Abstract
Arabidopsis and other Brassicaceae produce an enormous diversity of aliphatic glucosinolates, a group of methionine (Met)-derived plant secondary compounds containing a β-thio-glucose moiety, a sulfonated oxime, and a variable side chain. We fine-scale mapped GSL-ELONG, a locus controlling variation in the side-chain length of aliphatic glucosinolates. Within this locus, a polymorphic gene was identified that determines whether Met is extended predominantly by either one or by two methylene groups to produce aliphatic glucosinolates with either three- or four-carbon side chains. Two allelic mutants deficient in four-carbon side-chain glucosinolates were shown to contain independent missense mutations within this gene. In cell-free enzyme assays, a heterologously expressed cDNA from this locus was capable of condensing 2-oxo-4-methylthiobutanoic acid with acetyl-coenzyme A, the initial reaction in Met chain elongation. The gene methylthioalkylmalate synthase1 (MAM1) is a member of a gene family sharing approximately 60% amino acid sequence similarity with 2-isopropylmalate synthase, an enzyme of leucine biosynthesis that condenses 2-oxo-3-methylbutanoate with acetyl-coenzyme A.
Glucosinolates are a prominent and diverse class of plant secondary metabolites in Arabidopsis and other genera of the Brassicaceae (Halkier, 1999; Rask et al., 2000). Their general structure has two common domains, a β-thio-Glc moiety and a sulfonated oxime, plus a variable side chain derived from one of several amino acids. When plant tissue is damaged, glucosinolates come into contact with endogenous β-thioglucosidases, known as myrosinases. The resulting aglycones are unstable and rearrange to form isothiocyanates, nitriles, thiocyanates, epithionitriles, and other substances (Bones and Rossiter, 1996).
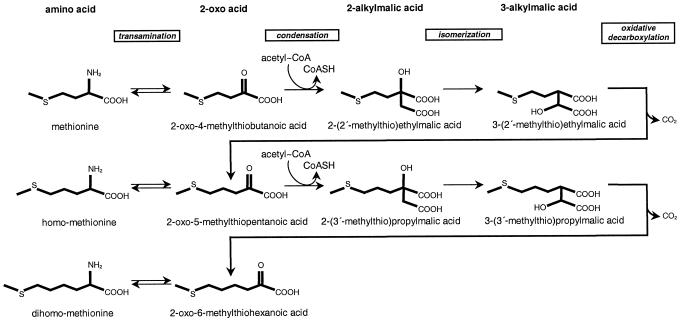
In Arabidopsis, most glucosinolates contain a variable side chain derived from chain-elongated derivatives of Met containing one to six additional methylene groups (Hogge et al., 1988). Although these additional methylene groups appear to arise from acetyl-coenzyme A (CoA), the side chain is not formed by direct fusion of C2 acetate units as in fatty acid biosynthesis but by an acetyl-CoA condensation/decarboxylation cycle that results in the net addition of one methylene group for each turn of the cycle (Graser et al., 2000). Based largely on in vivo studies with isotopically labeled tracers (Chisholm and Wetter, 1964; Matsuo and Yamazaki, 1964; Chapple et al., 1988; Haughn et al., 1991; Graser et al., 2000), the Met chain elongation pathway is thought to be initiated by the transamination of Met to form the corresponding 2-oxo acid (Chapple et al., 1990). This is followed by a three-step chain elongation cycle consisting of condensation of the 2-oxo acid with acetyl-CoA, isomerization, and oxidative decarboxylation (Fig. 1). The net result is a 2-oxo acid extended by one methylene group. The newly formed 2-oxo acid can be transaminated to form homo-Met or undergo additional cycles of chain elongation (Fig. 1). After elongation, the extended Met homologs enter the reaction sequence that creates the core structure of glucosinolates, i.e. conversion of the amino acid to an oxime, addition of a thiol moiety, formation of a β-thioester bond with Glc, and sulfonation of the oxime group (Halkier and Du, 1997). Finally, further side-chain modifications can occur.
Figure 1.
Postulated Met chain elongation pathway in glucosinolate biosynthesis. Shown are reaction steps and chemical structures for the first two rounds of chain elongation. The 2-oxo acid products are substrates for either transamination or subsequent condensation reactions.
Several Met-derived glucosinolates have significance in human health and agriculture. For instance, 4-methylsulfinylbutyl glucosinolate is hydrolyzed by myrosinase action to the isothiocyanate sulforaphane [1-isothiocyanato-4-(methylsulfinyl)butane], which is a strong inducer of phase II detoxification enzymes known to be of major importance in cancer protection (Zhang et al., 1992; Nastruzzi et al., 1996; Fahey et al., 1997). 2-Hydroxybut-3-enylglucosinolate, whose main hydrolysis product is 5-vinyloxazolidine-2-thione, has been identified as the goitrogenic component of rapeseed meal (Bradshaw et al., 1984). Glucosinolates and their breakdown products also may protect plants against pathogens (Doughty et al., 1991), regulate the feeding behavior of herbivores (Blau et al., 1978; Bodnaryk, 1991; Brown and Morra, 1995; Donkin et al., 1995; Giamoustaris and Mithen, 1995), and influence the oviposition preferences of insects (Reed et al., 1989; Du et al., 1995; Städler et al., 1995; Justus and Mitchell, 1996).
Arabidopsis ecotypes vary remarkably in their composition of Met-derived glucosinolates (Magrath et al., 1994; Kliebenstein et al., 2001) One major locus responsible for this variation was termed GSL-ELONG due to its ability to determine the degree of elongation of the side chain of Met-derived glucosinolates (Magrath et al., 1994). The predominant aliphatic glucosinolate in leaves of Landsberg erecta (Ler) is 3-hydroxypropyl glucosinolate, whereas in Columbia (Col-0) the leaves accumulate primarily 4-methylsulfinylbutyl glucosinolate. Thus, most aliphatic leaf glucosinolates in Ler have a basic side chain with three methylene groups (derived from homo-Met) and are termed “C3” glucosinolates, whereas those in Col-0 have a basic side chain with four methylene groups (derived from dihomo-Met) and are termed “C4” glucosinolates. This represents one and two cycles of chain elongation, respectively. In addition, lesser amounts of longer chain aliphatic glucosinolates are detectable in the leaves of both of these ecotypes. Six Col-0 mutants have been isolated that are altered in glucosinolate accumulation and two of these mutant lines, TU1 and TU5, were deficient in C4 glucosinolates (Haughn et al., 1991).
GSL-ELONG recently was mapped to a region of approximately 140 kb near the top of chromosome V (Campos et al., 2000). This region has two candidate genes with similarity to genes encoding 2-isopro-pylmalate synthase (IPMS), an enzyme that catalyzes a condensation reaction in Leu biosynthesis analogous to the condensation reactions between 2-oxo acids and acetyl-CoA occurring in Met chain elongation. Here, we use fine-scale mapping to identify which member of this gene family is responsible for the C3/C4 chain length variation of Met-derived glucosinolates. We also demonstrate that the Col-0 allele of this gene, named MAM1, codes for a methylthioalkylmalate synthase capable of condensing 2-oxo-4-methylthiobutanoic acid (OMTB) with acetyl-CoA to form 2-(2′-methylthio)ethylmalic acid (MTEM), the initial reaction in Met chain elongation. Moreover, we show that the mutant lines TU1 and TU5 harbor missense mutations in MAM1, reinforcing the conclusion that MAM1 plays a vital role in Met chain elongation in Arabidopsis.
