documents the fluorescence intensity (approximate PCR product concentration) plotted against the number of PCR cycles. Bottom, Melting curve analysis of PCR products. The diagram documents the negative derivative fluorescence plotted against temperature, where the peak therefore highlights the melting of DNA.
Elip1 and Elip2 Have Different Dark Expression Patterns
The Elip2 cDNA was isolated from a cDNA library made from etiolated seedlings (see “Materials and Methods”). This led us to question the significance of the lack of Elip expression found in dark-grown seedlings (Fig. 2A; Heddad and Adamska, 2000). To further study Elip1 and Elip2 regulation, the experiments were repeated using the highly sensitive LightCycler method. LightCycler RT-PCR was performed with cDNA samples from 4-d-old dark-grown seedlings treated with 1 h of white light or kept in darkness. The amplification curves of this experiment are shown in Figure 2B (top). The mRNA levels of Elip1 and Elip2 in light-treated seedlings are similar, with fluorescence levels rising after cycle 16. However, the mRNA levels of Elip1 and Elip2 in dark-grown seedlings are different. The fluorescence signal of the Elip2 transcript starts to rise after 20 cycles, whereas no Elip1 product is detected even after 45 cycles. The melting curves analysis (Fig. 2B, bottom) of this experiment confirms that there is no Elip1 PCR product in the dark-grown seedlings, as opposed to Elip2 product at the same condition, or to Elip1 and Elip2 in the light-exposed seedlings. These data indicate that Elip1 and Elip2 are differentially regulated in darkness. The result further shows the advantage of using the LightCycler system in that it is highly sensitive and allows the detection of very small amounts of transcript, as compared with the conventional PCR experiments or RNA gel-blot experiments.
Phytochromes Regulate Elip Expression via Red and Far-Red Light
To examine the role of different phytochromes in the regulation of Elip expression, we used phyA, phyB, and phyA/phyB double mutants. Four-day-old dark-grown wild-type and mutant seedlings were kept in the dark or exposed to 1 h of red or far-red light. Total RNA was isolated and conventional PCR experiments were performed. Red and far-red lights have similar positive effects on Elips in wild-type seedlings (Table I). This effect of far-red light on Elip mRNA was lost in the phyA mutant, indicating that PhyA positively regulates Elips. The phyA mutant also showed reduced red-light induction of both Elips. It is surprising that in phyB, the red and far-red induction of Elips was not impaired. However, the absence of both phytochromes in the phyA/phyB mutant resulted in a loss of red and far-red induction of both Elips. This suggests an essential requirement of PhyA for red-light induction of Elips. Analysis with the LightCycler identified very low levels of Elip transcripts in the phyA/phyB mutant (not shown).
Table I.
Involvement of photoreceptors in the regulation of Elip1 and Elip2
| Strain | Red
|
Far-Red
|
Blue
|
||
|---|---|---|---|---|---|
| Elip1 | Elip2 | Elip1 | Elip2 | Elip1 | |
| w.t. | ++ | ++ | ++ | ++ | ++ |
| phyA | + | + | – | – | nd |
| phyB | ++ | ++ | ++ | ++ | nd |
| phyA/phyB | – | – | – | – | ++ |
| cry1 | nd | nd | nd | nd | ++ |
| cry2 | nd | nd | nd | nd | ++ |
| cry1/cry2 | nd | nd | nd | nd | ++ |
| nph1 | nd | nd | nd | nd | ++ |
RNA was extracted from 4-d-old dark-grown seedlings that were either kept in the dark or exposed to 1 h light as indicated. RT-PCR was according to conventional procedures and quantitation was based on conventional RT-PCR analysis. The analysis for Elip2 under blue light is shown in Figure 3. ++, RT-PCR product detected; +, some product detected (<50% of wild type); –, no RT-PCR product detected; nd, not done; w.t., wild type.
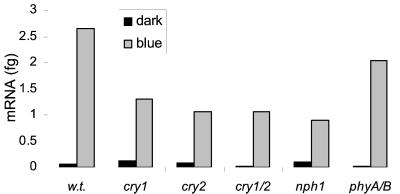
Cryptochrome1, Cryptochrome2, and Phototropin Are Not Vital for the Blue-Light Induction of Elip
To identify the blue-light photoreceptor(s) involved in Elip expression, we used mutants defective in photoreceptors for blue light. Four-day-old Arabidopsis dark-grown wild type, cry1, cry2, cry1/cry2 double mutant, nph1, and phyA/phyB double mutant seedlings were kept in the dark or exposed to 1 h of blue light before total RNA extraction. Blue light clearly results in an increase in Elip1 levels in wild type and in the mutants (Table I), indicating that none of these photoreceptors or pairs of photoreceptors have essential roles in the transcriptional regulation of Elip1.
To examine the expression level of Elip2 in these mutants, the experiments were continued using the LightCycler because conventional RT-PCR experiments yielded contradictory results (not shown). Figure 3 shows normalized initial amount of Elip2 mRNA. Like Elip1, Elip2 was induced by blue light in all mutants. However, Elip2 transcript levels were reduced in the cry1, cry2, cry1/cry2, and nph1 mutants relative to wild type, which may be indicative of a redundant function for these photoreceptors.
Figure 3.
RT-PCR analysis of Elip2 expression in blue-light receptor mutants. Four-day-old Arabidopsis dark-grown wild-type (w.t.), cry1, cry2, cry1/cry2, nph1, and phyA/phyB seedlings strains were kept in the dark or exposed to 1 h of blue light before total RNA extraction. One-tenth of the RT-reaction was amplified by the LightCycler using Elip2 and Ubq10 primers. The normalized initial amount of Elip2 mRNA is shown.
