Abstract
Gene expression in response to Fe deficiency was analyzed in Arabidopsis roots and shoots through the use of a cDNA collection representing at least 6,000 individual gene sequences. Arabidopsis seedlings were grown 1, 3, and 7 d in the absence of Fe, and gene expression in roots and shoots was investigated. Following confirmation of data and normalization methods, expression of several sequences encoding enzymes known to be affected by Fe deficiency was investigated by microarray analysis. Confirmation of literature reports, particularly for changes in enzyme activity, was not always possible, but changes in gene expression could be confirmed. An expression analysis of genes in glycolysis, the tricarboxylic acid cycle, and oxidative pentose phosphate pathway revealed an induction of several enzymes within 3 d of Fe-deficient growth, indicating an increase in respiration in response to Fe deficiency. In roots, transcription of sequences corresponding to enzymes of anaerobic respiration was also induced, whereas in shoots, the induction of several genes in gluconeogenesis, starch degradation, and phloem loading was observed. Thus, it seemed likely that the energy demand in roots required for the Fe deficiency response exceeded the capacity of oxidative phosphorylation, and an increase in carbon import and anaerobic respiration were required to maintain metabolism.
The role of Fe as an essential nutrient and its function in metabolism have been investigated in detail (Marschner, 1995; Fox and Guerinot, 1998). Fe is abundant in most soils, and plants can accumulate superoptimal levels of Fe and suffer from Fe toxicity when grown for example under hypoxia (Drew, 1997). However, under aerobic conditions the physical-chemical properties of Fe dictate the formation of highly insoluble Fe-oxides and -hydroxides, making Fe limiting for plant growth (Marschner, 1995). Mechanisms by which plants adapt to Fe deficiency have been frequently described in grasses (strategy II) and other plants (strategy I) and are the subject of comprehensive reviews (e.g. Guerinot and Yi, 1994; Moog and Brüggemann, 1994; Schmidt, 1999).
The primary visible symptom of Fe deficiency in the field is the development of intercostal chlorosis principally on young leaves. In this regard, Fe deficiency stress is correlated with changes in chloroplast ultrastructure (Spiller and Terry, 1980) and decreased expression of the small and large subunits of Rubisco, of chlorophyll a/b-binding proteins, and of chlorophyll, among other proteins (Spiller et al., 1987; Winder and Nishio, 1995). When grown under Fe-limiting conditions, plants could compensate for the lack of Fe through an increased Fe uptake capacity. These adaptive reactions in strategy I plants involved both morphological and metabolic changes in roots and shoots. Roots of Fe-deficient plants showed increased numbers and length of root hairs (Landsberg, 1986; Schmidt et al., 2000) and the formation of transfer cells (Landsberg, 1982; Schmidt and Bartels, 1996). In addition, strategy I plants showed increased rates of Fe reduction at the root surface (Chaney et al., 1972; Robinson et al., 1999), increased acidification of the rhizosphere (Römheld et al., 1984), secretion of riboflavin-like compounds (Susin et al., 1993), and increased capacity for Fe uptake (Chaney et al., 1972; Eide et al., 1996; Eckhardt, et al., 2001). These changes serve primarily to increase the surface area between the plant and the soil as well as the availability of Fe for uptake.
The biochemical and genetic components that are involved in the response to Fe deficiency have been identified only in isolated cases. A correlation exists between the development of Fe3+-chelate reductase activity, acidification of the rhizosphere, and the accumulation of citrate and malate in roots (Landsberg, 1986). In particular, the accumulation of malate has been interpreted in terms of the pH-stat theory (Sakano, 1998). As a consequence of apoplast acidification during the response to Fe deficiency, the pH of the root cytosol and vacuole increased (Espen et al., 2000). In support of the pH-stat, several investigations have established a correlation between Fe deficiency stress and increased non-photosynthetic carbon fixation and phosphoenolpyruvate (PEP) carboxylase activity (PEPCase; e.g. Rabotti et al., 1995; De Nisi and Zocchi, 2000). Increased activity of three glycolytic enzymes in cucumber (Cucumis sativus) roots, namely glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase, and Fru-6-phosphate kinase, indicated an elevated glycolysis activity in response to Fe deficiency (Espen et al., 2000). Glyceraldehyde-3-phosphate dehydrogenase was also shown to be increased in Fe-deficient tomato (Lycopersicon esculentum) roots (Herbik et al., 1996). Whether this increase is a result of decreased oxidative respiration or due to funneling of carbon out of glycolysis as a result of an increased PEPCase activity was not entirely clear, although increased formate dehydrogenase and ascorbate peroxidase in Fe-deficient tomato roots and superoxide dismutase and plastocyanin in leaves would support a decreased oxidative respiration and/or photosynthesis due to Fe deficiency (Herbik et al., 1996).
With the goal to better understand the processes involved in the adaptation to Fe deficiency stress, we have investigated gene expression using a large collection of Arabidopsis cDNA clones. Using this approach, we have been able to confirm and extend many of the observations made using enzymological and molecular biological methods.
RESULTS
Evaluation of the Response to Fe Deficiency
Induction of Fe3+-chelate reductase activity was shown to correlate with the Fe deficiency response (Schmidt, 1994; Moog et al., 1995) and was used in this study to determine the Fe nutritional status of Arabidopsis after growth in the absence of Fe. Compared with control plants (+Fe), the activity of reductase was induced 1.2-, 3.1-, and 6.3-fold in test plants (−Fe) after 1, 3, and 7 d of Fe-deficient growth, respectively. Root hair development has been described as a morphological response to Fe deficiency and was observed on roots after 3 d of growth but was less prominent after 7 d of growth in the absence of Fe (Schmidt et al., 2000).
Analysis of the Source of Variability in the Array Data
The signal intensities corresponding to individual cDNA clones were normalized to relative gene expression levels (REL; S. Kloska, B. Essigmann, and T. Altmann, unpublished data) to allow comparison between experiments as described (see “Materials and Methods,” Eqs. 1 and 2). Because the intensity of a signal depended on the efficiency of cDNA synthesis, labeling, and the amount of cDNA bound to the membrane (Schuchhardt et al., 2000), data were normalized with respect to these parameters. Prior to analysis of the effect of nutritional status on the expression of individual cDNA clones, the inherent variability of the data and the effect of normalization were analyzed in detail. In comparison with scatter plots of raw data, signal variation of normalized data was improved and allowed comparison of REL obtained from different hybridization experiments (data not shown).