RESULTS
IPMS-Like Genes in Arabidopsis
To aid in cloning the gene for the initial condensation step in the chain elongation of Met-derived glucosinolates (Fig. 1), the Arabidopsis genome was searched for genes with similarity to those encoding IPMS, which catalyzes a very similar condensation reaction in Leu biosynthesis. Four candidate genes were found, two of which are at the GSL-ELONG locus, a locus controlling glucosinolate side-chain length that maps near the top of chromosome V (Magrath et al., 1994; Campos et al., 2000). Based on the results described below, we have named these two genes MAM1 (encoding a methylthioalkylmalate synthase, see below) and MAM-L (MAM synthase-like). MAM1 and MAM-L are arranged head to tail on bacterial P1 clones of genomic Arabidopsis DNA T20O7 and MYJ24, respectively, and are separated by 11.5 kb of intervening sequence, as shown in Figure 2.
Figure 2.
Fine-scale mapping of GSL-ELONG. The DNA sequence of the GSL-ELONG region of chromosome V (outlined bar) was derived from the nonoverlapping sequences of K8E10, MRN17, T20O7, MYJ24, and MKD15 (GenBank accession nos. AB025618, AB005243, AB026660, AB006708, and AB007648). Vertical solid arrows labeled recMS1, rec6, recMS7, and recMS9 indicate the primer pair markers used for mapping the GSL-ELONG phenotype. The approximate sizes of intervals between these markers (horizontal arrows) are based on the Col-0 ecotype. Below, the region encompassing the MAM1 and MAM-L genes (thick horizontal arrows) is enlarged. Their approximate sizes and the size of the interval separating them are shown underneath. Additional primers and markers used to identify recombination points are indicated by vertical black arrows labeled Southex, 16Cm2, EDIT1-2a, 11t1, and EN-MS. The dashed vertical arrow indicates the recombination point which distinguishes functional effects of the MAM1 and MAM-L genes in family 5 (Table II).
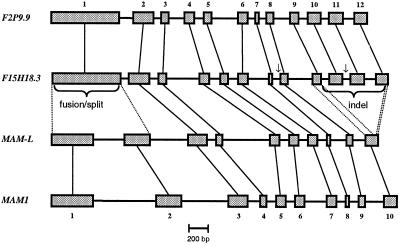
The other two sequences with high similarity to those encoding IPMS are on chromosome I. The first, designated F15H18.3 (accession no. AC013354.6), is located on top of chromosome I near the marker g15785. The second, F2P9.9 (accession no. AC016662.5), maps at the bottom of chromosome I close to the marker PAB5. The annotated exon-intron structure of these sequences was confirmed by sequence analysis of the respective cDNAs. Although the gene structure predicted by F2P9.9 was confirmed, two short insertions of 29 and 15 amino acids in the open reading frame (ORF) defined by F15H18.3 were shown to be artifacts of the gene prediction program. Thus, both the corrected F15H18.3 and F2P9.9 comprise 12 exons, whereas MAM1 and MAM-L consist of 10 exons as confirmed by sequencing of the corresponding cDNAs (Fig. 3). The MAM1 and MAM-L genes lack exons 10, 11, and part of 12 from F15H18.3 or F2P9.9. In addition, the first exon of the IPMS-like genes on chromosome I is homologous to both exons 1 and 2 of MAM1 and MAM-L on chromosome V (Fig. 3).
Figure 3.
Exon-intron structure of IPMS-like genes in Arabidopsis. Shown are exons (thick bars) and introns (thin bars) from start to stop of the ORF. Vertical thin lines connect homologous exons. Arrows indicate deviations from the predicted exon-intron structure of F15H18.3, as detected in the respective cDNAs.
The derived protein sequences from all four Arabidopsis IPMS-like genes contain an N-terminal extension of approximately 80 amino acids not present in bacterial IPMS sequences. The first 50 to 60 amino acids of this extension show the typical features of organellar targeting sequences, i.e. a high proportion of Ser and Thr residues and a low proportion of acidic residues (von Heijne et al., 1989; Emanuelsson et al., 1999). To better understand the evolutionary relationship of these genes, we performed a cluster analysis of the deduced amino acid sequences of IPMS and IPMS-like genes from Arabidopsis (four), tomato (Lycopersicon pennellii; two), and Synechocystis sp. PCC 6803 (one). The archaebacterial Methanococcus jannaschii sequence was used as an out-group. The analysis reveals that the Arabidopsis F15H18.3 and F2P9.9 are more closely related to one another and to the IPMSs from tomato than to either MAM1 or MAM-L. Because all plant species presumably possess at least one IPMS gene that encodes the enzyme functioning in Leu biosynthesis, F15H18.3 and/or F2P9.9 would appear to encode IPMS. Thus, MAM1 or MAM-L are the most likely candidates to be involved in glucosinolate chain elongation.
MAM1 Is Responsible for the C3/C4 Chain Length Variation of Met-Derived Glucosinolates
To determine which of the candidate sequences, MAM1 or MAM-L, controls C3/C4 chain length variation in the biosynthesis of Met-derived glucosinolates, a set of approximately 5,000 F2 progeny from a cross between the Col-0 wild type and the recombinant inbred line CL5 (Lister and Dean, 1993) was analyzed for recombination in the GSL-ELONG region. The CL5 line carries the Ler allele at GSL-ELONG (Mithen et al., 1995). Of approximately 4,600 plants (equal to 9,200 independent gametes) that were scorable, a total of 95 plants were recombinant between markers recMS1 and recMS9 (Fig. 2). These were further genotyped at markers rec6 and recMS7, which flank MAM1 and MAM-L. For 93 plants, the recombination point could be placed in intervals between recMS1 and rec6, rec6 and recMS7, or recMS7 and recMS9, as shown in Table I. On average, a recombination occurred every 2.2 kb. However, only two plants were recombinant in the 35.5-kb interval between markers rec6 and recMS7, indicating that recombination is suppressed around this region in Col-0 × CL5 (χ2 = 18.6851, degrees of freedom = 2, and P < 0.0001).
Table I.
Fine-scale mapping of the GSL-ELONG region
| Interval | Size | No. of Recombinants | Recombinants |
|---|---|---|---|
| kb | kb | ||
| recMS1–rec6 | 84.6 | 36 | 0.43 |
| rec6–recMS7 | 35.5 | 2 | 0.06 |
| recMS7–recMS9 | 89.1 | 55 | 0.62 |
| Undetermined | – | 2 | – |
| Total | 209.2 | 95 | 0.45 |
Approximately 4,600 F2 plants (equal to 9,200 independent gametes) were scored. The observed recombination frequency is 1%. Thus, the investigated interval equals 1 cM. Note that recombination in the interval comprising the GSL-ELONG locus (between markers rec6 and recMS7) is suppressed.