Downstream Regulators of Elip Expression
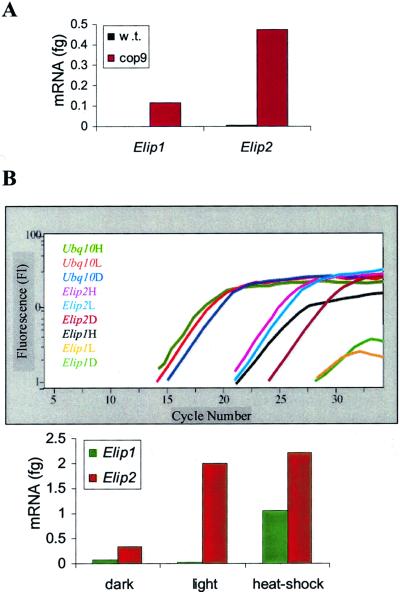
To determine the role of CSN in Elip regulation, RT-PCR was performed on dark-grown 4-d-old wild-type and CSN mutant cop9 seedlings. The normalized results in Figure 4A show that although in dark-grown wild-type seedlings Elip1 is not expressed and Elip2 is found in very low levels, transcripts of both genes accumulate at high levels in dark-grown cop9 seedlings. This indicates that the CSN fuctions in the repression of Elip expression in darkness.
Figure 4.
Analysis of Elip in downstream light-signaling mutants. A, RT-PCR analysis of Elip expression in cop9. RNA was isolated from 4-d-old dark-grown Arabidopsis wild-type (w.t.) and cop9 seedlings. One-tenth of the cDNA was amplified by the LightCycler using Elip1, Elip2, and ubq10 primers. The normalized initial amounts of Elip1 and Elip2 are shown. B, RT-PCR analysis of Elip expression in hy5. Four-day-old hy5 dark-grown seedlings were kept in the dark (D) or exposed to 1 h of white light (L) or to heat shock (H) before total RNA extraction. One-tenth of the cDNA was amplified by the LightCycler. Amplification curves of Elip1, Elip2, and Ubq10 are shown in the graph, with normalized initial amounts of Elip1 and Elip2 mRNA presented in the bar graph. Error bars represent sd based on three replicates of the same sample.
To identify a potential positive regulator of Elip expression, we next studied the hy5 mutant. HY5 is a light-regulated transcription factor for light-inducible genes (Oyama et al., 1997; Chattopadhyay et al., 1998). RNA was isolated from 4-d-old dark-grown hy5 mutant seedlings that were kept in the dark or exposed to 1 h of white light. As in the wild type (Fig. 2), Elip2 levels increase in the light in hy5 (Fig. 4B). In contrast, Elip1 levels do not increase in light in hy5. This suggests that HY5 is involved in the transcription of Elip1 gene directly, or promotes transcription of genes that are upstream of Elip1. To determine if Elip1 expression is totally silenced in hy5, or only in a light-dependant manner, we examined the effect of heat shock on Elips in this mutant. As shown in Figure 4B, both Elip1 and Elip2 are induced in a light-independent fashion by heat shock in hy5, indicating that HY5 acts downstream in a light signal transduction pathway that positively regulates Elip1, but that heat shock acts through independent signaling pathways.
Microarray Analysis of Elip1 Expression
Elip expression studies have concentrated primarily on studying Elip responses to singular phenomena, and until the present study, only by northern analysis. However, Elip1 was serendipitously included in the microarray distributed through the Arabidopsis Functional Genomics Consortium. Data from the publicly available experiments can help to clarify the factors involved in Elip regulation and its functions in Arabidopsis. Table II presents a summary of the relevant experiments having significant results for Elip1. The anatomical experiments show that Elip1 levels are higher in leaves relative to flowers, consistent with the chloroplast localization of ELIP (Kruse and Kloppstech, 1992). In addition to light, two other environmental stress situations, elevation of CO2 and aluminum, positively effect Elip expression.
Table II.
Summary of micro-array experiments with relevant Elip1 results
| Exp. No. | Description | Experimental Conditions
|
R/G Normalization | |
|---|---|---|---|---|
| Red channel (R) | Green channel (G) | |||
| Anatomical comparison | ||||
| 11333/11375 | Elip1 is transcriptionally regulated | Untreated | Cordycepin treated | 2.24 |
| 2370/2371 | Elip1 levels are higher in leaves than in flowers | Leaves | Flowers | 3.91 |
| 7197/7199 | Elip1 levels are higher in leaves relative to whole plant | Leaves | Whole-plant tissue | 1.26 |
| 7200/7201 | Elip1 levels are higher in flowers relative to whole plant | Whole-plant tissue | Flowers | 2.05 |
| Abiotic stress | ||||
| 4649/4650 | CO2 stress positively effects Elip1 | High CO2 | Control | 1.53 |
| 3749/3743 | Indole-3-acetic acid down-regulates Elip1 | Control | Indole-3-acetic acid treated | 3.37 |
| 7304/7305 | Aluminum stress positively effects Elip1 | Aluminum | Control | 1.67 |
| Light treatment | ||||
| 3610 | Elip1 levels are lower in etiolated seedlings than in adult tissue | Etiolated seedlings | Leaves | 0.857 |
| 6617 | Blue light induces Elip1 in etiolated seedlings | Etiolated plus blue light | Etiolated | 3.925 |
| 6619 | nph4-2 does not effect blue-light induction of Elip1 in etiolated seedlings | Wild-type etiolated seedlings exposed to blue light | nph4-2 etiolated seedlings exposed to blue light | 1.03 |
| 8266/7230 | Far-red-enriched light induces Elip1 in adult plants | Far-red-enriched light | White light | 3.07 |
Data were adapted from the Stanford Microarray Database (http://genome-www4.stanford.edu/Micro/Array/SMD). Only experiments giving reciprocal R/G results (when available as designated by two ID nos.) were chosen. R/G = 1, Elip1 levels in the two mRNA population are equal (i.e. no induction). R/G < 1, Elip1 is higher in the mRNA population that is marked by the green probes (G). R/G > 1, Elip1 is higher in the mRNA population that is marked by the red probes (R). Exp. No., Experiment identification no. according to Arabidopsis Functional Genomics Consortium.
