Abstract
Previous studies using purified RNA polymerase from mustard (Sinapis alba) chloroplasts showed control of transcription by an associated protein kinase. This kinase was found to respond to reversible thiol/disulfide formation mediated by glutathione (GSH), although at concentrations exceeding those thought to exist in vivo. In the present study, several lines of evidence are presented to substantiate the functioning of this regulation mechanism, also in vivo: (a) Studies on the polymerase-associated transcription kinase revealed that at appropriate ATP levels, GSH concentrations similar to those in vivo are sufficient to modulate the kinase activity; (b) GSH measurements from isolated mustard chloroplasts showed considerable differences in response to light intensity; (c) this was reflected by run-on transcription rates in isolated chloroplasts that were generally higher if organelles were prepared from seedlings incubated under high-light as compared with growth-light conditions; (d) the notion of a general transcriptional switch was strengthened by in vitro experiments showing that the kinase not only affects the transcription of a photosynthetic gene (psbA) but also that of a non-photosynthetic gene (trnQ); and (e) the polymerase-kinase complex revealed specific differences in the phosphorylation state of polypeptides depending on the light intensity to which the seedlings had been exposed prior to chloroplast isolation. Taken together, these data are consistent with GSH and phosphorylation-dependent regulation of chloroplast transcription in vivo.
Interest in chloroplast molecular biology has focused to a large extent on two main aspects: (a) These organelles are the important sites of photosynthesis and other biosynthetic key reactions (Aro and Andersson, 2001), and (b) they have their own genes and significantly contribute to cellular gene expression in response to environmental conditions (Bogorad and Vasil, 1991; Sugita and Sugiura, 1996). An integrating feature that has become increasingly recognized is the close physical proximity and functional relationship between photosynthetic electron transport and chloroplast gene expression. Both processes were found to be interdependent, suggesting two-way signaling mechanisms including sensor(s) of photosynthetic electron transport and redox regulation (Allen, 1993). This is strikingly evident under situations such as photostress at high-light (HL) intensity, which results in accelerated turnover rates of photosynthetic (reaction center) proteins (Aro et al., 1993). Usage of specific inhibitors of photosynthetic electron transport such as 3-(3,4-dichlorophenyl)-1,1-dimethylurea (Trebst, 1980) has provided further support for the notion that chloroplast gene expression is under redox control.
Redox regulation of plastid gene expression was first demonstrated to play a role in the case of translation initiation in Chlamydomonas reinhardtii. This process was shown to depend on the activity state of a redox-responsive oligomeric protein complex which is capable of binding to the 5′-untranslated region of psbA mRNA (Danon and Mayfield, 1994). With the identity and function of its components now becoming fully resolved, this mRNA binding complex can serve as a paradigm for redox-regulatory mechanisms in eukaryotic photosynthetic organisms (Bruick and Mayfield, 1999; Trebitsh et al., 2000). It has become clear more recently, however, that other steps in chloroplast gene expression are subjected to photosynthetic redox control as well. These steps include translation elongation (Kuroda et al., 1996; Kettunen et al., 1997; Zhang et al., 2000), RNA degradation (Liere and Link, 1997; Salvador and Klein, 1999), and RNA splicing (Deshpande et al., 1997).
Available evidence suggests that transcription, the very first step in chloroplast gene expression, is also under redox control. Pool sizes of specific plastid RNAs respond to light intensity in vivo (Kettunen et al., 1997). In addition, the transcription rate of chloroplast genes has been shown to be affected by the spectral quality of light (photosystem I versus photosystem II excitation; Deng et al., 1989; Pfannschmidt et al., 1999) and electron transfer inhibitors (Pfannschmidt et al., 1999).
Plastids contain two different types of RNA polymerase, a single-subunit (“phage-type”) enzyme of nuclear origin (NEP) and a multisubunit (“bacterial-type”) enzyme with chloroplast-encoded catalytic subunits (plastid-encoded polymerase [PEP]; Maliga, 1998). As shown for mustard (Sinapis alba), the latter exists in two subforms, PEP-A and PEP-B, which can be distinguished on the basis of their size and complexity, their biochemical properties and sensitivity to transcription inhibitors, and their relative abundance in different plastid types (Pfannschmidt and Link, 1994). Whereas the rifampicin-sensitive PEP-B form predominates in etioplasts (and perhaps other non-green plastid types), the rifampicin-resistant PEP-A form is the major chloroplast RNA polymerase in functional chloroplasts (Pfannschmidt and Link, 1997). This latter enzyme seems to be responsible for the transcription of most plastid genes, including those for proteins involved in photosynthesis (Hajdukiewicz et al., 1997; Maliga, 1998).