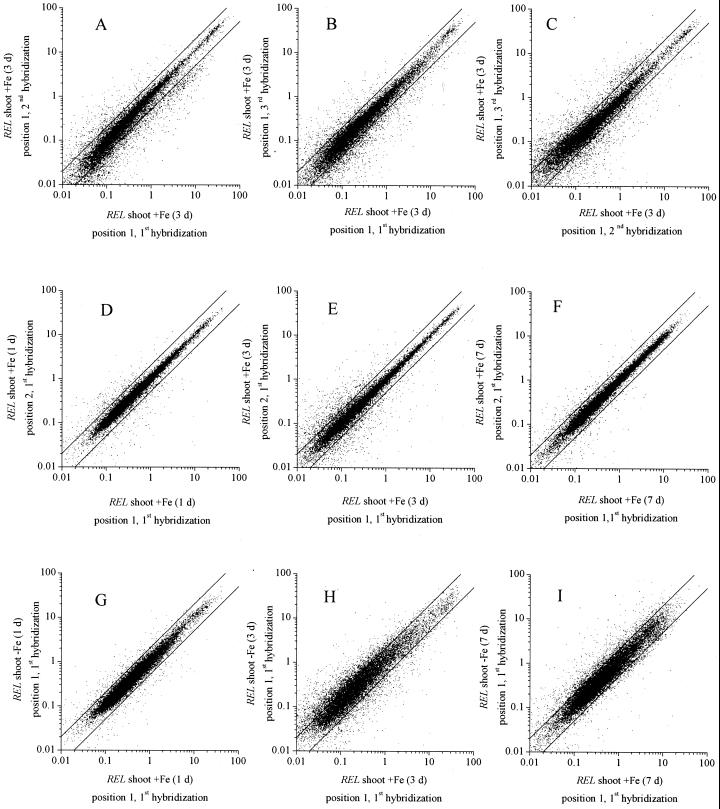
For each of the time points, three independent hybridization experiments were conducted. cDNA clones were spotted twice on the membrane. To evaluate the magnitude of hybridization effects on signal variation, REL was analyzed in sequential hybridizations at the same location on the filter. In Figure 1, A through C, an example is shown of such comparisons for REL of a clone at position 1 for control shoots (+Fe) after 3 d of treatment. Each clone was compared with itself at the same position on the filter for three hybridizations (i.e. first hybridization versus the second, the first versus the third, and the second versus the third). As is apparent, the variability between hybridizations was similar (compare with Fig. 1, A–C). Thus, variability inherent in the experimental procedure (e.g. labeling and hybridization conditions) was significant but relatively constant.
Figure 1.
Scatter plot analysis of signal variability. The effect of different factors on signal variation was analyzed: repetition of hybridization (A–C), position on the filter (D–F), and Fe deficiency (G–I). Quantified and normalized signals were expressed as REL of control shoot (+Fe) and test shoot (−Fe) arrays after 1 d −Fe (D and G), 3 d −Fe (A–C, E, and H), and 7 d −Fe (F and I). To analyze the influence of hybridization repetitions on signal variation, control shoot REL (3 d −Fe) of different hybridizations were scattered: first versus second (A), first versus third (B), and second versus third (C) hybridization. cDNA clones were doubly spotted on filters at position 1 and 2. To exclude position-induced signal variation, REL at position 1 were compared. Position-induced REL variation is shown in D through F. REL of doubly spotted cDNA clones of one filter were scattered: position 1 versus 2. Data of control shoot arrays after 1, 3, and 7 d of −Fe (D–F, respectively) of the first hybridization are given as an example. The influence of Fe deficiency on REL is presented in G through I (1, 3, and 7 d −Fe, respectively). Shoot control (+Fe) versus shoot test (−Fe) arrays were compared. REL variations due to hybridization repetition and position on the filter were excluded using REL of the first hybridization and at position 1. For comparison of signal variation, guide lines were added to figures (y = 2x, y = x/2).
In addition, the effect of the position of a cDNA clone on the filter on REL was determined for experiments after 1, 3, and 7 d of Fe-deficient growth, respectively (Fig. 1, D and E). Comparisons were made between the single REL of the doubly spotted cDNA clones at positions 1 and 2 on the same filter. Although position-related variation was somewhat greater at 3 d compared with 1 and 7 d, position-related signal variation was generally minor compared to variations induced by hybridization repetitions (compare with Fig. 1, A–F).
Finally, the normalized hybridization signals of cDNA clones for control (+Fe) and test shoots (−Fe) were compared with respect to hybridization and position on the membrane. Comparisons were made by scatter plot analysis of REL corresponding to clones of the first hybridization spotted at position 1 on the filter. Clear changes in expression levels could be observed for all experiments but were most prominent after 3 d and least prominent after 1 d of Fe deficiency (Fig. 1, G–I). Although the variability in REL related to Fe deficiency was greater than those related to hybridization or clone position, they were not great enough to neglect variability from other sources.
Because the variability was not distributed over the filter equally, common statistical methods (e.g. the Student's t test) were not suitable to select Fe-regulated clones. To take all sources of variations into consideration, an induction factor (IF) was calculated for each clone as described in “Materials and Methods” (Eq. 3). By this method, dimensionless values were obtained that allowed comparisons of changes in REL between different treatments and hybridization experiments. Throughout the investigation, only clones with a mean REL of > 0.1 were further analyzed. Clones with an IF > ±1 were defined initially as differentially expressed under Fe deficiency. In specific cases, clones encoding metabolic enzymes were analyzed regardless of the absolute value of the IF (see below).
General Changes in REL and IF Related to Fe Deficiency
A summary of the number of clones that met the criterion REL > 0 .1 and IF > ±1 is shown in Table I. In support of the results obtained from the scatter plot analyses (Fig. 1H), changes in REL under Fe deficiency were most prominent for both shoot and root after 3 d of Fe-deficient growth. Following 1 d Fe deficiency, the number of induced and repressed clones was approximately equal. Continued Fe deficiency stress resulted initially in a larger increase in the number of induced than repressed clones in shoots. In shoots after 3 d of Fe deficiency, 13.9% of the total cDNA clones (16,128 clones in total) were induced and 3.8% were repressed, whereas after 7 d these numbers were reduced to 5.2% induced and 1.9% repressed. A different trend was found in roots. After 1 and 3 d Fe deficiency, 0.5% and 4.8% of the clones were induced and an equal percentage repressed in roots, respectively. The percentage of repressed clones remained approximately constant (4.6%) after 7d of Fe deficiency, but the percentage of induced clones decreased to 0.9%. In light of the clearly visible Fe deficiency symptoms observed in shoots after 7 d Fe-deficient growth and the increased proportion of repressed clones in roots, it seems apparent that the plants after 7 d of treatment were severely stressed.