The phototropic stimulation experiments are compatible with our results provided above, and display a significant increase in Elip levels in the dark-grown wild type after exposure to 1 h blue-light illumination. In addition, the 1.03 R/G normalization ratio present in experiment number 6,619 confirms our result in Figure 2 that the Phototropin pathway has no effect on blue-light induction of Elip1 transcription. We demonstrate this by using the phototropin photoreceptor mutant, nph1, whereas in the microarray experiment it was shown by analysis of nph4. NPH4 functions downstream of phototropin (NPH1; Harper et al., 2000).
Experiment 7,230 shows that far-red light induces Elip1 in adult Arabidopsis plants. This information complements our result that far-red induces Elip1 via PhyA in de-etiolation, but does not support earlier findings in green pea in which no Elip transcript or protein could be detected under light of 480 to 780 nm (Adamska et al., 1992a).
The Development of PSII in hy5 Is Retarded
Because Elip1 could not be detected during greening of etiolated hy5 seedlings, further characterization of the hy5 phenotype may reveal potential functions of ELIP1. ELIP was shown to play a role during the greening process (for review, see Adamska, 1997); therefore, the functional state of PSII as measured by chlorophyll fluorescence induction was studied during greening of hy5 as compared with wild-type seedlings. Figure 5 shows the photosynthetic efficiency of 4-d-old wild-type and hy5 dark-grown seedlings that were exposed to light for increasing periods and under different light intensities. At both 25 and 100 μmol m−2 s−1 white light, the efficiency of the photosynthetic apparatus is lower in hy5 than in the wild type. However, the PSII efficiency values of 14-d-old light-grown wild-type and hy5 seedlings were essentially identical, indicating that hy5 does reach the same efficiency level as that of the wild type. These results suggest that the development of PSII activity in the mutant is temporally retarded relative to that of the wild type. The fact that this phenotype is expressed only for a limited time in the plastid development could be due to the expression of other ELIP family genes that are not affected by the mutation in hy5. Therefore, these results suggest that hy5 mutant seedlings display a slower formation of PSII activity.
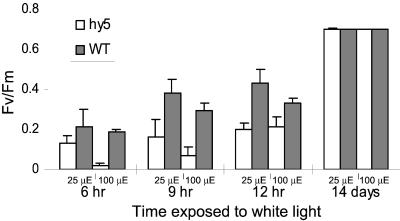
Figure 5.
Development of variable chlorophyll fluorescence during greening of hy5 and wild type. Four-day-old wild-type (WT) and hy5 dark-grown seedlings were exposed to 25 μmol m−2 s−1 or 100 μmol m−2 s−1 white light for different periods of time. A pulse amplitude-modulated fluorimeter (PAM) was used to calculate the maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) parameter. Error bars represent sd based on six different measurements.
DISCUSSION
The work presented here addresses the question how the two Elip genes in Arabidopsis are regulated at the genetic level during photomorphogenesis. For this purpose, we have investigated the transcript levels of these genes under different physiological conditions and genetic backgrounds by two quantitative RT-PCR techniques. It has been shown previously that steady-state levels of Elip mRNA correlate to changes in transcriptional activity (Adamska, 1995). This point is further emphasized in the microarray experiment where the transcriptional inhibitor cordycepin inhibited induction of Elip1 (Table II).
Our results that both Elips can be induced in dark-grown Arabidopsis seedlings by illumination with high irradiant red, far-red, and blue lights are consistent with earlier results showing Elip induction in etiolated pea seedlings (Adamska et al., 1992b). The low irradiance response was not addressed here. The microarray experiments present in Table II also confirm our results for the effects of blue light, far-red light, and the phototropin pathway on Elip1 expression. These far-red light results differ from those obtained in adult pea plants where only blue light was found to result in ELIP induction in adult plants (Adamska et al., 1992a).
The use of photoreceptor mutants provides direct evidence for the involvement of both PhyA and PhyB in Elip regulation. PhyA acts to induce Elip expression by HIR far-red light, but also has a role in perceiving HIR red light. In contrast, absence of PhyB by itself does not effect red and far-red induction of Elips. In this case, PhyA and maybe the other phytochromes (PhyC–E) compensate for the lack of PhyB. However, PhyB appears to work synergistically with PhyA in controlling the red-HIR induction of Elips as shown by the complete loss of red-light induction of both Elips in the phyA/phyB double mutant. Other phenomena are also known to be under HIR control of both PhyA and PhyB in Arabidopsis, including the control of hypocotyl elongation (Quail et al., 1995; Smith 1995) and expression of chlorophyll a/b binding protein (CAB) genes (Reed et al., 1994).