PEP-A shares a common catalytic core with PEP-B, but contains a number of accessory proteins, the identification of several of which has recently been achieved by using protein sequencing techniques (Pfannschmidt et al., 2000). One polymerase-associated protein, possibly with a central regulatory function, was independently purified from mustard cotyledons and characterized as a Ser-/Thr-type protein kinase related to the CK2 family (Baginsky et al., 1997, 1999). This enzyme, named plastid transcription kinase (PTK), was found to respond to changes in thiol/disulfide redox state mediated by glutathione (GSH), indicating that it may serve as a component of a signal transduction pathway that connects photosynthetic electron transport (via the production of reducing equivalents) with chloroplast transcription (Link et al., 1997). Consistent with this idea, in vitro transcription reactions performed with the kinase-polymerase complex and a cloned DNA template carrying the psbA promoter were specifically up-regulated both by Ser/Thr kinase inhibitors and reduced GSH (Baginsky et al., 1999).
Although these in vitro data seemed to imply that PTK might be a redox-regulatory component of chloroplast transcription also in vivo, the objective of the current study was to address this question further from both sides. The approaches that were taken include a search for in vitro conditions that more closely reflect the in vivo situation, as well as in organello run-on transcription and phosphorylation assays to monitor changes in the chloroplast transcription machinery and its activity, induced by redox changes in the organelle. In vivo studies were carried out using HL illumination conditions (Aro et al., 1993; Karpinski et al., 1997) as a means for shifting the chloroplast redox state.
RESULTS
In Vitro Assay Conditions under Which the PTK Is Sensitive to Physiological Concentrations of GSH
In previous work we found GSH to be an effective inhibitor of the PTK at a concentration of 20 to 30 mm (Baginsky et al., 1999). On the other hand, the organellar GSH concentration was estimated to be within a range of only 1 to 10 mm (Noctor and Foyer, 1998), i.e. a concentration too low to be significantly inhibitory in vitro under the previously used assay conditions. To help resolve this apparent discrepancy, we investigated conditions under which the in vitro assay would more closely resemble the in vivo situation.
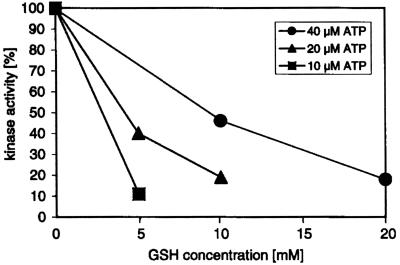
Of all parameters that were tested, alterations in the concentration of the ATP phosphodonor gave the most significant effect. As shown in Figure 1, using decreasing ATP levels (at a constant ratio of labeled to unlabeled ATP), we found an increasing inhibitory effect of GSH on the kinase activity. Approximately 90% inhibition was noticeable with 10 μm ATP in the presence of 5 mm GSH in this experiment, and at least 80% to 90% inhibition was found in two other independent experiments that were carried out with different PTK preparations (not shown). The GSH concentration of 5 mm that consistently gave strong inhibition of kinase activity in vitro is in good correlation with the expected in vivo concentration (1–10 mm; Noctor and Foyer, 1998). An inhibitory effect similar to that one found with casein (Fig. 1) was also observed with chloroplast proteins (Heparin-Sepharose column fractions; see “Materials and Methods”) as substrates for this kinase (data not shown). Taken together, these experiments are consistent with the notion that PTK could be a target for redox regulation in vivo at the GSH concentrations that exist in chloroplasts.
Figure 1.
Effect of reduced GSH on PTK activity in vitro. Heparin-Sepharose-purified PTK preparations were incubated with decreasing amounts of GSH at various ATP concentrations at 20°C for 20 min. Phosphorylation reactions were carried out using γ-32P-ATP and casein as a substrate, and the phosphorylated products were analyzed by SDS-PAGE and autoradiography. A representative experiment is shown and comparable data were obtained in two independent experiments with different PTK preparations.
GSH Content and Redox State in Chloroplasts from Mustard Seedlings Treated at Different Light Intensities
To further address the potential role of GSH as a regulator of chloroplast transcription in vivo, we sought conditions that would induce changes in the GSH concentration and/or redox state in the chloroplast. Because HL intensity is known to serve this function (Karpinski et al., 1997), mustard seedlings were exposed to 1,000 (HL) or only 50 (growth light [GL]) μmol photons m−2 s−1 for 3 h. Chloroplasts were then isolated, and the GSH concentrations and GSH to oxidized GSH (GSSG) ratios were determined. As judged from the equal chlorophyll (chl) content present in the HL and GL plastids on a similar plastid number basis, this treatment did not seem to result in appreciable photo-oxidative damage in the cotyledons. As can be seen in Table I, exposure to high irradiance resulted in a significant change in the GSH/GSSG ratio in chloroplasts, which was more than 3-fold higher upon the HL treatment of plants than in the GL conditions.
Table I.