Leaf glucosinolates were analyzed in a subset of recombinant F3 plants confirmed homozygous in the GSL-ELONG region. Plants carrying the Col-0 alleles at MAM1 and MAM-L accumulated C4 glucosinolates, whereas plants carrying the Ler alleles at these genes formed predominantly C3 glucosinolates (Table II, Fig. 4). Glucosinolate profiles were unaffected by recombinations in either the region 5′ of MAM1 between markers recMS1 and rec6 (Table II, Families 6, 7 and 8) or the region 3′ of MAM-L between recMS7 and recMS9 (Table II, Families 1–4 and 9–14).
Table II.
Col-0 (C) and Ler (L) allelic state and side-chain length of the predominant glucosinolates in recombinant F3 families
| F3 Family No. | Allelic State 5′ of GSL-ELONG | Allelic State at MAM1 | Allelic State at MAM-L | Allelic State 3′ of GSL-ELONG | Predominant Glucosinolate Type |
|---|---|---|---|---|---|
| 1 | C | C | C | L | C4 |
| 2 | C | C | C | L | C4 |
| 3 | C | C | C | L | C4 |
| 4 | C | C | C | L | C4 |
| 5 | C | C | L | L | C4 |
| 6 | L | C | C | C | C4 |
| 7 | C | L | L | L | C3 |
| 8 | C | L | L | L | C3 |
| 9 | L | L | L | C | C3 |
| 10 | L | L | L | C | C3 |
| 11 | L | L | L | C | C3 |
| 12 | L | L | L | C | C3 |
| 13 | L | L | L | C | C3 |
| 14 | L | L | L | C | C3 |
F3 plants derived from Col-0 × CL5 F2 plants with recombination events between recMS1 and recMS9 were confirmed for homozygosity, and glucosinolates were analyzed. In general, several plants were analyzed per family. L, Plants carried the Ler-0 allele at the respective locus; C, plants harbored the Col-0 allele. C3, Predominant glucosinolates are derived from homo-Met; C4, plants accumulate glucosinolates derived from dihomo-Met. Note that plants from family 5 harbor a recombination between MAM-1 and MAM-L.
Figure 4.
HPLC chromatograms of glucosinolate profiles from Col-0 × CL5 F3 recombinant plants. Glucosinolates were extracted and separated as described in “Materials and Methods,” and identified by their retention times and UV spectra. Shown are signals from 3.5- to 10-min retention time during which time all of the glucosinolates eluted that varied in these crosses. Amounts are given in relative absorption units. The allelic state at MAM1 and MAM-L is indicated in the upper left corners. Specific glucosinolates are identified by numbers that correspond to Table III.
The two F2 plants with recombinations in the interval flanked by the markers rec6 and recMS7 were subjected to further analysis. One of these plants was recombinant between markers EN-MSrev and recMS7 (Fig. 2), and the other had recombined between markers 16Cm2 and EDIT1-2a (Fig. 2). Only the latter plant harbors a recombination point separating the MAM1 and MAM-L candidate genes. This genotype is homozygous Col-0 at recMS1, rec6 ,and for at least 73% of the MAM1 gene (Fig. 2), but heterozygous at MAM-L, recMS7, and recMS9. Leaves of this plant form predominantly C4 glucosinolates. Also, its F3 progeny, verified to be homozygous for this recombination (i.e. homozygous Ler at MAM-L, recMS7, and recMS9), accumulate C4 glucosinolates (Fig. 4, Table II, Family 5). Thus, the allelic state of MAM1 is responsible for the C3/C4 chain length variation of Met-derived glucosinolates.
The Phenotypes of TU1 and TU5 Are Associated with Mutations in MAM1
The mutant lines TU1 and TU5, derived from the Col-0 ecotype, carry allelic gsm1 mutations (Haughn et al., 1991). To confirm their phenotype, leaf glucosinolate content was analyzed and compared with Col-0 wild type. The HPLC chromatogram of the TU1 line showed greatly reduced levels of 4-methyl-sulfinylbutyl glucosinolate (Table III), and an absence of 4-methylthiobutyl, 5-methylsulfinylpentyl, and 6-methylsulfinyl glucosinolates (Table III), consistent with the original description. Coincident with these losses was an increase in the C3 glucosinolates, 3-methylsulfinylpropyl and 3-methylthiopropyl (Table III). A qualitatively similar glucosinolate profile was obtained from TU5 (Table III). This pattern of changes in glucosinolate chain length would be expected from a defect in the conversion of a C3 to a C4 glucosinolate precursor. To establish whether the gsm1 mutants and the Ler MAM1 locus were allelic, the leaf glucosinolate profiles were determined in the F1 generation of crosses between Ler and either TU1 or TU5. The absence of C4 glucosinolates in F1 progeny indicated a lack of complementation. Thus, the biosynthetic lesions in the conversion of C3 to C4 glucosinolate precursors in the mutants are the same as in the Ler ecotype, indicating that gsm1 is probably allelic with MAM1.
Table III.
Glucosinolate content (micromole per gram dry wt) in mutant and wild-type Arabidopsis
| No. | Glucosinolatea | Col-0 | TU1 (gsm1-1) | TU5 (gsm1-2) | Ler | Ler × TU1 |
|---|---|---|---|---|---|---|
| 1 | 2-MSO-ethyl | 0.0 | 0.8 (0.1) | 0.1 | 0.0 | 0.0 |
| 2 | 3-OH-propyl | 0.0 | 0.0 | 0.0 | 63.3 (3.3) | 14.5 |
| 3 | 3-MSO-propyl | 3.8 (0.2) | 16.1 (0.8) | 25.5 (1.8) | 1.7 (0.3) | 0.0 |
| 4 | 3-MT-propyl | 0.0 | 0.2 (0.0) | 0.3 (0.1) | 0.1 (0.0) | 0.0 |
| 5 | 4-MSO-butyl | 22.6 (1.6) | 0.4 (0.0) | 2.7 (0.1) | 0.4 (0.0) | 0.0 |
| 6 | 4-MT-butyl | 2.8 (0.2) | 0.0 | 0.4 (0.2) | 0.0 | 0.0 |
| 7 | 5-MSO-pentyl | 0.6 (0.0) | 0.0 | 0.0 | 0.0 | 0.0 |
| 8 | 6-MSO-hexyl | 0.2 (0.0) | 0.0 | 0.0 | 0.0 | 0.0 |
| 9 | 7-MSO-heptyl | 0.7 (0.1) | 0.5 (0.1) | 0.3 (0.0) | 0.4 (0.0) | 0.1 |
| 10 | 7-MT-heptyl | 0.2 (0.0) | 0.1 (0.0) | 0.0 | 0.2 (0.0) | 0.0 |
| 11 | 8-MSO-octyl | 4.1 (0.2) | 5.9 (0.3) | 4.6 (0.4) | 7.8 (0.6) | 1.9 |
| 12 | 8-MT-octyl | 0.4 (0.2) | 1.1 (0.1) | 0.3 (0.1) | 1.3 (0.1) | 0.1 |
| 13 | 4-OH-indol-3-ylmethyl | 0.3 (0.1) | 0.3 (0.0) | 0.0 | 0.1 (0.0) | 0.0 |
| 14 | Indol-3-ylmethyl | 6.8 (0.9) | 11.0 (0.7) | 4.4 | 5.9 (0.7) | 1.5 |
| 15 | 4-MO-indol-3-ylmethyl | 0.5 (0.0) | 0.6 (0.0) | 0.9 | 0.5 (0.0) | 0.3 |
| 16 | 1-MO-indol-3-ylmethyl | 3.7 (1.2) | 3.1 (1.0) | 0.1 | 1.6 (0.3) | 0.1 |
| 17 | 4-OH-butyl | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
ses are given in parentheses. Expanding leaves were harvested just prior to bolting and extracted as described in “Materials and Methods.” Each analysis consisted of leaves from five pooled plants. All lines were analyzed three to four times, except for TU5 and Ler × TU1, which were analyzed only once.