The identity of the photoreceptor involved in blue-light induction of Elips remains cryptic. The blue-light-induced up-regulation of Elips was not silenced in cry1/cry2 double mutants, or in the nph1 mutants. Microarray experiment 6,619 also indicates that Elip1 expression is not effected by at least one of the pathways regulated by the phototropin receptor. While the present manuscript was in review, a fourth blue-light receptor, NPL1, was identified (Jarillo et al., 2001; Kagawa et al., 2001). To determine if NPL1 has an essential role in mediating the blue-light induction of Elips, we examined the npl1 mutant by RT-PCR. As seen in Figure 6, both Elip1 and Elip2 are induced by blue-light in npl1. These results lead to two hypotheses: (a) The known blue-light photoreceptors have redundant functions in regulation of Elips by blue light. To further test this hypothesis, Elip induction in an Arabidopsis quadruple mutant cry1/cry2/nph1/npl1 would need to be analyzed. (b) A novel blue-light photoreceptor is involved in the regulation of Elips. This photoreceptor would work either independently or in co-action with the four known blue-light photoreceptors. This last possibility needs further consideration because there is accumulating evidence for such a receptor. For example, the stomatal responses of light-grown cry1, cry2, cry1/cry2, nph1, and nph1/cry1 plants did not differ from those of wild type (Lasceve et al., 1999; Eckert and Kaldenhoff, 2000). The potential role of a carotenoid derivative as a blue-light chromophore has been controversial (Palmer et al., 1996; Frechilla et al., 1999; Lasceve et al., 1999; Tlalka et al., 2001; Eckert and Kaldenhoff, 2000; Jin et al., 2001). We attempted to dissect the role of carotenoids or xanthophylls in Elip regulation through the use of the carotnoid inhibiting herbicide noflurazon (Chamovitz et al., 1991). It is unfortunate that this norflurazon treatment itself induces Elips (not shown).
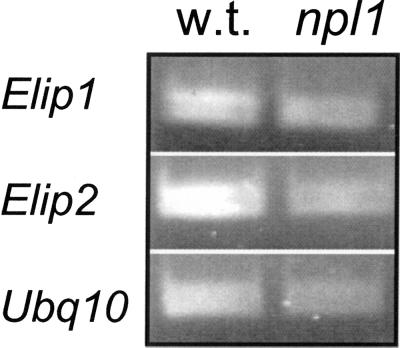
Figure 6.
NPL1 is not necessary for blue-light induction of Elip s. Four-day-old Arabidopsis dark-grown wild-type (w.t.) and cav1-1 (npl-1) seedlings strains exposed to 1 h of blue light before total RNA extraction. One-fifth of the PCR products with Elip1, Elip2, and Ubq10 primers were resolved in 1.7% (w/v) agarose gel.
Both Elip1 and Elip2 were induced by blue light in the phyA/phyB double mutant. This result appears to exclude the possibility that the blue-light induction of Elips is mediated by phytochrome. Other blue-light-regulated processes have also been shown to be independent of phytochrome in various systems. For example, a pea phyA/phyB double mutant was also recently reported to maintain normal blue-light responses, and CRY1 was shown to act independently of both PhyA and PhyB in tomato (Lycopersicon esculentum; Weller et al., 2001a, 2001b). However, the slight reduction in Elip2 levels in phyA/phyB could indicate a possible phytochrome involvement in blue-light regulation of Elips, similar to that reported for the dependence of Cry1 on phytochrome for regulating hypocotyl growth and anthocyanin accumulation (Ahmad and Cashmore, 1997).
In addition to light signals, Elips are also controlled by heat shock. It was shown previously that the accumulation of Elip transcript in etiolated barley and pea seedlings was induced by cyclic heat shock applied for several days (Beator et al., 1992; Otto et al., 1992). However, in other experiments performed with heat-treated etiolated pea, heat shock could not induce Elip without involvement of short illumination (Kloppstech et al., 1991). A recent study in adult light-grown Arabidopsis plants tested the possibility that Elips are induced by other environmental stresses other than light, but RNA gel-blot analysis could not detect induction of either Elip following heat shock (Heddad and Adamska, 2000).
Despite the similar light and heat shock regulation patterns of Elip1 and Elip2, the two genes have different accumulation patterns in the dark. Elip2 is expressed at low levels in dark-grown seedlings, whereas no Elip1 mRNA could be detected. This first evidence for presence of Elip transcript under dark conditions results from the sensitivity of the LightCycler system because no Elip transcript could be detected in the earlier studies performed by RNA gel blot, or in this study as shown in Figure 2, by conventional RT-PCR.
Our results demonstrate the positive role of HY5 on Elip1 transcription during photomorphgenesis. The absence of Elip1 mRNA in the hy5 light-treated seedlings is not surprising because this transcription factor has been already shown to bind directly to G-box DNA sequences, well-characterized light-responsive elements in light-responsive promoters (Chattopadhyay et al., 1998). A study in transgenic pea plants had demonstrated that two light-responsive elements are involved in light-regulated expression of Elip. One element is similar to the GT1 binding site and the other resembles a G-box-like ACGT element (Blecken et al., 1994).
The CSN mutant, cop9, which mimics light growth while grown in darkness, shows high expression levels of the classic light-inducible genes such as CAB, Chs, and PsbA when grown in a total darkness (Wei and Deng, 1992). This study shows that the light induction of both Elips is repressed in the dark by the CSN. From this result, together with the results of hy5 experiment, it can be concluded that light signals abolish the CSN-mediated degradation of the transcription factor HY5, and thus allow it to activate directly or indirectly the transcription of Elip1 gene. However, the reduction of Elip2 expression in the dark must be mediated by a different mechanism because HY5 does not regulate Elip2 expression. Based on earlier data, a working model for the signal transduction pathways that regulate Elip expression in Arabidopsis seedlings is suggested in Figure 7.