GSH to GSSG ratios and GSH content in chloroplasts from differentially light-treated mustard seedlings
| Light | GSH/GSSG | GSH |
|---|---|---|
| nmol (mg chl)−1 | ||
| GL | 20 ± 13 | 0.74 ± 0.03 |
| HL | 73 ± 4 | 0.48 ± 0.03 |
Run-On Transcription with Chloroplasts from HL and GL Seedlings
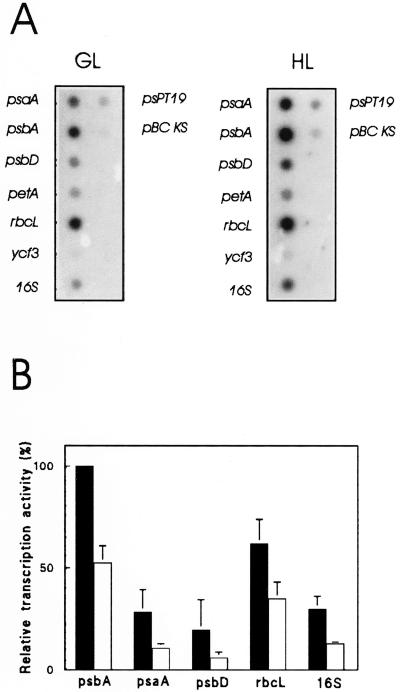
Next, we asked whether the light intensity-dependent changes in the plastid GSH/GSSG ratio were accompanied by changes in the transcriptional activity. To test this, we isolated chloroplasts from seedlings exposed for 3 h either to GL or HL illumination and carried out in organello run-on transcription in the presence of radiolabeled UTP. Chloroplast isolation typically yielded up to 90% intact plastids (not shown). Labeled transcripts were isolated and hybridized against chloroplast genes. Figure 2A shows a typical dot blot pattern obtained, and in Figure 2B the results from five independent run-on experiments are summarized. Based on equal amounts of total radioactivity used in each hybridization experiment, these data indicate a relative increase in transcriptional activity in HL- versus GL-treated seedlings for all the genes that were tested. It is interesting that this HL effect did not appear to be specific for photosynthesis-related genes because a similar induction of transcription was observed also for the 16S rRNA gene (Fig. 2) and for tRNA genes (trnS and trnG; data not shown).
Figure 2.
Chloroplast run-on transcription. A, Dot-blot autoradiograms following transcription using chloroplasts from 5-d-old mustard seedlings that were illuminated under GL or HL conditions for 3 h before isolation of chloroplasts. Newly synthesized transcripts of several representative genes for proteins related to photosynthesis as well as 16S ribosomal RNA sequences were analyzed. B, Relative transcription rates as measured by chloroplast run-on transcription assays (3 × 107 plastids per reaction). Gene-specific activities were calculated by normalizing the amounts of hybridized labeled run-on transcripts to the total transcriptional activity of the corresponding run-on reaction. The values are given as the percentage of the psbA transcription rate at HL (100%) in arbitrary units. The values are the mean of five independent experiments. Black bars represent samples isolated after HL and white bars those after GL treatment (3 h at 1,000 and 50 μmol photons m−2 s−1, respectively).
PTK Affects the PEP-A in Vitro Transcription Activity Driven by Promoters of Both Photosynthetic and Non-Photosynthetic Genes
We have shown previously that phosphorylation of the RNA polymerase leads to decreased transcriptional activity from the psbA promoter in vitro (Baginsky et al., 1999). To decide whether or not this inhibitory effect was restricted to this particular promoter, we tested the role of PTK in the same in vitro system, yet with the trnQ promoter, i.e. a chloroplast promoter for a non-photosynthetic gene (encoding tRNAGln; Sugita and Sugiura, 1996). The partially purified RNA polymerase was first treated with PTK in the presence of ATP and then re-isolated and further purified by glycerol gradient centrifugation. In control experiments, the polymerase was “mock” phosphorylated in the absence of ATP and then carried through the same additional purification. Silver staining revealed that the polypeptide patterns of the phosphorylated and mock-phosphorylated preparations were virtually identical, indicating that the PTK treatment did not affect the subunit composition of the RNA polymerase under these conditions (data not shown).
To investigate the effect of phosphorylation state on the transcriptional properties of the purified polymerase, we carried out in vitro transcription assays followed by nuclease S1 treatment to determine the amounts of correctly initiated transcripts at either the psbA (Fig. 3, lanes 1–5, left) or trnQ promoter (Fig. 3, lanes 6–10, right). In both sets of experiments, phosphorylation of the polymerase prior to RNA synthesis resulted in a decrease of specific transcript formation as indicated by the loss of correctly sized S1-resistant material (arrowheads). Although this effect was most pronounced for the trnQ transcripts (Fig. 3, lanes 9 and 10), the amount of specific psbA transcripts also was reduced when the polymerase was phosphorylated (Fig. 3, lanes 4 and 5). Taken together, the results obtained with the two different promoters support the notion (Fig. 2) that PTK-mediated control of plastid transcription is not restricted to a single class of chloroplast genes, but applies to both photosynthetic (psbA) and non-photosynthetic (trnQ) genes.
Figure 3.