MSO, Methylsulfinyl; OH, hydroxy; MT, methylthio; MO, methoxy.
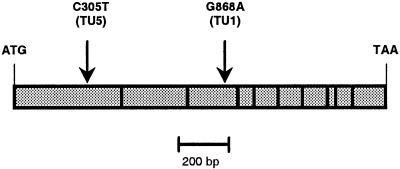
To confirm the allelism, cDNAs corresponding to the MAM1 and MAM-L ORFs of Col-0, TU1, and TU5 were sequenced. Although no differences were detected in the MAM-L gene, both of the mutant lines exhibited a single transition mutation in MAM1 (Fig. 5). The TU1 line had a base substitution of adenine for the wild-type guanine at position 868 relative to the start of the ORF (G868A). The TU5 line had a transition from cytosine to thymine at position 305 relative to the start (C305T). These base substitutions are consistent with the mutagenic properties of the alkylating reagent ethylmethane sulfonate used to generate the mutants.
Figure 5.
Missense mutations in MAM1 from mutant lines TU1 and TU5. The ORF of MAM1 is presented as a bar. Vertical lines within this bar show exon-exon borders. Nucleotide substitutions causing missense mutations are shown above the bar. Numbering refers to the nucleotide sequence.
Both base changes are predicted to cause missense mutations. In the gsm1-1 allele from TU1, the nucleotide change G868A would convert an Ala codon to a Thr (A290T) and introduce a polar residue within a stretch of hydrophobic amino acids. The gsm1-2 allele from TU5 is predicted to cause a change from a wild-type Ser to a Phe at position 102 (S102F). The fact that the gsm1 mutants with altered levels of C3/C4 glucosinolates both possess a nonconservative amino acid substitution in MAM1 supports the hypothesis that this gene controls the variation between C3 and C4 glucosinolates.
The Col-0 MAM1 Polypeptide Is Capable of Condensing a 2-Oxo Acid with Acetyl-CoA to Form a 2-Alkylmalate Derivative
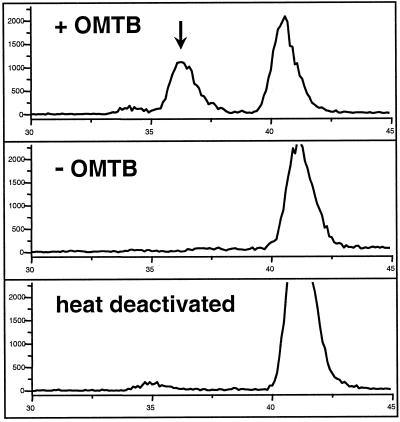
The initial reaction of the Met chain elongation pathway involves the condensation of a 2-oxo-acid derived from Met with acetyl-CoA to form a 2-methylthioalkylmalate derivative (Fig. 1). To demonstrate directly that the MAM1 gene encoded an enzyme capable of catalyzing such a reaction, the Col-0 allele was expressed in Escherichia coli. Bacterial cells containing an overexpression construct were disrupted by sonication and the resulting supernatant was assayed with [1-14C]acetyl-CoA and OMTB, the oxo-acid derived from the deamination of Met (Fig. 6). A novel peak was detected that was coincident with the retention time of an authentic standard of MTEM, the expected product of the condensation reaction (Fig. 1). This novel peak was absent in assays with heat-denatured enzyme or without the 2-oxo acid cosubstrate, and was also not detected in extracts from E. coli lacking the expression construct. Activity was maximal at pH 8.5 and required a divalent metal ion, such as Mn2+. The identity of the biosynthetic product was confirmed by liquid chromatography-mass spectrometry (LC-MS) analyses that gave a mass spectrum identical to that of the authentic standard. Assays of the MAM1 protein expressed from both Ler and the mutant line TU1, performed under identical conditions as assays for Col-0 MAM1, did not show this condensation activity. Thus, bacterial expression of MAM1 cDNAs showed that expression constructs with the sequence from Col-0, but not from Ler or TU1, encoded a protein capable of using acetyl-CoA and the 2-oxo-derivative of Met to catalyze the initial condensation reaction in the chain elongation of glucosinolates. This activity was also absent when MAM-L from Ler was expressed and assayed under the same conditions.
Figure 6.
Cell-free enzyme assay of E. coli expressing the MAM1 cDNA from Col-0. Cells were grown, extracted, and assayed with acetyl-CoA as described in “Materials and Methods.” Pictured are radioactivity detector traces of the HPLC separations of assay products from 30 to 45 min. The upper trace represents the complete assay, including [1-14C]acetyl-CoA and OMTB, and shows the production of MTEM (36.5 min), indicated by an arrow, and acetic acid (41 min), resulting from the hydrolysis of acetyl-CoA. The middle trace represents an assay without the cosubstrate OMTB, and the lower trace represents an assay with heat-denatured enzyme.
DISCUSSION
The Role of MAM1 in Met Chain Elongation
In recent years, an effort to map genes controlling glucosinolate variation in Arabidopsis has identified a locus on the upper arm of chromosome V that regulates the side chain length of Met-derived glucosinolates (Magrath et al., 1994). This locus, GSL-ELONG, controls the accumulation of glucosinolates with C3 versus C4 side chains. It was mapped to a 140-kb region encompassing two genes that had approximately 60% identity at the amino acid level with enzymes, such as isopropylmalate synthase and homocitrate synthase, that condense acetyl-CoA with 2-oxo acids (Campos et al., 2000). These two genes in the GSL-ELONG region (MAM1 and MAM-L) were found to be highly homologous and within 11.5 kb of each other. In this report, we have determined that one of these genes (MAM1) controls side-chain length variation in glucosinolate biosynthesis and encodes a condensing activity using acetyl-CoA and the 2-oxo derivative of Met as substrates. This is the first reaction of the Met chain elongation cycle and is closely analogous to the condensation of acetyl-CoA and 2-oxo-3-methylbutanoate in Leu biosynthesis.