Table I.
Differentially expressed cDNA clones in Arabidopsis shoots and roots after 1-, 3-, and 7-d Fe deficiency
| Fe Deficiency | 1 d
|
3 d
|
7 d
|
|||
|---|---|---|---|---|---|---|
| Induced | Repressed | Induced | Repressed | Induced | Repressed | |
| Shoot | 143 | 143 | 2,240 | 620 | 847 | 299 |
| Root | 84 | 76 | 776 | 767 | 142 | 735 |
Differential expression was determined by comparison of quantified and normalized signals from control (+Fe) and test (−Fe) arrays expressed as IF. cDNA clones were defined as induced or repressed with a median IF > ±1. Only cDNA clones with a mean REL > 0.1 were used.
In an attempt to identify changes in REL associated with the Fe deficiency response, the published expressed sequence tag (EST) sequences of approximately 2,800 cDNA clones from all experiments were compared with databases as described in “Materials and Methods” to verify their identity and then grouped into eight functional categories (Fig. 2). The largest group, “unknown,” contained between 27% and 43% of all cDNA clones. The distribution of the induced clones in the seven remaining groups is summarized for the three time points in Figure 2. The categories summarize cDNA clones as follows: metabolism (amino acid, carbon, lipid, nucleic acid, nucleotide sugar and secondary metabolism, glycolysis, and respiration- and microbody-associated clones), protein (protein modification and catabolism), transport (nutrient uptake and homeostasis), signaling (DNA binding, RNA binding, signal perception, and transduction cDNA clones), photosynthesis, cellular organization (cell wall and development, cytoskeleton, and intracellular trafficking), and stress.
Figure 2.
Distribution of induced clones. Alteration of REL was determined by IF > ±1 for each detectable clone (REL > 0.1). Clones were compared to online database to predict function by homology and were grouped to eight functional categories: metabolism (amino acid, carbon, lipid, nucleic acid, nucleotide sugar and secondary metabolism, glycolysis, and respiration- and microbody-associated clones), protein (protein modification and catabolism), transport (nutrient uptake and homeostasis), signaling (DNA binding, RNA binding, signal perception, and transduction cDNA clones), photosynthesis, cellular organization (cell wall and development, cytoskeleton, and intracellular trafficking), and stress. Shown are data of three experiments (1, 3, and 7 d Fe deficiency) in shoot and root. The number of clones in each category is expressed in percent (%) of the cDNA clones analyzed (ranging from 85–363).
In roots, the proportion of cDNA clones coding for proteins involved in metabolism and stress reactions increased with increasing Fe-deficient growth, whereas in shoots, the proportion of induced clones in these categories remained relatively constant. Particularly noteworthy is the relatively large proportion of cDNA clones for signaling in root that were induced already after 1 d. With increased Fe deficiency growth, the proportion of induced signaling clones decreased. In shoots, the number of clones in the signaling category was not changed with increasing Fe deficiency stress.
cDNA clones that were repressed were also analyzed, although except for clones encoding enzymes involved in photosynthesis, which were increasingly repressed with increasing stress treatment, no significant trends could be identified (data not shown). The large number of repressed cDNA clones encoding photosynthetic enzymes in roots was somewhat surprising. We have no explanation for this observation. High-resolution, electrophoretic analyses of all fractions showed no chloroplast rRNA in the root RNA preparations (data not shown; Agilent 2100 Bioanalyzer, Waldbronn, Germany) The presence of photosynthetic enzymes in roots has been reported previously (Silverthorne and Tobin, 1990).
A table containing the cDNA clones with the greatest IF in each category is available from our website (http://www.biologie.hu-berlin.de/˜botanik).
Specific Changes in IF in cDNA Clones for Reference Genes
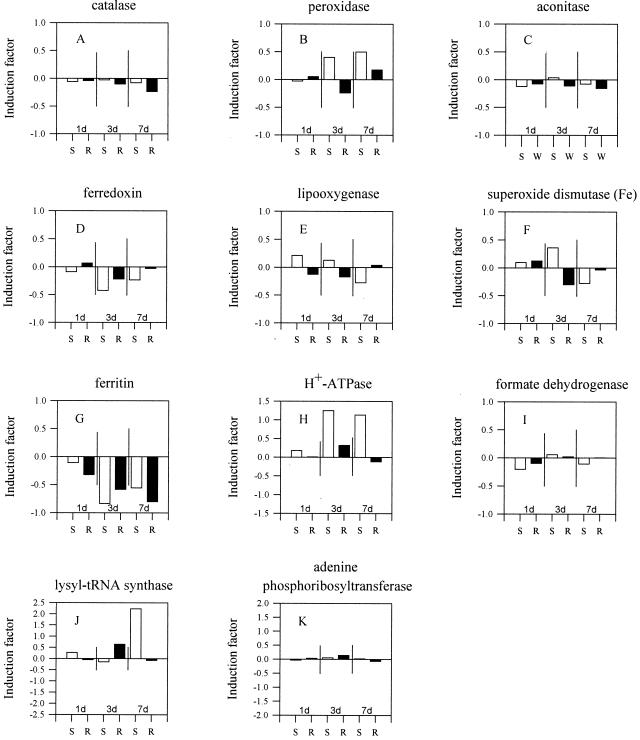
Changes in expression were investigated in cDNA clones for enzymes whose activity, transcription, or translation have been previously reported to be influenced by Fe nutritional status. Decreased enzyme activity under Fe deficiency was reported for the heme proteins, catalase and peroxidase (Machold, 1968) and the Fe-sulfur proteins, ferredoxin (Alcaraz et al., 1986), aconitase (De Vos et al., 1986), and we assumed also for Fe-SOD as a Fe-containing protein (Sevilla et al., 1984). Fe-containing lipoxygenase (Hildebrand, 1989) and the storage protein ferritin (Gaymard et al., 1996) were also decreased in Fe-deficient plants. In contrast, H+-ATPase (Dell'Orto et al., 2000), formate dehydrogenase (Suzuki et al., 1998), lysyl-tRNA synthetase (Giritch et al., 1997), and adenine phosphoribosyltransferase (Itai et al., 2000) were shown to be increased in expression or enzyme activity under Fe deficiency.