Effect of phosphorylation on in vitro transcription of psbA (D1 reaction center protein of photosystem II; left) and trnQ (Gln-specific tRNAGln; right) genes by chloroplast PEP-A RNA polymerase. Heparin-Sepharose-purified fractions containing both the polymerase and associated kinase (PTK) activity were first incubated in phosphorylation buffer in the presence (○P; lanes 3, 5, 8, and 10) or absence of ATP (lanes 2, 4, 7, and 9). The polymerase was then further purified by glycerol gradient centrifugation and used to study the in vitro effect of PEP-A phosphorylation on the intensity of specific transcript formation from the chloroplast psbA and trnQ promoters. Following RNA synthesis, transcripts were hybridized to a 5′-labeled DNA probe containing the respective promoter and a portion of the transcribed region (see “Materials and Methods”). The hybrids were challenged with nuclease S1 and the S1-protected products were analyzed on sequencing gels (f, full-length DNA probe; s, transcripts of sizes expected for initiation at the corresponding promoters). Controls lacked either DNA (−DNA; lanes 2 and 3 and 7 and 8) or proteins (−pol; lanes 1 and 6).
Both PTK Activity and the Phosphorylation State of Polymerase-Associated Polypeptides Undergo Changes during HL Treatment
We next addressed the question of whether the HL activation of chloroplast transcription was correlated with a change in PTK activity, and with an altered phosphorylation state of the RNA polymerase.
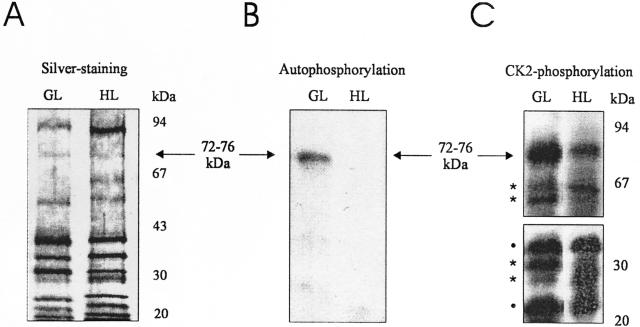
To compare PTK from chloroplasts of HL versus GL plants, we purified the enzyme up to the phosphocellulose step (see “Materials and Methods”). This intermediate purification step was chosen because on one hand, this material lacks many contaminating proteins still present after the preceding heparin-Sepharose step, and on the other hand, it still contains both the polymerase-bound (kinase-polymerase complex) and “free” (kinase complex) forms of PTK (Baginsky et al., 1997). The latter would have been lost in the final (glycerol gradient) step of the standard procedure for preparation of the highly purified kinase-polymerase complex, which therefore was omitted here. The phosphocellulose-bound material was eluted and analyzed for stained polypeptides and autophosphorylation activity in the presence of γ-32P-ATP using the endogenous chloroplast proteins as substrates (Fig. 4, A and B).
Figure 4.
Phosphorylation of chloroplast RNA polymerase-associated proteins from seedlings illuminated under HL and GL conditions. The enzyme complex was partially purified by heparin-Sepharose, followed by phosphocellulose chromatography. A, Polypeptides of chloroplast RNA polymerase preparations isolated from GL and HL seedlings, as revealed by SDS-PAGE and silver staining. B and C, In vitro phosphorylation assays in the presence of γ-32P-ATP. Polymerase-associated polypeptides from GL or HL plants were subjected to phosphorylation reactions using either the endogenous PTK activity (autophosphorylation; B), or by adding exogenous CK2 from rat liver (CK2-phosphorylation; C). The kinase-treated polypeptides were separated thereafter by SDS-PAGE and the dried gel was exposed to x-ray film. The horizontal arrows indicate the band in the 72- to 76-kD region, which is visible in A through C. Asterisks denote bands that differ in their intensity and dots denote bands with similar intensity in the GL and HL lanes in C.
As shown in Figure 4A, the polypeptide composition of the PTK preparations from HL as compared with GL seedlings was very similar, the only difference being the accumulation of an about 94-kD protein in HL-treated seedlings. Phosphorylation activity, however, could only be observed in the GL preparation (Fig. 4B), with no radioactive signal in the HL sample, even in overexposed films (not shown). In the GL sample, a single band was phosphorylated by the endogenous kinase following incubation of the preparation with labeled ATP (Fig. 4B). The size range of this band at approximately 72 to 76 kD matches that of the polymerase subunit(s) previously shown to be preferred PTK substrates in highly (glycerol gradient-) purified kinase-poly-merase complexes (Baginsky et al., 1999).
The apparent lack of 32P-incorporation into the HL sample suggested a lower or entirely lost PTK activity relative to that in the GL sample. However, an alternative explanation for this result was that the HL protein(s) might have existed e.g. in a more highly phosphorylated form prior to the labeling reaction and therefore were not accessible to in vitro phosphorylation by PTK. To distinguish between these two possibilities, the phosphocellulose-purified preparations were incubated with γ-32P-ATP in the presence of an exogenously supplied protein kinase. Because of the known similarity of PTK with CK2-type kinases (Baginsky et al., 1999), a purified CK2 from rat liver was chosen.
As is evident from the autoradiogram in Figure 4C, a labeled 72- to 76-kD band was generated both in the GL and HL samples following phosphorylation by the heterologous kinase. Although the intensity of this band was lower in the HL lane, its presence indicated that phosphorylation sites were accessible in the HL sample. Hence, the absence of labeled products in the HL autophosphorylation assay (Fig. 4B) seemed to reflect inactivation of PTK as a result of the HL treatment of seedlings.