Evidence for the role of MAM1 in Met chain elongation is based on mapping experiments in which an individual genotype was identified that had a recombination between the two IPMS-like genes in the GSL-ELONG region. Glucosinolate profile was controlled by the MAM1 locus rather than MAM-L. This finding was confirmed by the presence of independent missense mutations at the MAM1 locus of the two allelic mutant lines, TU1 and TU5, which also possessed altered glucosinolate chain length profiles. In both of these mutants, the MAM-L locus had a wild-type sequence. In addition, the mutant lines were unable to complement the naturally occurring lack of C4 glucosinolates in crosses with the Ler ecotype, suggesting that chain length control in both Ler and these mutants is mediated by a common locus.
Heterologous expression of the Col-0 MAM1 allele in E. coli revealed that this gene encoded an enzyme capable of condensing acetyl-CoA with the Met derivative, OMTB, to produce MTEM, the first step in the C2 to C3 elongation cycle of Met chain elongation. Although we surmise that the MAM1 enzyme can also catalyze the condensation step in the C3 to C4 extension cycle, the lack of commercial availability of the requisite substrate, 2-oxo-5-methylthiopentanoic acid, has so far precluded its direct demonstration. Nevertheless, the molecular and genetic evidence for the role of the Col-0 MAM1 allele in regulating the abundance of both C3 and C4 glucosinolates is compelling.
To date, our results do not yet suggest a candidate gene for the condensation step in the C2 to C3 elongation cycle in Ler and the TU1 and TU5 mutants, both of which accumulate C3 glucosinolates and so must perform this reaction. Heterologously expressed MAM1 from Ler, TU1 and TU5, along with MAM-L from Ler were not capable of catalyzing this activity when tested with the same substrates and under identical conditions to those that promoted the condensation reaction of MAM1 from Col-0. The other two IPMS-like genes in Arabidopsis have high similarity to IPMS-like genes from non-glucosinolate-forming species and are expected to be involved in Leu formation. However, these may also encode proteins capable of performing the initial condensation step in the C2 to C3 elongation cycle of glucosinolate biosynthesis, a possibility presently under investigation.
The accumulation of Met-derived C4 glucosinolates in Col-0 versus the accumulation of C3 glucosinolates in Ler is a result of a polymorphism in a gene encoding the condensation step of Met chain elongation. In principle, such a metabolic difference could also result from differences in other aspects of the biosynthetic pathway, such as the substrate specificity of the aminotransferase responsible for converting chain-extended 2-oxo acids to their corresponding amino acids after elongation, or the transport of intermediates (Haughn et al., 1991). However, the three independent experimental approaches taken here (mapping, mutant analysis, and heterologous expression) all indicate that the GSL-ELONG polymorphism in Arabidopsis is due solely to the presence or absence of the Col-0 MAM1 allele.
It has been assumed that a single series of chain extensions generates all of the chain-elongated variants of Met (Haughn et al., 1991). However, despite the dramatic change in C3, C4, C5, and C6 glucosinolates in the TU1 and TU5 mutant lines, the levels of C7 and C8 glucosinolates in these plants are largely unaffected. Thus, C7 and C8 Met-derived glucosinolates are likely to be formed by a pathway that is at least partially independent from that forming shorter chain glucosinolates. MAM-L and/or other, undescribed loci may encode the condensing activities involved in the formation of the longer chain glucosinolates.
Side-Chain Elongation and Glucosinolate Diversity
Met-derived glucosinolates in Arabidopsis can undergo one to six cycles of chain elongation resulting in the formation of C3 through C8 glucosinolates. Chain elongation greatly increases the range and chemical properties of possible glucosinolate structures and their hydrolytic derivatives. For example, upon myrosinase action longer chain glucosinolates might be expected to produce hydrolysis products that are much less polar and less volatile than those from shorter chain glucosinolates. Later side-chain modifications, including thiol oxidation, desulfation, hydroxylation, and esterification further increase the chemical diversity of glucosinolates produced from Met. Diversity might play a role in reducing the impact of herbivores that have evolved tolerance to a particular glucosinolate profile. Thus, the MAM1 locus and other genes that function in chain elongation may have evolutionary and ecological significance.
Recruitment of Glucosinolate Biosynthetic Genes from Primary Metabolism
Cluster analysis of IPMS-like sequences reveals that MAM1 and MAM-L are probably derived from ancestral IPMS genes. Therefore, following an initial gene duplication of IPMS, one of the resulting copies may have acquired a new capacity in secondary metabolism, whereas the other locus retained its function in Leu biosynthesis. Subsequent gene duplications gave rise to F15H18.3 and F2P9.9, on one hand, and MAM1 and MAM-L, on the other.
After IPMS, three additional enzymes are involved in the later steps of Leu biosynthesis: 2-isopropyl-malate isomerase (dehydratase), 3-isopropylmalate dehydrogenase, and Leu aminotransferase. These catalyze reactions analogous to the remaining steps in Met chain elongation: isomerization, oxidative decarboxylation, and transamination (Fig. 1). It is tempting to speculate that duplication of the remaining Leu biosynthetic genes gave rise to copies that acquired an analogous function in Met chain elongation. Leu biosynthesis in higher plants takes place in the chloroplast, and IPMS is situated in the thylakoids (Hagelstein and Schultz, 1993; Hagelstein et al., 1997). Because all of the IPMS-like sequences, F15H18.3, F2P9.9, MAM1, and MAM-L, have putative chloroplast targeting sequences and, furthermore, because biochemical studies have demonstrated that methylthioalkylmalate synthase activity is localized in the chloroplast (K. Falk and J. Gershenzon, unpublished data), it is likely that Met chain elongation also occurs at this subcellular site.
Fine-Scale Mapping as a Means to Dissect Function within Gene Clusters
When gene families are arranged in tightly linked clusters, identifying the function of individual genes is a major challenge. Members of gene families often share sufficient sequence similarity to prevent antisense or overexpression studies from successfully distinguishing between them. Although T-DNA or transposon-tagged mutants can provide ready access to individual members of a gene family, in the case of GSL-ELONG, screening of T-DNA insertion lines using populations available from the stock centers did not identify mutations in either MAM1 or MAM-L. Therefore, we chose to exploit naturally occurring variation between Arabidopsis ecotypes to map GSL-ELONG at a fine scale.
In a cross between the Col-0 wild type and the recombinant inbred line CL5, a set of 4,600 F2 plants (9,200 gametes) were scorable for recombination in the GSL-ELONG region. Because 1 cM equals approximately 200 kb in Arabidopsis, we expected to detect approximately one crossover per 2.3 kb in the 209-kb assayed region. The observed number of 95 recombinants (one crossover per 2.2 kb or 0.45 recombinants kb−1) is almost exactly the number expected from the average relationship between physical distance and recombination frequency, indicating that fine-scale mapping can be a powerful tool to separate functional effects of tightly linked members of gene families, especially in the era of high-throughput genetics and genomics. However, crossover density varied significantly across the assayed interval, indicating recombination suppression near GSL-ELONG (Table I). This might be caused by differences in the organization of this region in the respective ecotypes and will be the subject of subsequent studies.