REL for all cDNA clones encoding the enzymes mentioned above were extracted from the microarray data, and an IF was calculated as described below. Reference clone data for 1-, 3-, and 7-d Fe deficiency experiments in shoots and roots are shown in Figure 3. Changes in expression were presented as a mean change in IF for all cDNA clones with homology to reference genes. In general, changes in REL did not consistently agree with changes in enzyme activity reported in the literature. Whereas the REL for ferritin and ferredoxin was decreased (Fig. 3, D and G), in agreement with published reports, on average the IF for catalase, aconitase, and lipoxygenase varied little with the length of Fe deficiency (Fig. 3, A, C, and E). Changes of ±25% were considered of marginal significance. The expression patterns of other groups of cDNA clones were complex. For example, in early stages of Fe deficiency, Fe-SOD (Fig. 3E) was induced in shoots and repressed in roots, whereas after 7 d shoot expression was repressed and no change was detected in roots. Fe deficiency stress induced REL of peroxidase in shoots, although depressed enzyme activities were reported by Machold (1968). Induction was found in cDNA clones for H+-ATPase and lysyl-tRNA synthetase, in shoots after 3 and 7 d of Fe deficiency (Fig. 3, H and J, respectively) but not in roots. cDNA clones for the formate dehydrogenase and adenine phosphoribosyltransferase (Fig. 3, I and K, respectively) revealed no significant changes of REL. In summary, agreement between changes in REL and reported enzyme activity was not consistent.
Figure 3.
Array data of 1, 3, and 7 d of Fe deficiency for shoots (S) and roots (R). Extracted were data of clones with significant homology to reference clones that were described as repressed (A–G) or induced (H–K) in gene expression, enzyme activity, or translational level by Fe deficiency. Shown are means of IF of all detectable cDNA clones (REL > 0.1) as a value for Fe deficiency-induced change of expression level. Vertical lines separate different experiments (1, 3, and 7 d of −Fe) and indicate a change in expression of ±50%.
Specific Changes in IF in cDNA Clones Glycolysis, Tricarboxylic Acid Cycle (TCA), and Oxidative Pentose Phosphate (OPP) Pathways
The results presented here indicated Fe deficiency induced changes in root metabolism (Fig. 2). To gain further insight into the nature of these changes, three pathways were analyzed in detail. As with the reference cDNA clones investigated above, all cDNA clones with homology to genes for glycolysis, citric acid cycle, and oxidative pentose phosphate cycle were extracted from microarray data. Clones with IF < ±1 were also selected from data, but clones were excluded with a REL mean < 0.1. In the Figures 4 through 6, regulation of genes is shown as mean of IF for all homolog clones. Data from 1-, 3-, and 7-d experiments is shown individually and separated for shoots and roots. Glycolytic reactions, including PEPCase and pyruvate dehydrogenase, and fermentation were also shown in Figure 4. In general, no changes in REL were found following 1 d of Fe-deficient growth. However, following 3 d the REL for cDNA clones encoding several glycolysis enzymes increased. In roots, these changes were found primarily in clones homologous to enzymes in fermentation (lactate dehydrogenase, pyruvate decarboxylase, and alcohol dehydrogenase), and in shoots the REL of clones for hexokinase, Glc-6-phosphate isomerase, Fru bisphosphate aldolase, and triose phosphate isomerase were increased. In addition, genes encoding α-amylase, the phosphate translocator, and the H+-Suc symporter were induced after 3 d of Fe deficiency; although no induction was observed for the Suc phosphate synthase.
Figure 4.
Fe deficiency-induced changes in expression of genes involved in glycolysis. Shown are means of IF of all detectable cDNA clones (REL > 0.1) with homology to the genes shown. Presentation of the data is shown in the standard graph to the left: IF means of 1, 3, and 7 d of Fe deficiency for roots (R) and shoots (S). Vertical lines separate different experiments and indicate a change in expression of ±50%.
Figure 6.
Fe deficiency-induced changes in expression of genes involved in oxidative pentose phosphate cycle. Shown are means of IF of all detectable cDNA clones (REL > 0.1) with homology to the genes shown. Presentation of the data is shown in the standard graph to the left: IF means of 1, 3, and 7 d of Fe deficiency for roots (R) and shoots (S). Vertical lines separate different experiments and indicate a change in expression of ±50%.
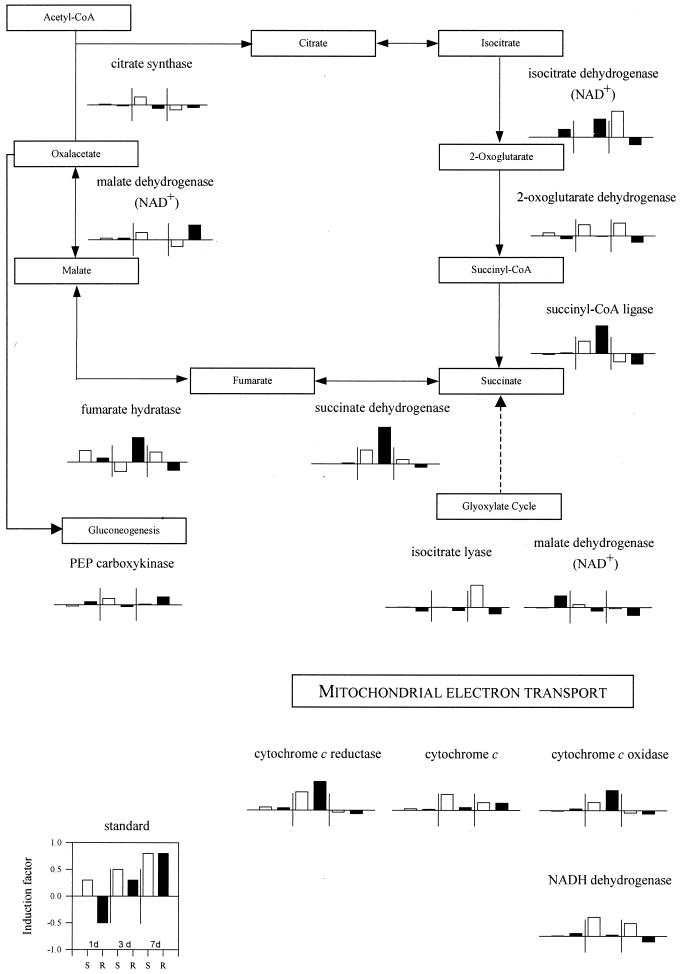
Changes in REL for enzymes in the citrate cycle, including the available glyoxylate cycle enzymes, isocitrate lyase, malate dehydrogenase, electron transport chain, and cytochrome c oxidoreductases were summarized in Figure 5. As for glycolysis, no changes in REL were observed after 1 d of Fe-deficient growth. The increased REL in roots for succinate dehydrogenase, cytochrome c reductase/oxidase, and to a lesser extent fumarate dehydratase and succinyl-CoA ligase were noteworthy. The REL for shoot enzymes shown in Figure 5 was largely unchanged with the exception of succinate dehydrogenase, cytochrome c, and cytochrome c reductase/oxidase after 3 d of Fe-deficient growth and isocitrate dehydrogenase after 7 d. Two available glyoxylate cycle enzymes and the anaplerotic enzyme, PEP carboxykinase, were largely unchanged throughout the analysis period.