In addition to the band at 72 to 76 kD, several other labeled bands were visible following phosphorylation by heterologous CK2 (Fig. 4C). A number of them had about equal intensity in both the HL and GL samples (e.g. those marked by dots at approximately 35 and 20 kD), whereas others (marked by asterisks) revealed reduced intensity in either the GL (at 65 and 29 kD) or HL samples (at 62 and 30 kD). These additional bands indicate a more stringent substrate specificity of the endogenous PTK as compared with the heterologous CK2 under the experimental conditions used here. The (differentially labeled) extra bands generated by CK2 may represent additional targets for phosphorylation control in vivo.
DISCUSSION
Although chloroplast transcription is considered not to be regulated to the same extent as various posttranscriptional steps in plastid gene expression (Deng and Gruissem, 1987; Stern et al., 1997), this view has become gradually modified as more molecular details have emerged (Baumgartner et al., 1993; DuBell and Mullet, 1995; Pfannschmidt et al., 1999). Perhaps one of the strongest arguments in favor of redox regulation of chloroplast transcription was initially based on prokaryotic systems, with focus on two-component (Stock et al., 2000) signaling mechanisms involving (His kinase) sensors and transcriptional regulators in bacteria (Allen, 1993). Despite demonstrations of two-component-type regulatory systems in eukaryotes (Chang and Stewart, 1998), however, this model has not yet been experimentally confirmed for higher plant chloroplasts.
Instead of the bacterial two-component His kinase system, there is now accumulating evidence supporting a scenario in which the major chloroplast RNA polymerase PEP-A is controlled by an associated Ser/Thr kinase of the CK2 type (Baginsky et al., 1997; K. Ogrzewalla and G. Link, unpublished data). The identified enzyme, named PTK, controls chloroplast transcription via phosphorylation of sigma-like transcription factors and several other polypeptides that are associated with the plastid transcription apparatus (Baginsky et al., 1999; Pfannschmidt et al., 2000).
PTK has been shown previously to be subject to thiol (SH) group regulation by GSH, i.e. a major redox mediator in chloroplasts (Karpinski et al., 1997; Noctor and Foyer, 1998). In the present work, the inhibitory effect of GSH on PTK activity was further investigated by searching for conditions in which PTK would respond to physiological concentrations of GSH. When the ATP concentration was reduced to 10 μm, PTK activity was severely inhibited by 5 mm GSH, a concentration that is within the range measured in the organelle (1–10 mm; Noctor and Foyer, 1998). Although ATP concentrations as low as 10 μm are seldom measured in chloroplasts (Stitt et al., 1980; Kuroda et al., 1992), it must be kept in mind that our experiments were performed in vitro, and that ATP is not likely to be the only factor that modulates the GSH effect in intact chloroplasts.
To examine the GSH effect in vivo, we chose HL treatment of seedlings as a tool to induce reduction of the chloroplast GSH pool (Table I). In addition of triggering changes in the organellar redox state, HL irradiance is also an elicitor of gene expression responses, and this system has been well characterized in terms of the associated metabolic changes, including those that involve redox-reactive compounds (Noctor and Foyer, 1998). In the present study, we have investigated transcription in chloroplasts isolated from HL- versus GL-treated plants. The in organello run-on assays (Fig. 2) suggest that the transcriptional activity of all genes investigated was increased during the HL treatment when compared with the GL controls. The observation of a global activation of transcription in response to HL is in line with a possible stress-related signaling within the chloroplast. To address the question of whether PTK might be involved in a global regulation of plastid transcription, we tested the effect of PTK-mediated phosphorylation on in vitro transcription using two different chloroplast promoters. As shown in Figure 3 for both the psbA and trnQ promoters, despite quantitative differences, PTK seems capable of affecting the transcription of more than a single class of plastid genes. This would be consistent with a global up-regulation of transcription under HL conditions in vivo, when PTK is inhibited by elevated GSH levels (Table I, Figs. 1–3).
In an attempt to obtain a link between redox conditions (Fig. 1, Table I), PTK activity (Fig. 1), and transcription rates (Fig. 2), we searched for possible targets for PTK among the polymerase-associated polypeptides. As shown in Figure 4A, a single labeled band in the 72- to 76-kD region was visible upon autophosphorylation of the partially purified kinase-polymerase complex. In highly purified PEP-A preparations (Pfannschmidt et al., 2000), this region contains at least three different polypeptides, one of which has recently been identified as the β′-core subunit of the polymerase (K. Ogrzewalla, A. Sickmann, S. Jung, H.E. Meyer, and G. Link, unpublished data). It remains to be clarified whether this one and/or any of the other polymerase-associated polypeptides in this region is the preferred substrate for PTK.