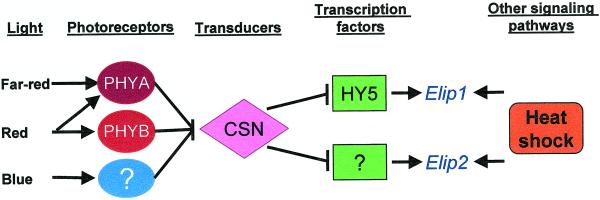
Figure 7.
Working model for the signal transduction pathways that regulate the expression of the Elip genes in Arabidopsis seedlings. Different light qualities are sensed by at least three different photoreceptors, PhyA, PhyB, and a novel blue-light receptor, to initiate a signal transduction cascade that abolishes the repressory action of the CSN, leading to the expression of both Elip1 and Elip2. The expression of Elip1 is positively effected by HY5, whereas other transcription factors regulate Elip2. Heat shock stimulates the expression of Elip through an independent signaling pathway.
Our results that the dark levels of Elip2 are higher than the dark level of Elip1, together with the result that HY5 is not involved in the regulation of Elip2, hint that the light regulation of Elip1 occurs at the level of transcription, whereas other mechanisms may be involved in the regulation for Elip2. These two differences in regulation imply different function. Because Elip1 is strictly light induced and apparently responds as a light stress protein, Elip2 may be a “housekeeping gene” that is constantly expressed at low levels, ready to be translated under stress conditions.
The microarray data further indicate that the ELIPs are stress-related proteins. This is consistent with previous evidence that Elip steady-state levels are regulated by other environmental stresses. Low temperature, for example, positively regulated Elip transcription and stabilization (Adamska and Kloppstech, 1994). Because low temperature noticeably increases excitation accumulation of PSII, it was proposed that this cold-induction accumulation of Elip mRNA could be interpreted in terms of redox control of gene expression (Montane et al., 1998). In Crateostigma plantagineum, an ELIP-related Dsp-22 protein is induced during desiccation; in this case, abscisic acid (ABA) and light were simultaneously required for Elip induction (Bartels et al., 1992). In a similar experiment when 6-d-old green barley plants were treated with a combination of high light and ABA, Elip level increased in comparison with light-treated control. However, no effect was observed with just ABA (Potter and Kloppstech, 1993). This environmental induction of Elip1 transcription implicates additional functions that are controlled by signal transduction pathways other than those described here.
Although accumulating correlative evidence indicates that ELIPs are involved in protection of the photosynthetic apparatus, the elucidation of the physiological role of the ELIPs has been hampered by lack of a genetic system. The discovery that hy5 lacks Elip1 expression provides a preliminary model system to study the role of ELIP1. A defect in PSII activity displayed by hy5 seedlings may result from the loss of Elip1 expression in this mutant. The development of photosynthetic activity in hy5 is retarded relative to that of the wild type. The low Fv/Fm ratio in the hy5 seedlings seems to be due mostly to a relatively high initial (minimum) PSII fluorescence in the dark-adapted state (F0) level. This could be due to PSII centers in which electron flow to the plastoquinone pool is partially inhibited (that is, closed PSII centers), a situation that can be induced by light stress. The fact that the Fv/Fm ratio in the mutant seedlings exposed to 100 μmol m−2 s−1, as compared with those exposed to 20 μmol m−2 s−1, reached lower levels supports this suggestion. However, as the time of illumination and thus of the development of the thylakoids continues, hy5 seedlings recover from this initial light stress and the Fv/Fm ratio reaches the same values in mutant seedlings exposed to both low and high light. Following prolonged illumination, hy5 completely recovers from the light stress and the Fv/Fm ratio is similar in the mutant to the wild type. We hypothesize that the phenotype exhibited by hy5 during the early phase of the greening process results from a lack of Elip1 transcription. Further study is needed to validate this hypothesis.
MATERIALS AND METHODS
Plant Materials, Growth, and Illumination Conditions
Arabidopsis seedlings were grown for 4 d on Murashhige and Skoog medium (Sigma, St. Louis) with 1% (w/v) agar, in darkness, at 22°C. Different light qualities were obtained by using a cool-white fluorescent light (100 μmol m−2 s−1; OSRAM, Munich) with the filters (Chris James, London): blue (380–500 nm, 40 μmol m−2 s−1), red (600–700 nm, 50 μmol m−2 s−1), or with far-red enriched lights with a far-red filter (700–780 nm, 2 μmol m−2 s−1). For heat shock treatment, the seedlings were incubated at 37°C in the dark. Seedlings were harvested and frozen in liquid nitrogen after 1 h of light or heat shock treatment. All subsequent manipulations were done under a green safe light.
The mutants used in this work were: cry1-304 (Ahmad and Cashmore, 1993), cry2-1 (Guo et al., 1998), cry1-304/cry2-1 (Guo et al., 1998), nph1-5 (Liscum and Briggs, 1995), phyA (Reed et al., 1993), phyB (Koornneef et al., 1980), phyA/phyB (Reed et al., 1993), hy5-1 Koornneef et al., 1980), cop9-1 (Wei and Deng, 1992), and cav1-1 (npl1; Kagawa et al., 2001).