Figure 5.
Fe deficiency-induced changes in expression of genes involved in citrate cycle. Shown are means for IF of all detectable cDNA clones (REL > 0.1) with homology to the genes shown. Presentation of the data is shown in the standard graph to the left: IF means of 1, 3, and 7 d of Fe deficiency for roots (R) and shoots (S). Vertical lines separate different experiments and indicate a change in expression of ±50%.
Finally, the expression of enzymes of the OPP pathway was investigated (Fig. 6). NADPH was reported to be the source of reductant for the chelated Fe3+ at the plasma membrane. An increased flow of carbon through the oxidative pentose phosphate pathway has been observed in Fe-deficient Phaseolus vulgaris roots, and interpreted in terms of synthesis of NADPH for Fe3+-chelate reduction (Lubberding et al., 1988). Although cDNA clones for both the Glc-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase were induced after 3 d of Fe deficiency, this increase only occurred in shoots. Thus, either regulation of the OPP pathway occurred at a level other than gene expression, or due to lack of Fe in the nutrient media, there was no increased demand for NADPH in the roots used in this study.
DISCUSSION
To estimate the reliability of the microarray system used in this study, an extensive investigation of the variability of signal intensity and source of errors was conducted. To visually estimate signal variability, scatter plot analysis proved valuable. As a result of these investigations, the variability due to cDNA synthesis, labeling, or hybridization predominated. In fact, this variability in the data equaled that induced by changes in REL due to Fe nutritional status (compare Fig. 1, A–C, with G–I). To circumvent these errors, IF was calculated. IF was useful for comparison of changes in REL between hybridizations and experiments but give no estimation of the level of REL. Using this approach, we have been able to identify steps in metabolic pathways where induction or repression of specific cDNA clones occurred (see below). In cases where several clones are present (e.g. alcohol dehydrogenase), the IF represents an average of the behavior of all clones.
To aid in evaluation of the data, induced or repressed cDNA clones were identified and categorized (Fig. 2). Using this approach, it was evident that the proportion of metabolic cDNA clones that were induced increased with increasing Fe stress in roots, whereas in shoots little change in distribution over time was observed. Although this general trend held true, individual clones did not always follow this pattern (e.g. hexokinase; see below). Data analysis was hampered by the general disarray of the EST database. A large number of cDNA clones were incorrectly annotated. Sequencing of approximately 200 cDNA clones of interest revealed that at least 15% of the sequences obtained did not correspond to those reported in the EST databases. Therefore, drawing conclusions from single clones would only be possible after confirming identity. Sequencing has been done only for selected cDNA clones that were shown in Figures 4 through 6.
Behavior of Reference cDNA Clones to Fe Deficiency
To verify the reliability of the microarray analysis, it would have been desirable to investigate cDNA clones such as IRT1 and FRO2, which encode an Fe transporter and the Fe3+-chelate reductase, respectively, and whose expression is induced under Fe deficiency (Eide et al., 1996; Robinson et al., 1999). However, these cDNA clones were not included in the collection available for this study. The clones 117P14T7 and 221A15T7 were identified as FRO2 homolog and FRO1/FRO2-like protein. The expression levels of these clones were found to be induced in Fe-deficient roots after 1 d of −Fe with an IF of 0.79 and 0.22, respectively. After 7 d of −Fe, the clone 117P14T7 showed an IF of 0.26 for shoots, whereas 221A15T7 was not expressed. As an alternative, reference clones were selected from the data that have been reported to be induced or repressed by Fe deficiency (Fig. 3). In most cases, these changes were determined for enzyme activity. In some examples, the behavior of clones selected did not correspond to the expected results, based on the literature. Catalase, peroxidase, and aconitase have been reported to decrease in activity and expression of formate dehydrogenase shown to increase Fe deficiency; however, the REL of these cDNA clones remained unchanged or, in the case of peroxidase, increased over the period investigated. Other cDNA clones did behave in whole or in part as expected, particularly when the response to Fe deficiency had progressed.
Ferritin was of particular interest. After 24 h of Fe-deficient growth, the REL for ferritin already was decreased in roots, and repression was significant in both roots and shoots after 3 and 7 d of Fe-deficient growth. This result confirms the publication of Gaymard et al. (1996). Thus, for ferritin the response to Fe deficiency developed first in the roots and subsequently in the shoots.
Acidification of the rhizosphere was often correlated with the response to Fe deficiency in several plants. Acidification of the apoplast was presumed to increase the solubility of Fe3+ and increase its availability. However, an induction of the plasma membrane H+-ATPase was observed primarily in shoots and only weakly after 3 d in roots. This finding indicated that the increase in apoplast acidification in roots was not due to increased ATPase synthesis. Expression of lysyl-tRNA synthetase was found to be increased in Fe-deficient tomatoes (Giritch et al., 1997), an observation supported in the present study (Fig. 3K). However the significance of this increase remains obscure. The expression of adenine phosphoribosyltransferase, an enzyme in the Yang cycle, was increased in barley but only weakly in tobacco (Itai et al., 2000). Its increased expression in grasses has been interpreted in the framework of phytosiderophore synthesis. Nicotianamine is thought to share the same biosynthetic pathway in dicotyledons as phytosiderophores in grasses. In agreement with findings in tobacco, adenine phosphoribosyltransferase was induced in roots and not shoots under Fe deficiency stress.
The fact that the behavior of the reference clones did not agree in every case with reports in the literature may not be surprising. Expression of changes in REL in the form of an IF offered no insight into the level of expression. Weakly expressed cDNA clones, whose expression changed dramatically, would only slightly affect the mean IF as calculated here. In addition, transcriptional regulation is only one form of control for enzyme activity. Posttranscriptional modification and allosteric enzyme regulation represent equally important mechanisms for control of activity and flux through an enzyme pathway.