Phosphorylation of additional bands was noticeable in Figure 4C after incubation of the partially purified PEP-A fraction with exogenous CK2 from rat liver. This may mean that the substrate specificity of the heterologous kinase was less restrictive than that of the authentic enzyme. In an alternate manner, the latter may have lost the capacity to use these additional substrates, e.g. as a result of conformational changes and/or removal of additional factor(s) during the isolation procedure. The additional phosphorylation bands would also be consistent with the idea that, besides PTK, another (CK2-type) kinase also might be involved in vivo but was purified away in vitro. It is interesting to note that one of the labeled bands in Figure 4C migrates at 29 kD, i.e. the expected size of one of the chloroplast sigma-like factors (SLF29; Tiller et al., 1991), which was earlier shown to be subject for phosphorylation/dephosphorylation in vitro (Tiller and Link, 1993). Whether or not the plastid sigma factors are under SH group redox control in vivo is currently unknown. Further progress in addressing this question thus will be tightly linked to the identity and function of the transcription regulatory kinase(s) involved.
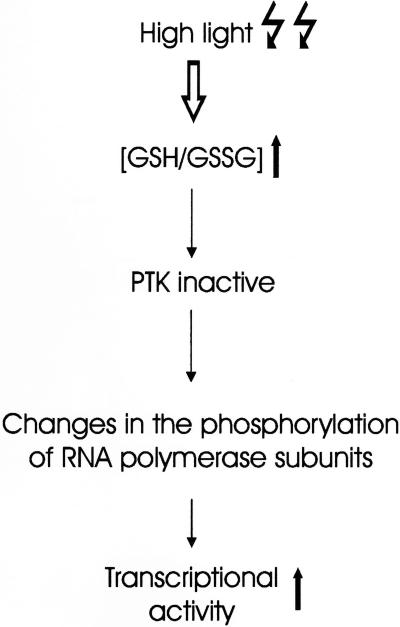
One important objective of the current work was to unravel the details of the signaling network connecting photosynthesis and gene expression, and to help in bridging gaps between the transcription data mostly obtained by in vitro experiments and the physiological situation in vivo. As is summarized in Figure 5, by combining the results from in vitro experiments with those obtained from intact chloroplasts, we suggest a mechanism by which changes in the SH group redox state of the organelle could be transduced into transcriptional responses. The use of transgenic plants with a modified GSH pool (Creissen et al., 1999) might prove very helpful for future experiments aiming to test the present model. A central component in this scheme is the redox-responsive transcription kinase PTK, the activity of which seems to reflect the irradiation conditions (HL versus GL in Fig. 4). The cloning and targeted manipulation of this enzyme in transgenic backgrounds might help open up new avenues in our understanding of redox regulation of chloroplast gene expression.
Figure 5.
Hypothetical model suggesting a mechanism connecting light-mediated changes in thiol redox state to transcriptional responses of chloroplast gene expression.
MATERIALS AND METHODS
Plant Material and Light Treatments
Mustard (Sinapis alba) seedlings were grown in continuous light at 50 μmol photons m−2 s−1 (GL) for 5 d. Thereafter, they were either maintained under GL conditions, or they were transferred to 1,000 μmol photons m−2 s−1 for the HL treatment for 3 h. The cotyledons were harvested on ice and then quickly processed as outlined below.
Assay for Protein Kinase Activity
PTK activity was assayed as described (Baginsky et al., 1999), except that hydrolized and partially dephosphorylated bovine casein (C-4765, Sigma, St. Louis) was used as an efficient protein substrate.
Chloroplast run-on Transcription
Chloroplast isolation for in organello run-on transcription was performed as described (Mullet and Klein, 1987) using 3 × 107 plastids per assay. DNA probes representing mustard chloroplast genes for detection of labeled transcripts were psaAB (insert of plasmid pSA244-EBH1.9; Dietrich et al., 1987), psbA (pSA452a; Link, 1981), psbD (H. Summer and G. Link, unpublished data; GenBank accession no. AF209094), petA (pSA120a; Dietrich et al., 1987), rbcL (pSA204; Link, 1981), ycf3 (pSAYCF3; Summer et al., 2000), rrn16 (pBSH895; Pfannschmidt and Link, 1997), trnS (pSA364-EX0.9; Neuhaus and Link, 1990), and trnG (pSA364 derivative pBSE996; Liere and Link, 1994). Gene-specific activities were quantified with a phosphoimager by subtracting the background signal of the corresponding plasmid vector, and by normalizing the value to the total transcriptional activity of the corresponding run-on reaction.
Determination of GSH Concentration from Chloroplasts
Chloroplasts were isolated and resuspended in 7.5 mm Na-ascorbate (in 125 mm Na-phosphate and 6.3 mm EDTA, pH 7.5). Chl was extracted from the chloroplast samples in 80% (v/v) buffered acetone {25 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH, pH 7.5}, quantitated according to Porra et al. (1989), and the samples were adjusted to the same chl concentration. All steps were done in a cold room (+4°C) using ice-cold solutions. Proteins were removed from the samples by centrifugation at maximum speed in a bench centrifuge (0°C; 15 min), boiling at 100°C for 4 min, and final recentrifugation of 15 min. The supernatant was used for determination of GSH concentration (Griffith, 1980). For measurements of total GSH amounts, 200 μL of sample (corresponding to 0.5 mg chl) were mixed with 700 μL of 0.3 mm NADPH and 100 μL of 6 mm 5–5′-dithiobis-(2-nitrobenzoic acid). The mix was warmed to 30°C, the reaction was started by adding 10 μL of GSH reductase (50 units mL−1), and the kinetics of A412 changes were recorded.