Measurement of Transcript Levels by RT-PCR
RNA Preparation and RT Reaction
Total RNA or DNA was isolated using the SV RNA isolation kit (Promega, Madison, WI) according to the manufacturer's protocol. RNA concentrations were measured using a GeneQuont spectrophotometer (Pharmacia Biotech, Uppsala), with concentration of each sample calculated from the average of six measurements. RT of total RNA was carried out using oligo(dT) as a primer. Each sample contained 1 μg of total RNA. The reaction mixture included: 500 ng of oligo(dT), 10 mm each dNTPs, 0.2 m dithiothreitol, 5× RT buffer, and 200 units of SuperScript II RT (GibcoBRL, Carlsbad, CA) in a total reaction volume of 20 μL. The reaction was incubated at 70°C for 10 min, 42°C for 50 min, and then inactivated at 70°C for 15 min.
The following primers were used for amplification by PCR: Ubq10, 5′-cgattactcttgaggtggag-3′ (forward) and 5′-agaccaagtgaagtgtggac-3′ (reverse); Elip1, 5′-gcttaaagttctgtaacctaagcg-3′ (forward) and 5′-ttaggtttcataggaggaggagg-3′ (reverse); and Elip2, 5′-cagtgttcgctgctccttcc-3′ (forward) and 5′-tcgatgccaacgtcaacaac-3′ (reverse). The Elip primers are around introns of 84 bp (Elip1), and 92 and 213bp (Elip2), yielding cDNA products of 330 bp (Elip1) and 400 bp (Elip2).
Conventional PCR Followed by Hybridization
The PCR mixture contained 0.625 μm of each oligonucleotide primer, 40 mm each dNTPs (Roche, Mannheim, Germany), 10× Taq polymerase buffer, 1.8 units of Supertherm DNA polymerase (Promega), and 4 μL of the RT reaction mixture (cDNA) in a total volume of 50 μL. The samples were amplified: 94°C/2 min, cycled at 94°C/1 min, 55°C/1 min, and 72°C/40 s in a PTC-100 (MJ Research, Clearwater, MN). PCR products were separated by electrophoresis through a 1.7% (w/v) agarose gel and transferred to Hybond N+ membrane, or were loaded directly on the membrane (dot blot). The probes were labeled by random priming with “DIG High Prime” (Boehringer Mannheim) according to the manufacturer's protocol.
Membranes were prehybridized in 5× SSC (850 mm NaCl, 85 mm trisodium citrate-2H2O, ph 7.0), 0.1% (w/v) N-lauroylsarcosine, 0.02% (w/v) SDS, and 1% (w/v) blocking reagent for 2 h at 68°C followed by hybridization in the same solution containing denatured DIG-labeled probe at 68°C overnight. The membranes were washed to a final stringency of 0.1% (w/v) SSC and 0.1% (w/v) SDS at 68°C. Disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-{5′-chloro}tricyclo{3.3.1.13,7}decan]4-yl) was used as a chemilumenscent detection substrate. The membranes were exposed to x-ray film to record the chemilumenscent light-signals that were analyzed using the Scion Image software (Frederick, MD). The RT-PCR results of Elip1 and Elip2 were corrected according to the relative quantity of the RT-PCR product of Ubq10 mRNA.
Real-Time PCR by LightCycler
The LightCycle System (Roche) provides simultaneous PCR amplification and product analysis. The double-stranded DNA (dsDNA) SYBR Green I stain (Wittwer et al., 1997) is included in the PCR mixture, allowing template quantification during amplification. Fluorescence is monitored once each cycle after product extension and increases above background fluorescence at a cycle number that is dependent on initial template concentration. Because this dye detects all dsDNA, including primer dimers and other undesired products, sequence confirmation for the amplified product is provided through a function termed “melting curve analysis.” Melting curve analysis is performed after the amplification cycles are completed and a PCR product is formed. During this process, the temperature is slowly raised to 95°C and the fluorescence in each tube is measured every 0.2°C. As the DNA starts to denature, the SYBR Green I dye is released from the dsDNA, resulting in a decrease in fluorescence. Fluorescence data were converted into melting peaks by software that removes background fluorescence and the effect of temperature (T) on fluorescence (F), then plotted as the negative derivative of fluorescence with respect to temperature (−dF/dT versus T). Each dsDNA product has is own specific melting temperature, which is defined as the temperature at which 50% becomes single stranded, and 50% remains double stranded. Because the melting curve of the products is dependent on GC content, length and sequence, specific PCR products can be distinguished from nonspecific products by their melting curves without the necessity of electrophoretic analyses. The software allows an additional step in each PCR cycle, in which the LightCycler instrument is programmed to increase the temperature before measurement. Measurement at the elevated temperature instead of measurement at the elongation temperature increases specificity. PCR product levels were recorded at the end of each cycle at 84°C, where all nonspecific products of Elip1, Elip2, and Ubq10 primers pairs were denatured and thus not detected.
The initial amount of cDNA before the amplifiction for a particular template in the cDNA mixture was extrapolated from a standard curve with external standards. The standards run in parallel with the samples under identical PCR conditions. Elip1 (10−3 to 102 ng) was used as a quantification standard each experiment. This amount was corrected according to the relative amount of Ubq10.
The reaction mixture of the LightCycler PCR contained: 2 μL of the RT reaction mixture as a template, 4 μL of MgCl2, 1× LightCycler-FastStart DNA Master SYBR Green Ι, and 0.5 μm each primers. The reaction condition was: 95°C/10 min (activation of the FastStart Taq DNA polymerase), amplification: 95°C/10 s, 62°C to 55°C/10 s, 72°C/22 s, and detection at 85°C.