REL in Specific Metabolic Pathways
Because of the proportional increase in expression of metabolic clones during Fe deficiency stress, the microarray data obtained in this study were used to analyze the changes in REL in three central metabolic pathways. In glycolysis, as for OPP and TCA cycles, no clear changes in REL were observed after 1 d. However, after 3 d of Fe-deficient growth, several steps in these pathways showed significant changes.
In 3-d Fe-deficient roots, the enzymes associated with anaerobic metabolism, namely lactate dehydrogenase, pyruvate decarboxylase, and alcohol dehydrogenase, were induced. In addition, an induction in roots for several enzymes in the TCA cycle (isocitrate dehydrogenase, succinyl CoA ligase, succinate dehydrogenase, and fumarate hydratase) and in the mitochondrial electron transport chain (cytochrome c reductase and oxidase) were observed. An enhancement of respiration rate has also been reported in Fe-deficient roots (Espen et al., 2000). Because the hydroponic cultures used in this study were continuously aerated, the transcriptional induction of enzymes associated with anaerobic metabolism was probably not a result of hypoxia. Rather, these results were interpreted as an attempt to increase energy production through oxidative phosphorylation. Because heme synthesis in the absence of Fe was not possible, regulation by de novo synthesis of electron transport proteins could not occur in roots. Induction of fermentation reactions would lead to an increased NADH consumption and maintain carbon flow and energy production by glycolytic reactions.
In Fe-deficient roots, an increase in dark CO2 fixation has been reported frequently, beginning with Rhoads and Wallace (1960). PEP carboxylase activity has also been shown to be induced in Fe-deficient roots (De Nisi and Zocchi, 2000; Lopéz-Millán et al., 2000). Increased PEP carboxylase activity would lead to increased synthesis of oxalacetate and malate that has been shown to occur in roots of Fe-deficient strategy I plants. The microarray data presented here confirmed these reports (Fig. 3). The PEPCase was shown to be regulated posttranscriptionally in C4 and CAM plants (Chollet et al., 1996). This may explain an IF of only 0.46 in roots after 3 d of −Fe, whereas enzyme activities were induced about 4-fold in Fe-deficient cucumber roots (De Nisi and Zocchi, 2000) and up to 60-fold in sugar beet root tips after 10 d of Fe deficiency (Lopéz-Millán et al., 2000).
Despite the induction of PEPCase expression in roots, ultimately the root must import carbon for energy. This increased demand can only be met by export from the shoots. In shoots, the IF in the activation phase of glycolysis was increased for hexokinase, Glc-6-phosphate isomerase, aldolase, and triose phosphate isomerase. In principle, these changes are consistent with either an increased carbon flow toward gluconeogenesis or glycolysis. The lack of change in IF for the anaplerotic step catalyzed by the PEP carboxykinase (Fig. 5) and in the IF for Suc phosphate synthase does not argue for an increased flow through gluconeogenesis in shoots. In fact, the development of chlorosis after 3 d of Fe-deficient growth and the repression of numerous photosynthesis-related EST sequences might indicate an increased respiration in shoots. However, the induction of cDNA clones encoding the UDP-Glc pyrophosphorylase, the H+-Suc symporter, the H+-ATPase (Fig. 3 H), the phosphate translocator, and α-amylase would lead to mobilization of starch and to export of carbon from shoots. This export would be needed to fuel the increased carbon demand in roots resulting from an anaerobic respiration.
CONCLUSIONS
Microarray techniques revealed changes of expression level due to Fe deficiency and have allowed insights into the transcriptional regulation of most enzymes in glycolysis, the TCA cycle, and the OPP pathway. These data indicated that previously observed increase in respiration activity in response to Fe deficiency involved transcriptional regulation of several genes encoding metabolic enzymes. The dramatic induction in expression for enzymes usually involved in anaerobic respiration was interpreted as an effect of Fe deficiency on energy production through oxidative phosphorylation. To maintain carbon flow through glycolysis under conditions of decreased electron transport, oxidation of NADH would be necessary.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis cv Landsberg erecta was grown under controlled conditions in a growth chamber (day/night regime of 10 h, 21°C/14 h, 18°C, photon flux density of approximately 300 μmol−2 s−1, relative humidity 75%). The nutrient solution was composed of KNO3 (3 mm), MgSO4 × 7 H2O (0.5 mm), CaCl2 × 6 H2O (1.5 mm), K2SO4 (1.5 mm), NaH2PO4 × 2 H2O (1.5 mm), H3BO3 (25 μm), MnSO4 × 4 H2O (1 μm), ZnSO4 × 7 H2O (0.5 μm), (NH4)6Mo7O24 (0.05 μm), CuSO4 × 5 H2O (0.3 μm), and Fe-EDTA (40 μm) with pH adjusted to 6 with KOH (Schmidt, 1994). Thirty or 32 d after sowing, Fe was removed from the media for 1, 3, or 7 d. Shoots and roots were separated when harvesting the plants at the end of the light period. From Fe-sufficient and -deficient treatments, a minimum of 40 plants was pooled for RNA preparation to minimize biological variation. The Fe3+-chelate reductase activity was determined as a measure of the response to Fe deficiency as described by Moog et al. (1995).
RNA Preparation and Radiolabeling
Total RNA was extracted from shoots and roots (RNeasy Midi Kit, Qiagen GmbH, Hilden, Germany). For reverse transcription and radiolabeling, 10 μg total RNA was used (SuperScript II, GibcoBRL, Karlsruhe, Germany; [33P]CTP, Amersham Pharmacia, Freiburg, Germany). After transcription, RNA was hydrolyzed with NaOH (0.25 N) and neutralized with HCl (0.2 N) and sodium phosphate buffer (40 mm, pH 7.2). Labeling efficiency was controlled by scintillation countering (LS6500, Beckman, Munich) after removal of unincorporated oligonucleotides by Sephadex G-50 chromatography (NICK Columns, Amersham Pharmacia).