For measurements of GSSG, 200 μL of sample were thoroughly mixed with 700 μL of 0.3 mm NADPH, 100 μL of 6 mm 5–5′-dithiobis-(2-nitrobenzoic acid), and 4 μL of 2-vinylpyridine. The mix was kept at 25°C, and the kinetics of A412 changes were recorded at 0-, 20-, and 40-min time points, by warming the sample to 30°C and adding 10 μL of GSH reductase (50 units mL−1). All solutions were prepared in a buffer containing 125 mm Na-phosphate and 6.3 mm EDTA, pH 7.5. Standard curves (0, 10, 50, 100, and 200 ng) were done for GSSG (for GSSG measurements) and GSH (for total GSH measurements), and the results were fitted to the corresponding standard curve with the linear regression model of the FigP program (Biosoft, Cambridge, UK). The results are the average of two independent measurements for each light treatment.
In Vitro Transcription and S1 Analysis
Transcription reactions were performed with highly purified PEP-A RNA polymerase from mustard (Baginsky et al., 1999). Prior to the final glycerol gradient centrifugation step, the polymerase complex was phosphorylated by the endogenous PTK activity in the presence of unlabeled ATP, or it was mock phosphorylated in the absence of ATP. The DNA templates used in the transcription reactions were circular plasmids that carry chloroplast promoters and flanking sequences: pSA05/H120 carrying the psbA promoter (Baginsky et al., 1999), and pSA364-ET0.2 containing the trnQ promoter from mustard (Neuhaus and Link, 1990). RNA isolation, DNA-RNA solution hybridization, treatment of hybrids with nuclease S1 (Roche, Basel), and subsequent analysis of S1-resistant products on 6% (w/v) sequencing gels were as previously described (Baginsky et al., 1999). 5′-End-labeled DNA probes were generated by treatment with calf intestinal alkaline phosphatase (Roche), followed by incubation with γ-32P-ATP and polynucleotide kinase (New England Biolabs, Beverly, MA). The 120-bp BamHI/EcoRI fragment of pSA05/H120 served as the psbA probe and the 200-bp EcoRI/HindIII fragment of pSA364-ET02 as the trnQ probe (Pfannschmidt and Link, 1997).
Purification of RNA Polymerase and Phosphorylation Assays
Chloroplasts were isolated, purified by Suc density gradient centrifugation, and lysed as described (Tiller et al., 1991). In brief, the lysate was adjusted to 200 mm ammonium sulfate and then subjected to heparin-Sepharose chromatography. The column was washed with three volumes of the same buffer, bound proteins were subsequently eluted with a linear 0.2 to 1.6 m ammonium sulfate gradient, and the polypeptide composition of the fractions was analyzed by SDS-PAGE and silver staining. The fractions containing RNA polymerase were pooled, dialyzed against the column buffer containing 10% (v/v) glycerol, adjusted to 50 mm ammonium sulfate, and subjected to phosphocellulose chromatography. Following washing of the column with 3 volumes of the same buffer, bound proteins were eluted using a linear 0.05 to 1.6 m ammonium sulfate gradient. Fractions containing RNA polymerase were pooled and dialyzed against the column buffer containing 50% (v/v) glycerol.
For the autophosphorylation of polymerase-associated polypeptides by the endogenous PTK activity, samples after the phosphocellulose step were incubated in kinase buffer in the presence of γ-32P-ATP at 30°C for 30 min before they were subjected to SDS-PAGE and autoradiography as described (Baginsky et al., 1999). For the assessment of phosphorylation state, fractions were incubated with added CK2 from rat liver. Fifty-microliter reactions containing 60 units of enzyme (Promega, Madison, WI) in the buffer provided by the same supplier were used.
ACKNOWLEDGMENTS
We thank Karsten Ogrzewalla for initial help and discussion, and Silvia Hester for expert technical assistance.
Footnotes
This work was supported by the Academy of Finland (grant to E.-M.A.), by the Deutsche Akademische Austausdienst, by the Deutsche Forschungsgemeinschaft, and by the Fonds der Chemischen Industrie (to G.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010168.