Due to this recombination suppression, approximately 5,000 F2 plants were necessary to identify one recombinant separating the two adjacent genes of interest. Nevertheless, careful choice of F1 plants generated a whole set of nearly isogenic lines harboring recombinations in the vicinity of GSL-ELONG. These lines can be used for fine quantitative trait locus mapping of many related traits in laboratory or field conditions, including glucosinolate quantity and insect resistance. Fine-scale mapping alone reduced the interval for candidate genes to less than 35 kb. In combination with heterologous expression in E. coli and the analysis of appropriate mutants, the gene responsible for C3/C4 chain length variation was unequivocally identified.
MATERIALS AND METHODS
Fine-Scale Mapping of GSL-ELONG
Arabidopsis ecotype Col-0 was crossed with the recombinant inbred line CL5 (Lister and Dean, 1993). CL5 is Col-0 WT for about 70% of the genome but harbors the Ler allele at the GSL-ELONG locus. F1 plants were verified for heterozygosity. F2 plants were raised in 53 flats each containing 94 plants, in addition to Col-0 and Ler WT plants, grown at a density of 337 plants/m2 under 11.5-h/12.5-h light/dark cycles on a 1:3 (v/v) vermiculite:standard soil (Einheitserdenwerk, Fröndenberg, Germany) mix.
Total leaf DNA was isolated following a protocol modified from Edwards et al. (1991). Recombinants in the 209-kb region surrounding GSL-ELONG were identified with the primer pairs recMS1f/recMS1r and recMS9f/recMS9r using 3 μL from the DNA extract, 0.75 units of Taq polymerase (Qiagen, Hilden, Germany), 2 pmol of each primer of the first and 1 pmol of each primer of the second pair, and 5 nmol dNTPs in 23 μL of 1× Qiagen PCR buffer supplemented with MgCl2. Cycling conditions were 94°C for 2 min followed by 38 cycles of 94°C for 15 s, 53°C for 15 s, and 72°C for 30 s, followed by 72°C for 5 min. The PCR fragments were fractionated on 6% (w/v) Metaphor agarose (Biozym, Hess. Oldendorf, Germany) gels or on an ABI 3700 DNA sequencer (PE Applied Biosystems, Sunnyvale, CA). In the latter case, recMS1f was 5′ labeled with 7′,8′-benzo-5′-fluoro-2′,4,7-trichloro-5-carboxyfluorescein, and recMS9f with 6-hexachlorofluorescein. Primers were obtained from MWG (Ebersberg, Germany), Metabion (Martinsried, Germany), or PE Applied Biosystems. Additional primer pairs used for fine-scale mapping were recMS2f/recMS2r, recMS5f/recMS5r, rec6f/rec6r, and recMS7f/recMS7r using reaction conditions similar to above. Finally, plants recombinant between rec6 and recMS7 were also analyzed with primer pairs EN-MSfor/EN-MSrev, Southex/16Cm2, and EDIT1-2a/11t1. Primers sequences were chosen from the Arabidopsis Col-0 GSL-ELONG region. Primer pairs recMS1f/recMS1r, recMS2f/recMS2r, recMS5f/recMS5r, recMS7f/recMS7r, recMS9f/recMS9r, and EN-MSfor/EN-MSr amplify polymorphic microsatellites. PCR products amplified from Col-0 and Ler with rec6f/rec6r can be distinguished by a 72-bp insertion/deletion polymorphism. Col-0 and Ler EDIT1 2a/11t1 and Southex/16Cm1 PCR products differ by several SNPs and 1-bp insertion/deletion polymorphisms that can be distinguished by sequencing. All primer sequences and marker sizes are listed in Table IV.
Table IV.
Primer sequences used in this study
| Primer Name | Sequence (5′ → 3′) | Size (Col-0) | Size (Ler) |
|---|---|---|---|
| bp | |||
| recMS1f | GTGTGTCTATTAATATATCAAGAC | 125 | ∼120 |
| recMS1r | GTTGTTTTATCTATGTTTATCAAG | ||
| recMS2f | CCATTGGAGATGGTCTTACAC | 107 | ∼90 |
| recMS2r | CTTTGTCACCAAGTCTTGTATAG | ||
| recMS5f | GAATCAGATAATTAAGTTAGATAG | 106 | No product |
| recMS5r | GGTTCTTCATAAAAACAAAGCAC | ||
| rec6f | GTGTAGTTTCATTTGTGCTATC | 75 | 147 |
| rec6r | CTTTAAAACTCTAAGCCTAGAC | ||
| recMS7f | CATAAGATGGAAGCGTTAGAC | 132 | ∼150 |
| recMS7r | GATGGTAAACTACTTTGTGAAC | ||
| recMS9f | GGTTCCAAGTTACAAACTTTAAC | 105 | ∼90 |
| recMS9r | CGACCTAACGGCAAGTGAAAG | ||
| EN-MSfor | CATGAGAGCGGCATTCACCAG | 100 | 88 |
| EN-MSrev | GGACAATTATACACATACAACGC | ||
| 16Cm2 | GGGCTTATGTGCTTGAAC | 430 | 430 |
| Southex | GAAATGTCGAGGGGCATATG | SNPs, 1-bp indels | SNPs, 1-bp indels |
| EDIT1-2a | GTACCACAGTAATATCAATTTCC | 207 | 208 |
| 11t1 | GTTCATGAGATATGAGTAAGTGG | SNPs, 1-bp indel | SNPs, 1-bp indel |
| C/L-ESfEE | ATATGAATTCAACTATGTGCGTGTATTCGAC | – | – |
| CESrEE | ATATCTCGAGCACATTCGATGAAACCTGAGG | – | – |
| L-ENfEE | ATATGGATCCTATGTGCGTGTATTAGACAC | – | – |
| L-ENrEE | ATATCTCGAGTACAACAGCGGAAATCTGAGG | – | – |
| IPMS0 | CACCGTCACGTTGAAACCTTC | – | – |
| E116r | TTTTTAATTCAGGCAGCGACTCT | – | – |
| 1MAM1a | ACTCCATAGTGATGGCTTCATCGCTTCTG | – | – |
| 2MAM1b | AGAGATACAACGTGTTTACACATTC | – | – |
| 1MAMLa | TATAGTAATGGCTTCGTTACTTCTC | – | – |
| 2MAMLb | TCTTCCCAAACTTATACAACAGCG | – | – |
Size is indicated for PCR products used as genetic markers.