Reference and Complex Hybridization
A set of 16,128 cDNA clones from the Michigan State University (East Lansing) collection, characterized by EST analysis (Newman et al., 1994), was provided by the Arabidopsis Biological Resource Center (Columbus, OH). The cDNA was amplified by PCR using LacZ-specific primers (forward LacZ1 5′ GCTTCCGGCTC GTATGTTGTGTG 3′ and reverse LacZ2 5′ AAAGGGGGATGTGCTGCAAGGCG 3′). The PCR products were spotted automatically onto nylon membranes (Biogrid, Biorobotics, Cambridge, UK; Nytran Supercharge, 22.2 × 22.2 cm, Schleicher and Schüll, Dassel, Germany). PCR products were not checked on an agarose gel for contamination before spotting. To normalize the amount of spotted cDNA, a reference hybridization for each filter was carried out using [33P]-labeled PCR product-specific primer (T4 polynucleotide kinase, New England Biolabs, Beverly, MA; [33P]ATP, Amersham Pharmacia; 5′ TTCCCAGTCACGA 3′). The filters were hybridized at 5°C overnight and washed for 40 min at 5°C in SSarc (4× SSC, 7% [v/v] Sarcosyl NL30, and 4 μm EDTA). Filters were exposed for 16 h on imaging plates and detected with a phosphorimager (BAS-1800, Fuji, Tokyo). Radioactivity was removed from filters by washing two times in SSarc at 65°C for 30 min. After prehybridization for 2 h at 65°C in Church buffer (7% [w/w] SDS, 1 mm EDTA, pH 8.0, and 0.5 m sodium phosphate, pH 7.2) containing salmon sperm DNA (100 ng ml−1, Roth, Carl GmbH & Co, Karlsruhe, Germany), filters were hybridized with the labeled cDNA probe at 65°C for 36 h. Washing steps were carried out at 65°C for 20 min each with 1× SSC, 0.1% (w/v) SDS, 4 mm Na2PO4 (pH 7.2); 0.2× SSC, 0.1% (w/v) SDS, 4 mm Na2PO4 (pH 7.2); and 0.1× SSC, 0.1% (w/v) SDS, 4 mm Na2PO4 (pH 7.2). The filters were exposed on imaging plates for 16 h and signals were detected using a phosphorimager (BAS-1800 II, Fuji) followed by stripping for 1 h at 65°C (0.1% [w/v] SDS and 5 mm Na2PO4, pH 7.2) as above. Hybridization of each filter was repeated three times with a newly synthesized and labeled cDNA probe of the corresponding RNA pool.
Data Analysis
For data analysis, the signal intensities of the reference and test hybridization were quantified using the software Arrayvision (Imaging Research Inc., Haverhill, UK). A predefined grid, determining the area of signal quantification, was manually optimized to ensure correct signal recording. The quantified signal, defined as photostimulated luminescence mm−2, was linked to the corresponding cDNA clone ID in the database.
The cDNA on the filter were arranged as 4 × 4 arrays, each containing seven doubly spotted clones, a human gene (desmin) to control nonspecific hybridization (not used in this study), and an empty field to determine specific local background (LBx). In this manner, LB was determined at 2,304 positions on each filter. To improve comparability of different hybridizations, the quantified signals were normalized. Normalization of the total radioactivity bound to the nylon membrane was conducted according to the formula (S. Kloska, B. Essigmann, and T. Altmann, unpublished data):
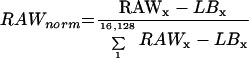 |
1 |
where RAWx was the activity detected at a specific position “x” on the filter and corresponded to a unique cDNA clone and LBx was the local background corresponding to RAWx.
To correct for the amount of spotted cDNA (REF) on the filter andfor the spotting and PCR efficiency, the signals were evaluated as follows:
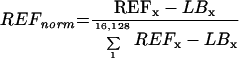 |
2 |
where REFx is the activity of a unique clone derived from the reference hybridization and LBx is the local background for the reference hybridization corresponding to REFx. The quotient of RAWnorm and REFnorm was defined as REL.
To identify cDNA clones that were differentially expressed under Fe deficiency, the REL from control (+Fe) and test (−Fe) roots and shoots were compared at 1, 3, and 7 d of Fe-deficient growth. To determine differential expression, an IF for each cDNA clone was calculated from the REL in control (RELT) and test (RELC) arrays:
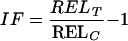 |
3 |
An IF > 1 indicates induction and IF < 0 indicates repression. Single values of the doubly spotted cDNA clones as well as repetition of hybridization were treated individually to minimize signal variation. Thus, six IF were obtained, from which a median was calculated. Means of the REL were used to confirm expression levels; cDNA clones with a REL mean < 0.1 were not analyzed further.
Possible functions of ESTs were obtained via clone identification numbers or sequences from online databases (TIGR Arabidopsis Gene Index, Institute for Genomic Research, Rockville, MD; The Arabidopsis Information Resource, Stanford, CA; and BLAST, National Center for Biotechnology Information, Bethesda, MD).
Throughout this manuscript, mention of EST clone ID has not been made. RAW data of all experiments as well as most induced clones shown in Figure 2 are available from our website (http://www.biologie.hu-berlin.de/˜botanik).
ACKNOWLEDGMENTS
We gratefully acknowledge Prof. H.-P. Herzel and Dieter Beule for their advice, encouragement, and support with the analysis of the data. We thank Sabine Fischer, Jeane Heyd, Peggy Lange, and Susanne Olstowski-Jacoby for their expert technical assistance.
Footnotes
This work was supported in part by the Deutsche Forschungsgemeinschaft (to T.A. and T.J.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010191.