LITERATURE CITED
- Allen JF. Redox control of gene expression and the function of chloroplast genomes: a hypothesis. Photosynth Res. 1993;36:95–102. doi: 10.1007/BF00016274. [DOI] [PubMed] [Google Scholar]
- Aro E-M, Andersson B. Regulatory Aspects of Photosynthesis. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2001. [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Link G. Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba) Plant Mol Biol. 1997;34:181–189. doi: 10.1023/a:1005802909902. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol Biol. 1999;39:1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development: evidence for selective stabilization of psbA mRNA. Plant Physiol. 1993;101:781–791. doi: 10.1104/pp.101.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L, Vasil IK. The Molecular Biology of Plastids. San Diego: Academic Press; 1991. [Google Scholar]
- Bruick RK, Mayfield SP. Light-activated translation of chloroplast mRNAs. Trends Plant Sci. 1999;4:190–195. doi: 10.1016/s1360-1385(99)01402-8. [DOI] [PubMed] [Google Scholar]
- Chang C, Stewart RC. The two-component system: regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Wellburn A. Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell. 1999;11:1277–1292. doi: 10.1105/tpc.11.7.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987;49:379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Tonkyn JC, Thornber JP, Gruissem W. Posttranscriptional control of plastid mRNA accumulation during adaptation of chloroplasts to different light quality environments. Plant Cell. 1989;1:645–654. doi: 10.1105/tpc.1.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande NN, Bao Y, Herrin DL. Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts. RNA. 1997;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- Dietrich G, Detschey S, Neuhaus H, Link G. Temporal and light control of plastid transcript levels for proteins involved in photosynthesis during mustard (Sinapis alba L.) seedling development. Planta. 1987;172:393–399. doi: 10.1007/BF00398669. [DOI] [PubMed] [Google Scholar]
- DuBell AN, Mullet JE. Differential transcription of pea chloroplast genes during light-induced leaf development transcription: continuous far-red light activates chloroplast transcription. Plant Physiol. 1995;109:105–112. doi: 10.1104/pp.109.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem. 1980;106:207–212. doi: 10.1016/0003-2697(80)90139-6. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen R, Pursiheimo S, Rintamäki E, Van Wijk KJ, Aro E-M. Transcriptional and translational adjustments of psbA gene expression in mature chloroplasts during photoinhibition and subsequent repair of photosystem II. Eur J Biochem. 1997;247:441–448. doi: 10.1111/j.1432-1033.1997.00441.x. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Inagaki N, Satoh K. The level of stromal ATP regulates translation of the D1 protein in isolated chloroplasts. Plant Cell Physiol. 1992;33:33–39. [Google Scholar]
- Kuroda H, Kobashi K, Kaseyama H, Satoh K. Possible involvement of a low redox potential component(s) downstream of photosystem I in the translational regulation of the D1 subunit of the photosystem II reaction center in isolated pea chloroplasts. Plant Cell Physiol. 1996;37:754–761. [Google Scholar]
- Liere K, Link G. Structure and expression characteristics of the chloroplast DNA region containing the split gene for tRNAGly (UCC) from mustard (Sinapis alba L.) Curr Genet. 1994;26:557–563. doi: 10.1007/BF00309950. [DOI] [PubMed] [Google Scholar]
- Liere K, Link G. Chloroplast endoribonuclease p54 involved in RNA 3′-end processing is regulated by phosphorylation and redox state. Nucleic Acids Res. 1997;25:2403–2408. doi: 10.1093/nar/25.12.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. Cloning and mapping of the chloroplast DNA sequences for two messenger RNAs from mustard (Sinapis alba L.) Nucleic Acids Res. 1981;9:3681–3694. doi: 10.1093/nar/9.15.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G, Tiller K, Baginsky S. Glutathione, a regulator of chloroplast transcription. In: Hatzios KK, editor. Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 125–137. [Google Scholar]
- Maliga P. Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci. 1998;3:4–6. [Google Scholar]
- Mullet JE, Klein RR. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus H, Link G. The chloroplast psbK operon from mustard (Sinapis alba L.): multiple transcripts during seedling development and evidence for divergent overlapping transcription. Curr Genet. 1990;18:377–383. doi: 10.1007/BF00318220. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol. 1994;25:69–81. doi: 10.1007/BF00024199. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet. 1997;257:35–44. doi: 10.1007/s004380050621. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.): integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur J Biochem. 2000;267:253–261. doi: 10.1046/j.1432-1327.2000.00991.x. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b with four different solvents: verification of the concentration of chlorophyll by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Salvador ML, Klein U. The redox state regulates RNA degradation in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol. 1999;121:1367–1374. doi: 10.1104/pp.121.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- Stitt M, Wirtz W, Heldt HW. Metabolite levels during induction in the chloroplast and extrachloroplast compartments of spinach protoplasts. Biochim Biophys Acta. 1980;593:85–102. doi: 10.1016/0005-2728(80)90010-9. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- Summer H, Pfannschmidt T, Link G. Transcripts and sequence elements suggest differential promoter usage within the ycf3-psaAB gene cluster on mustard (Sinapis alba L.) chloroplast DNA. Curr Genet. 2000;37:45–52. doi: 10.1007/s002940050007. [DOI] [PubMed] [Google Scholar]
- Tiller K, Eisermann A, Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.): evidence for three different transcription factors which resemble bacterial sigma factors. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Levitan A, Sofer A, Danon A. Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol Cell Biol. 2000;20:1116–1123. doi: 10.1128/mcb.20.4.1116-1123.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A. Inhibitors in electron flow. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- Zhang L, Paakkarinen V, Van Wijk KJ, Aro E-M. Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell. 2000;12:1769–1781. doi: 10.1105/tpc.12.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]