Extraction and Identification of Glucosinolates
Glucosinolates from Col-0 and Ler wild-type, from the gsm1 mutant lines TU1 and TU5 (Nottinhgam Arabidopsis Stock Centre [Nottingham, UK]/Arabidopsis Biological Resource Center, Ohio State University, Columbus; stock nos. CS2226 and CS2227, respectively) and Ler × TU1 F1 plants were extracted using boiling water as previously described (Graser et al., 2000). Plant material consisted of 0.05 g of lyophilized juvenile leaves of 5-week-old plants (approximately 10 plants). Samples were separated by HPLC (HP1100 Series, Agilent, Palo Alto, CA) fitted with a C-18 reverse-phase column using a water (Solvent A)-acetonitrile (Solvent B) gradient at a flow rate of 1 mL min−1 and at ambient room temperature. The 42-min run consisted of 1.5% (v/v) B (1 min), 1.5% to 5.0% (v/v) B (5 min), 5.0% to 7.0% (v/v) B (2 min), 7.0% to 21.0% (v/v) B (10 min), 21.0% to 29.0% (v/v) B (5 min), 29.0% to 43.0% (v/v) B (7 min), 43.0% to 93.0% (v/v) B (0.5 min), a 4-min hold at 93.0% (v/v) B, 93.0% to 1.5% (v/v) B (0.5 min), and a 7-min hold at 1.5% (v/v) B. Eluent was monitored by diode array detection between 190 and 360 nm (2-nm interval). Glucosinolates were identified by retention time and UV spectra as compared with those of purified standards and quantified by A229 nm relative to an internal standard (2-propenyl glucosinolate) included in the extraction. Response factors determined from pure desulfoglucosinolates for all compounds except 2-methylsulfinylethyl and 7-methylsulfinylheptyl glucosinolates (response factors assumed to be 1) were used to calculate molar concentrations of individual glucosinolates. Glucosinolates were extracted from the recombinant plants in a similar fashion except for modifications to facilitate high-throughput analyses. Lyophilized leaf samples (10 mg) were extracted in methanol and then treated essentially as described (Graser et al., 2000). The HPLC separation was carried out at 1 mL min−1 and the solvent program was modified as follows to accommodate high throughput analyses: 1.5% to 5.0% (v/v) B (6 min), 5.0% to 7.0% (v/v) B (2 min), 7.0% to 25.0% (v/v) B (7 min), 25.0% to 92.0% (v/v) B (2 min), 92.0% (v/v) B (6-min hold), 92.0% to 1.5% (v/v) B (2 min), and 1.5% (v/v) B (5 min).
Amplification of cDNAs
Total plant RNA was isolated with Trizol (Gibco-Invitrogen, Karlsruhe, Germany) according to the manufacturer's recommendations. First strand cDNA was synthesized following a protocol modified from Frohman et al. (1988). A cDNA corresponding to F15H18.3 was amplified using 1 μL of first strand product derived from Col-0 using primers IPMS0 and E116r (Table IV). The cDNAs corresponding to the entire ORFs of MAM1 and MAM-L from both TU1 and TU5 were synthesized in a 33-μL reaction mix consisting of 1× TaKaRa (TaKaRa, Japan) reaction buffer, 8.5 nmol of dNTPs, and 10 pmol of the primer pairs 1MAM1-a/2MAM1-b and 1MAMLa/2MAMLb, respectively. Reaction products were gel purified (QiaQuick, Qiagen). In each case, reactions were performed three times, and PCR products were sequenced on an ABI 3700 DNA sequencer with Big Dye Terminators (PE Applied Biosystems). Assembly and comparison of DNA sequence data was done using LASERGENE software (DNASTAR, Inc., Madison, WI) or GCG (version 10).
Expression Constructs
MAM1 cDNAs from both Col-0 and Ler were amplified with the primer pair C/L-ESfEE/CESrEE (Table IV). These primers were chosen to truncate the first 246 nucleotides that appear to represent an N-terminal signal sequence and thus could hamper expression in a prokaryotic system. The PCR products were gel purified (Qiagen) and cloned into pCR2.1-TOPO (Invitrogen, Breda, The Netherlands). Insert sequences were verified by sequencing. Suitable MAM1 inserts were excised with EcoRI and XhoI and recloned directionally into a pET-28a expression vector (Novagen, Madison, WI). An expression construct encoding the mutant allele from TU1 was obtained by site-directed mutagenesis of the functional MAM1 expression construct from Col-0 using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Expression constructs of MAM-L were prepared by similar methods with cDNAs amplified by the primer pair l-ENfEE/l-ENrEE.
cDNA Expression in Escherichia coli
The MAM1 and MAM-L constructs were expressed in E. coli strain BL21(DE) (F−ompT rB−mB−; (Studier et al., 1990) grown in M9 medium with acid-hydrolyzed casein to an optical density at 600 nm of 0.6 and then induced with 1 mm isopropylthio-β-galactoside for 2 h. Cells were harvested by centrifugation at 6,500g. After resuspension in 50 mm Tris, pH 8.0, with 1 mm dithiothreitol, cells were sonicated with a Sonoplus HD2070 sonicator (Bandeln, Berlin) twice with a microprobe at 55% full power for a 5-min −20% cycle. Cell debris was precipitated by centrifugation at 8,500g and the supernatant used for the enzyme assays.
Enzyme Assay
The assay mixture contained 100 mm AMPSO, pH 9.0, 4 mm MnCl2, 1 mm [1-14C]acetyl-CoA (14.8 GBq mol−1), 20 mm ATP, 3 mm OMTB, and 150 μL of enzyme preparation in a final volume of 250 μL. After an incubation period of 16 h at 32°C, the reaction was stopped by the addition of 750 μL of ethanol and denatured protein precipitated by centrifugation. The supernatant was concentrated to a volume of about 50 μL for HPLC analysis. An ion exclusion column (Nucleogel ion-300 OA, Macherey and Nagel, Düren, Germany) was run isocratically with 0.005 n H2SO4 at a flow rate of 0.25 mL min−1, 60°C, for 45 min. Detection employed a flow-through radioactivity monitor (Radiomatic 500TR, Packard, Dreieich, Germany) with a 0.5-mL flow cell and Ultima-Flo AP scintillation fluid (Packard) in a ratio of 4:1 (v/v) to column eluent. The counting efficiency for 14C was 55% to 60%. Under these conditions, the reaction product, MTEM, eluted at approximately 36.5 min, well separated from the substrates, acetyl-CoA (16.5 min) and OMTB (28 min), and from free acetate (41 min).
Product Identification
The assay product was identified with a liquid chromatography-MS system using a Quattro II (Micromass, Altrincham, UK) tandem quadrupole mass spectrometer equipped with an electrospray interface (capillary, 2.5 kV; sample cone, 12 V; and desolvation temperature, 375°C). The HPLC conditions were as described above except that the solvent was 0.03% (v/v; aqueous) CF3COOH and the flow rate was 0.2 mL min−1. The MS-MS spectra were recorded by fragmentation of the [M + H]+ parent ion at m/z 209. Argon was used as the collision gas at 1.4 × 10−3 mbar, and a collision energy of 7 eV was employed to achieve fragmentation. The scanned mass range was m/z 50 to m/z 215 and the scan time 1 s. The biosynthetic product gave a mass spectrum identical to that of synthetic MTEM prepared by methods previously described (Chapple et al., 1988).
Cluster Analysis of IPMS-Like Sequences in Arabidopsis
Alignments of IPMS-like sequences were carried out with DNASTAR (Lasergene), and corrected manually. A neighbor-joining tree (Saitou and Nei, 1987) was constructed with TREECON (van de Peer and de Wachter, 1994) taking into account residues 27 through 392 of the derived IPMS protein sequence from Methanococcus sp. and the corresponding amino acids from other IPMS-like proteins, and using an algorithm for distance estimation developed by Tajima and Nei (1984). Reliability of the branching order was estimated by bootstrapping (100 replicates; Felsenstein, 1985). GenBank accessions numbers are: AAB99199 (Methanococcus jannaschii), BAA10079 (Synechocystis sp.), AF004165 (tomato [Lycopersicon pennellii] IPMSa), AF004166 (tomato IPMSb), AC016662 (F2P9.9), AC013354 (F15H18.3), AB006708 (MAM-L), and AB026660 (MAM1).