LITERATURE CITED
- Alcaraz CF, Martinez-Sanchez F, Sevilla F, Hellin E. Influence of ferredoxin levels on nitrate reductase activity in iron deficient lemon leaves. J Plant Nutr. 1986;9:1405–1413. [Google Scholar]
- Chaney RL, Brown JC, Tiffin LO. Obligatory reduction of ferric chelates in iron uptake by soybeans. Plant Physiol. 1972;50:208–213. doi: 10.1104/pp.50.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- Dell'Orto M, Santi S, De Nisi P, Cesco S, Varanini Z, Zocchi G, Pinton R. Development of Fe-deficiency responses in cucumber (Cucumis sativus L.) roots: involvement of plasma membrane H(+)-ATPase activity. J Exp Bot. 2000;51:695–701. [PubMed] [Google Scholar]
- De Nisi P, Zocchi G. Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. J Exp Bot. 2000;51:1903–1909. doi: 10.1093/jexbot/51.352.1903. [DOI] [PubMed] [Google Scholar]
- De Vos CR, Lubberding HJ, Bienfait HF. Rhizosphere acidification as a response to iron deficiency in bean plants. Plant Physiol. 1986;81:842–846. doi: 10.1104/pp.81.3.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:223–250. doi: 10.1146/annurev.arplant.48.1.223. [DOI] [PubMed] [Google Scholar]
- Eckhardt U, Mas Marques A, Buckhout TJ. Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol. 2001;45:437–448. doi: 10.1023/a:1010620012803. [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Feit J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espen L, Dell'Orto M, De Nisi P, Zocchi G. Metabolic responses in cucumber (Cucumis sativus L.) roots under Fe-deficiency: a 31P-nuclear magnetic resonance in-vivo study. Planta. 2000;210:985–992. doi: 10.1007/s004250050707. [DOI] [PubMed] [Google Scholar]
- Fox TC, Guerinot ML. Molecular biology of cation transport in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:669–696. doi: 10.1146/annurev.arplant.49.1.669. [DOI] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF. Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem J. 1996;15:67–73. doi: 10.1042/bj3180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritch A, Herbik A, Balzer H-J, Ganal M, Stephan UW, Bäumlein H. A root-specific iron-regulated gene of tomato encodes a lysyl-tRNA-synthetase-like protein. Eur J Biochem. 1997;244:310–317. doi: 10.1111/j.1432-1033.1997.00310.x. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious and not readily available. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A, Giritch A, Horstmann C, Becker R, Balzer H-J, Bäumlein H, Stephan UW. Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol. 1996;111:533–540. doi: 10.1104/pp.111.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand DF. Lipoxygenases. Physiol Plant. 1989;76:249–253. [Google Scholar]
- Itai R, Suzuki K, Yamaguchi H, Nakanishi H, Nishiyawa H-K, Yoshimura E, Mori S. Induced activity of adenine phosphoribosyltransferase (APRT) in iron-deficient barley roots: a possible role for phytosiderophore production. J Exp Bot. 2000;51:1179–1188. [PubMed] [Google Scholar]
- Landsberg EC. Transfer cell formation in the root epidermis: a prerequisite for Fe-efficiency? J Plant Nutr. 1982;5:415–432. [Google Scholar]
- Landsberg EC. Function of rhizodermal transfer cells in the Fe stress response mechanism of Capsicum annuum L. Plant Physiol. 1986;82:511–517. doi: 10.1104/pp.82.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopéz-Millán AF, Morales F, Andaluz S, Gogorcena Y, Abadía A, De Las Rivas J, Abaía J. Responses of sugar beet roots to iron deficiency: changes in carbon assimilation and oxygen use. Plant Physiol. 2000;124:885–897. doi: 10.1104/pp.124.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberding HJ, de Graf FHJM, Bienfait HF. Ferric reducing activity in roots of Fe-deficient Phaseoulus vulgaris: source of reducing equivalents. Biochem Physiol Pflanzen. 1988;183:271–276. [Google Scholar]
- Machold O. Einfluss der ernährungsbedingungen auf den zustand des eisens in den blättern, den chlorophyllgehalt und die katalase-, sowie peroxidaseaktivität. Flora. 1968;159:1–25. [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. London: Academic Press; 1995. [Google Scholar]
- Moog PR, Brüggemann W. Iron reductase systems on the plant plasma membrane: a review. Plant Soil. 1994;165:241–260. [Google Scholar]
- Moog PR, van der Kooij TAW, Brüggemann W, Schiefelbein JW, Kuiper PJC. Responses to iron deficiency in Arabidopsis thaliana: the turbo iron reductase does not depend on the formation of root hairs and transfer cells. Planta. 1995;195:505–513. doi: 10.1007/BF00195707. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabotti G, De Nisi P, Zocchi G. Metabolic implications in the biochemical responses to iron deficiency in cucumber (Cucumis sativus L.) roots. Plant Physiol. 1995;107:1195–1199. doi: 10.1104/pp.107.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads WA, Wallace A. Possible involvement of dark fixation of CO2 in lime-induced chlorosis. Soil Sci. 1960;89:248–256. [Google Scholar]
- Robinson NJ, Procter CM, Conolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Römheld V, Müller C, Marschner H. Localization and capacity of proton pumps in roots of intact sunflower plants. Plant Physiol. 1984;76:603–606. doi: 10.1104/pp.76.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano K. Revision of biochemical pH-stat: involvement of alternative pathway metabolism. Plant Cell Physiol. 1998;39:467–473. [Google Scholar]
- Schmidt W. Root-mediated ferric reduction: responses to iron deficiency, exogenously induced changes in hormonal balance and inhibition of protein synthesis. J Exp Bot. 1994;45:725–731. [Google Scholar]
- Schmidt W. Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol. 1999;141:1–26. [Google Scholar]
- Schmidt W, Bartels M. Formation of root epidermal transfer cells in Plantago. Plant Physiol. 1996;110:217–225. doi: 10.1104/pp.110.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Tittel J, Schikora A. Role of hormones in the induction of Fe deficiency responses in Arabidopsis root. Plant Physiol. 2000;122:1109–1118. doi: 10.1104/pp.122.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchhardt J, Beule D, Malik A, Wolski E, Eickhoff H, Lehrach H, Herzel H. Normalization strategies for cDNA microarrays. Nucleic Acids Res. 2000;28:E47. doi: 10.1093/nar/28.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla F, del Rio LA, Hellin E. Superoxide dismutases from a citrus plant: presence of two iron-containing isoenzymes in leaves of lemon trees (Citrus limonum L.) J Plant Physiol. 1984;116:381–387. doi: 10.1016/S0176-1617(84)80130-3. [DOI] [PubMed] [Google Scholar]
- Silverthorne J, Tobin EM. Post-transcriptional regulation of organ-specific expression of individual rbc mRNAs in Lemna gibba. Plant Cell. 1990;12:1181–1190. doi: 10.1105/tpc.2.12.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller SC, Kaufman LS, Thompson WF, Briggs WR. Specific mRNA and rRNA levels in greening pea leaves during recovery from iron stress. Plant Physiol. 1987;84:409–414. doi: 10.1104/pp.84.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller SC, Terry N. Limiting factors of photosynthesis II: iron stress diminishes photochemical capacity by reducing the number of photosynthetic units. Plant Physiol. 1980;65:121–125. doi: 10.1104/pp.65.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin S, Abián J, Sánchez-Baeza F, Peleato JL, Abadia A, Gelpi E, Abadia J. Riboflavin 3′- and 5′- sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris) J Biol Chem. 1993;268:20958–20965. [PubMed] [Google Scholar]
- Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa N-K, Yoshimura E, Mori S. Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol. 1998;116:725–732. doi: 10.1104/pp.116.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder TL, Nishio JN. Early iron deficiency stress response in leaves of sugar beet. Plant Physiol. 1995;108:1487–1494. doi: 10.1104/pp.108.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]