Plasmids Used in This Work
The cDNA clones for Elip1 (clone Id174P5T7) and Elip2 (clone IdVCVCD09) were obtained through AIMS. Elip1 was isolated from the mixed tissue cDNA library Lambda PRL2 (Newman et al., 1994). Elip2 was isolated from a cDNA library made from 5-d-old etiolated seedlings (T. Desprez, J. Amselem, H. Chiapello, P. Rouze, M. Caboche, and H. Hofte, unpublished data).
Chlorophyll Fluorescence Measurements
Wild-type and mutant seedlings were grown on agar plates and sowed in such a way as to form clusters of several seedlings so one could measure simultaneously the fluorescence emission of at least five to seven seedlings, thus obtaining an average result. Several clusters were measured on each plate. Variable fluorescence was measured using a Pulse Modulated Fluorimeter (PAM-101, Waltz, Germany). The modulated beam (650 nm) intensity at the seedlings level was about 1 mmol m−2 s−1 at 1.5 kHz and the intensity of the saturation light pulse was 3,000 mmol m−2 s−1 for 1-s duration. The variable fluorescence, Fv, was calculated as (Fm − F0). F0 is the minimal fluorescence and was determined with the modulated beam. Fm is the maximal fluorescence and was determined with the saturation light pulse. The ratio Fv/Fm is normalized with the concentration of chlorophyll and interpreted as functional state of PSII. The plants were dark adapted for several minutes before the onset of measurements and maintained thereafter in dim-green light.
ACKNOWLEDGMENTS
We thank Dr. Blanca Shanitzki (Dyn Diagnostics, Israel) for guidance in setting up the LightCycler system; Profs. Chentao Lin (UCLA, Los Angeles), Timothy Short (Queens College, CUNY, Flushing, NY), Winslow Briggs (Carnegie Institute, Palo Alto, CA), and Kiyotaka Okada (Kyoto University, Kyoto) for providing seeds for photoreceptor mutants; Arabidopsis Biological Resource Center (Columbus, OH) for providing the cDNA clones; Dr. Nir Keren (Washington University, St. Louis) for critical reading of the manuscript; the Interdepartmental Equipment Center at the Tel Aviv University Faculty of Medicine for use of the LightCycler; and Shirley Kadouri for assistance in the early stages. This work was carried out as the Master of Science thesis of O.H.-S.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010270.
LITERATURE CITED
- Adamska I. Regulation of early light inducible-protein gene expression by blue and red in etiolated seedlings involves nuclear and plastid factors. Plant Physiol. 1995;107:1167–1175. doi: 10.1104/pp.107.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska I. ELIPs: light-induced stress proteins. Physiol Plant. 1997;100:794–805. [Google Scholar]
- Adamska I, Kloppstech K. Low temperature increases the abundance of early light-inducible transcript under light stress conditions. J Biol Chem. 1994;269:30221–30226. [PubMed] [Google Scholar]
- Adamska I, Kloppstech K, Ohad I. UV light stress induces the synthesis of the early light-inducible protein and prevents its degradation. J Biol Chem. 1992a;267:24732–24737. [PubMed] [Google Scholar]
- Adamska I, Kloppstech K, Ohad I. Early light-inducible protein in pea is stable during light stress but is degraded during recovery at low light intensity. J Biol Chem. 1993;268:5438–5444. [PubMed] [Google Scholar]
- Adamska I, Ohad I, Kloppstech K. Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA. 1992b;89:2610–2613. doi: 10.1073/pnas.89.7.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska I, Roobol-Boza M, Lindahl M, Andersson B. Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem. 1999;260:453–460. doi: 10.1046/j.1432-1327.1999.00178.x. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR. HY4 gene of Arabidopsis thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR. The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J. 1997;3:421–427. doi: 10.1046/j.1365-313x.1997.11030421.x. [DOI] [PubMed] [Google Scholar]
- Bartels D, Hanke C, Schneider K, Michel D, Salamini F. A desiccation-related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. Embo J. 1992;11:2771–2778. doi: 10.1002/j.1460-2075.1992.tb05344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschauer A. Photoreceptors of higher plants. Planta. 1998;206:479–492. doi: 10.1007/s004250050425. [DOI] [PubMed] [Google Scholar]
- Beator J, Potter E, Kloppstech K. The effect of heat shock on morphogenesis in barley. Plant Physiol. 1992;100:1780–1786. doi: 10.1104/pp.100.4.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecken J, Weisshaar B, Herzfeld F. Two distinct cis-acting elements are involved in light-dependent activation of the pea elip promoter. Mol Gen Genet. 1994;245:371–379. doi: 10.1007/BF00290118. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- Chamovitz D, Pecker I, Hirschberg J. The molecular basis of resistance to the herbicide norflurazon. Plant Mol Biol. 1991;16:967–974. doi: 10.1007/BF00016069. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert M, Kaldenhoff R. Light-induced stomatal movement of selected Arabidopsis thaliana mutants. J Exp Bot. 2000;51:1435–1442. [PubMed] [Google Scholar]
- Frechilla S, Zhu J, Talbott LD, Zeiger E. Stomata from npq1, a zeaxanthin-less Arabidopsis mutant, lack a specific response to blue light. Plant Cell Physiol. 1999;40:949–954. doi: 10.1093/oxfordjournals.pcp.a029627. [DOI] [PubMed] [Google Scholar]