ACKNOWLEDGMENTS
We thank Susanne Ring, Domenica Schnabelrauch, Antje Figuth, Einar Stauber, Natascha Sandoval, and Nadine Gallischke for excellent technical assistance; Jaqueline Fritsche for careful plant work; Michael Reichelt, Paul Brown, and Daniel Kliebenstein for help with the HLPC analyses; and Neil Oldham for mass spectral analyses. We also acknowledge the helpful comments of several anonymous reviewers and the resources and friendly support of the Nottingham Arabidopsis Stock Centre and the Arabidopsis Biological Resources Center.
Footnotes
This work was supported by the Max-Planck-Gesellschaft, by the U.S. National Science Foundation (grant no. DEB–9527725 to T.M.-O.), by the European Union, and by the German Science Foundation (grant to J.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010416.
LITERATURE CITED
- Blau PA, Feeny P, Contardo L. Allylglucosinolate and herbivorous caterpillars: a contrast in toxicity and tolerance. Science. 1978;200:1296–1298. doi: 10.1126/science.200.4347.1296. [DOI] [PubMed] [Google Scholar]
- Bodnaryk RP. Developmental profile of sinalbin (p-hydroxybenzyl glucosinolate) in mustard seedlings, Sinapis albaL., and its relationship to insect resistance. J Chem Ecol. 1991;17:1543–1556. doi: 10.1007/BF00984687. [DOI] [PubMed] [Google Scholar]
- Bones AM, Rossiter JT. The myrosinase-glucosinolate system, its organization and biochemistry. Physiol Plant. 1996;97:194–208. [Google Scholar]
- Bradshaw JE, Heaney RK, Smith WHM, Gowers S, Gemmell DJ, Fenwick GR. The glucosinolate content of some fodder brassicas. J Sci Food Agric. 1984;35:977–981. [Google Scholar]
- Brown PD, Morra MJ. Glucosinolate-containing plant tissues as bioherbicides. J Agric Food Chem. 1995;43:3070–3074. [Google Scholar]
- Campos H, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Mithen R. α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet. 2000;101:429–437. [Google Scholar]
- Chapple CC, Decicco C, Ellis BE. Biosynthesis of 2-(2′-methylthio) ethylmalate in Brassica carinata. Phytochemistry. 1988;27:3461–3463. [Google Scholar]
- Chapple CC, Glover JR, Ellis BE. Purification and characterization of methionine-glyoxylate aminotransferase from Brassica carinata and Brassica napus. Plant Physiol. 1990;94:1887–1896. doi: 10.1104/pp.94.4.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm MD, Wetter LR. Biosynthesis of mustard oil glucosides: IV. The administration of methionine-C14and related compounds to horseradish. Can J Biochem. 1964;42:1033–1040. doi: 10.1139/o64-114. [DOI] [PubMed] [Google Scholar]
- Donkin SG, Eiteman MA, Williams PL. Toxicity of glucosinolates and their enzymatic decomposition products to Caenorhabditis elegans. J Nematol. 1995;27:258–262. [PMC free article] [PubMed] [Google Scholar]
- Doughty KJ, Porter AJR, Morton AM, Kiddle G, Bock CH, Wallsgrove R. Variation in the glucosinolate content of oilseed rape (Brassica napus L.) leaves: response to infection by Alternaria brassicae(Berk.) Sacc. Ann Appl Biol. 1991;118:469–477. [Google Scholar]
- Du YJ, van Loon JJA, Renwick JAA. Contact chemoreception of oviposition-stimulating glucosinolates and an oviposition-deterrent cardenolide in two subspecies of Pieris napi. Physiol Entomol. 1995;20:164–174. [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Prot Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Zhang Y, Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamoustaris A, Mithen R. The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica napus ssp. oleifera) on its interaction with specialist and generalist pests. Ann Appl Biol. 1995;126:347–363. [Google Scholar]
- Graser G, Schneider B, Oldham NJ, Gershenzon J. The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa(Brassicaceae) Arch Biochem Biophys. 2000;378:411–419. doi: 10.1006/abbi.2000.1812. [DOI] [PubMed] [Google Scholar]
- Hagelstein P, Schultz G. Leucine synthesis in spinach chloroplasts: partial characterization of 2-isopropyl-malate synthase. Biol Chem Hoppe Seyler. 1993;374:1105–1108. doi: 10.1515/bchm3.1993.374.7-12.1105. [DOI] [PubMed] [Google Scholar]
- Hagelstein P, Sieve B, Klein M, Jans H, Schultz G. Leucine synthesis in chloroplasts: leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J Plant Physiol. 1997;150:23–30. [Google Scholar]
- Halkier BA, Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;11:425–431. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Halkier BA. Glucosinolates. In: Ikan R, editor. Naturally Occurring Glycosides: Chemistry, Distribution and Biological Properties. Chichester, UK: John Wiley and Sons; 1999. pp. 193–223. [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: the glucosinolates. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thalianaand their identification using thermospray liquid chromatography-mass spectrometry. J Chromatogr Sci. 1988;26:551–556. [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis thalianaglucosinolate accumulation. Plant Physiol. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justus KA, Mitchell BK. Oviposition site selection by the diamondback moth, Plutella xylostella(L.) (Lepidoptera: Plutellidae) J Insect Behav. 1996;9:887–898. [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers. Plant J. 1993;4:745–750. [Google Scholar]
- Magrath R, Bano F, Morgner M, Parkin I, Sharpe A, Lister C, Dean C, Turner J, Lydiate D, Mithen R. Genetics of aliphatic glucosinolates: I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity. 1994;72:290–299. [Google Scholar]
- Matsuo M, Yamazaki M. Biosynthesis of sinigrin. Chem Pharmacol Bull. 1964;12:1388–1389. doi: 10.1248/cpb.12.1388. [DOI] [PubMed] [Google Scholar]
- Mithen R, Clarke J, Lister C, Dean C. Genetics of aliphatic glucosinolates: III. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity. 1995;74:210–215. [Google Scholar]
- Nastruzzi C, Cortesi R, Esposito E, Menegatti E, Onofrio L, Iori R, Palmieri S. In vitrocytotoxic activity of some glucosinolate-derived products generated by myrosinase hydrolysis. J Agric Food Chem. 1996;44:1014–1021. [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Reed DW, Pivnick KA, Underhill EW. Identification of chemical oviposition stimulants for the diamondback moth, Plutella xylostella, present in three species of Brassicaceae. Entomol Exp Appl. 1989;53:277–286. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Städler E, Renwick JAA, Radke CD, Sachdev-Gupta K. Tarsal contact chemoreceptor response to glucosinolates and cardenolides mediating oviposition in Pieris rapae. Physiol Entomol. 1995;20:175–187. [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorf JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- van de Peer Y, de Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;189:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Zhang GY, Talalay P, Cho C, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]