- Grimm B, Kloppstech K. The early light-inducible proteins of barley: characterization of two families of 2-h-specific nuclear-coded chloroplast proteins. Eur J Biochem. 1987;167:493–509. doi: 10.1111/j.1432-1033.1987.tb13364.x. [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Adamska I. Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA. 2000;97:3741–3746. doi: 10.1073/pnas.050391397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- Jin X, Zhu J, Zeiger E. The hypocotyl chloroplast plays a role in phototropic bending of Arabidopsis seedlings: developmental and genetic evidence. J Exp Bot. 2001;52:91–97. [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- Karniol B, Chamovitz DA. The COP9 signalosome: from light signaling to general developmental regulation and back. Curr Opin Plant Biol. 2000;3:387–393. doi: 10.1016/s1369-5266(00)00101-1. [DOI] [PubMed] [Google Scholar]
- Kloppstech K, Otto B, Sierralta W. Cyclic temperature treatments of dark-grown pea seedlings induce a rise in specific transcript levels of light-regulated genes related to photomorphogenesis. Mol Gen Genet. 1991;225:468–473. doi: 10.1007/BF00261689. [DOI] [PubMed] [Google Scholar]
- Kolanus W, Scharnhorst C, Kuhne U, Herzfeld F. The structure and light-dependent transient expression of a nuclear-encoded chloroplast protein gene from pea (Pisum sativum L.) Mol Gen Genet. 1987;209:234–239. doi: 10.1007/BF00329648. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolf E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kruse E, Kloppstech K. Integration of early light-inducible proteins into isolated thylakoid membranes. Eur J Biochem. 1992;208:195–202. doi: 10.1111/j.1432-1033.1992.tb17174.x. [DOI] [PubMed] [Google Scholar]
- Lasceve G, Leymarie J, Olney MA, Liscum E, Christie JM, Vavasseur A, Briggs WR. Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol. 1999;120:605–614. doi: 10.1104/pp.120.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Chan J, Cashmore A. CRY2: a second member of the Arabidopsis cryptochrome gene family. Plant Physiol. 1996;110:1047. [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montane MH, Tardy F, Kloppstech K, Havaux M. Differential control of xanthophylls and light-induced stress proteins, as opposed to light-harvesting chlorophyll a/b protein, during photosynthetic acclimation of barley leaves to light irradiance. Plant Physiol. 1998;118:227–235. doi: 10.1104/pp.118.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovici-Kadouri S, Chamovitz DA. Chamracterization of a cDNA encoding the early light-inducible protein (ELIP) from Arabidopsis. Plant Physiol. 1997;115:1287. [Google Scholar]
- Mustilli AC, Bowler C. Tuning in to the signals controlling photoregulated gene expression in plants. Embo J. 1997;16:5801–5806. doi: 10.1038/sj.emboj.7590554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kircher S, Schafer E. Nucleo-cytoplasmic partitioning of the plant photoreceptors phytochromes. Semin Cell Dev Biol. 2000;11:505–510. doi: 10.1006/scdb.2000.0202. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Newman T, deBruijn FJ, Raikhel N, Somervilles S, Thomashow M, Retzel E, Somerville C. Gene galore: a summary of methods for accessing results from larg-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto B, Ohad I, Kloppstech K. Temperature treatments of dark-grown pea seedlings cause an accelerated greening in the light at different levels of gene expression. Plant Mol Biol. 1992;18:887–896. doi: 10.1007/BF00019203. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Warpeham KMF, Briggs WR. Evidence that zeathanthin is not the photoreceptor for phototropism in maize. Plant Physiol. 1996;110:1323–1328. doi: 10.1104/pp.110.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter E, Kloppstech K. Effects of light stress on the expression of early light-inducible proteins in barley. Eur J Biochem. 1993;214:779–786. doi: 10.1111/j.1432-1033.1993.tb17980.x. [DOI] [PubMed] [Google Scholar]
- Quail P, Boylan M, Parks BM, Short T, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnhorst C, Heinze H, Meyer G, Kolanus W, Bartsch K, Heinrichs S, Gudschun T, Moller M, Herzfeld F. Molecular cloning of a pea mRNA encoding an early light induced, nuclear coded chloroplast protein. Plant Mol Biol. 1985;4:241–245. doi: 10.1007/BF02418242. [DOI] [PubMed] [Google Scholar]
- Smith H. Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:289–315. [Google Scholar]
- Sun CW, Callis J. Independent modulation of Arabidopsis thaliana polyubiqutin mRNA in different organs and in response to environmental changes. Plant J. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- Tlalka M, Runquist M, Fricker M. Light perception and the role of the xanthophyll cycle in blue-light-dependent chloroplast movements in Lemna trisulca L. Plant J. 2001;20:447–459. doi: 10.1046/j.1365-313x.1999.00614.x. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. COP9: a new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell. 1992;4:1507–1518. doi: 10.1105/tpc.4.12.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Beauchamp N, Kerckhoffs LHJ, Platten JD, Reid JB. Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 2001a;26:283–294. doi: 10.1046/j.1365-313x.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- Weller JL, Perrotta G, Schreuder ME, van Tuinen A, Koornneef M, Giuliano G, Kendrick RE. Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J. 2001b;25:427–440. doi: 10.1046/j.1365-313x.2001.00978.x. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–131. doi: 10.2144/97221bi01. , 134–138. [DOI] [PubMed] [Google Scholar]
- Zeiger E, Zhu J. Role of zeaxanthin in blue light photoreception and the modulation of light-CO2 interactions in guard cells. J Exp Bot. 1998;49:433–442. [Google Scholar]