Abstract
Tocopherols, collectively known as vitamin E, are lipid-soluble antioxidants synthesized exclusively by photosynthetic organisms and are required components of mammalian diets. The committed step in tocopherol biosynthesis involves condensation of homogentisic acid and phytyl diphosphate (PDP) catalyzed by a membrane-bound homogentisate phytyltransferase (HPT). HPTs were identified from Synechocystis sp. PCC 6803 and Arabidopsis based on their sequence similarity to chlorophyll synthases, which utilize PDP in a similar prenylation reaction. HPTs from both organisms used homogentisic acid and PDP as their preferred substrates in vitro but only Synechocystis sp. PCC 6803 HPT was active with geranylgeranyl diphosphate as a substrate. Neither enzyme could utilize solanesyl diphosphate, the prenyl substrate for plastoquinone-9 synthesis. In addition, disruption of Synechocystis sp. PCC 6803 HPT function causes an absence of tocopherols without affecting plastoquinone-9 levels, indicating that separate polyprenyltransferases exist for tocopherol and plastoquinone synthesis in Synechocystis sp. PCC 6803. It is surprising that the absence of tocopherols in this mutant had no discernible effect on cell growth and photosynthesis.
Tocopherols are a group of amphipathic compounds synthesized only by photosynthetic organisms. The best characterized and probably most important function of tocopherols is to act as recyclable chain reaction terminators of polyunsaturated fatty acid free radicals generated by lipid oxidation. Tocopherols have a well-documented role in mammals both as an essential nutrient (vitamin E) and general antioxidant (Fryer, 1993; Liebler, 1998; Brigelius-Flohe and Traber, 1999). A similar though less well-documented antioxidant role is also proposed for tocopherols in photosynthetic organisms (Fryer, 1992; Niyogi, 1999).
From a biosynthetic perspective, tocopherols are members of a large, multifunctional family of lipid-soluble compounds called prenylquinones that also include tocotrienols, plastoquinones, and phylloquinones (vitamin K1). Structural features shared by all prenylquinones include hydrophobic prenyl tails of various lengths attached to aromatic head groups that can reversibly change their redox states. Tocopherols contain a chromanol head-group and lipophillic tail derived from the 20-carbon alcohol phytol, whereas plastoquinones contain a quinone head group and isoprenoid tails of 40, 45, or 50 carbons. Such structural features are essential for the diverse biochemical and physiological roles fulfilled by various prenylquinones.
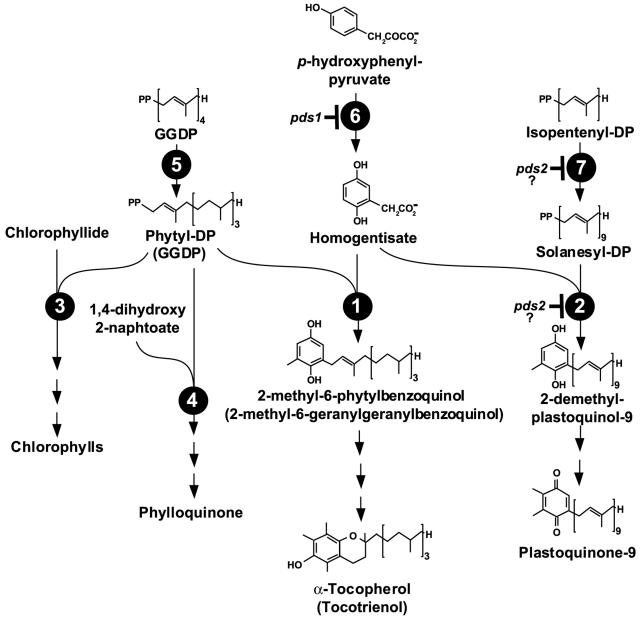
The committed step in the synthesis of all prenylquinones is the condensation of various aromatic precursors and prenyl-diphosphate (DP) substrates in reactions catalyzed by a small family of related polyprenyltransferases (Lopez et al., 1996). Most aromatic and prenyl-DP substrates are utilized by more than one polyprenyltransferase (Fig. 1). For example, the aromatic compound homogentisic acid (HGA) is used for condensation with phytyl DP (PDP), geranylgeranyl DP (GGDP), or solanesyl DP (SDP) in tocopherol, tocotrienol, and plastoquinone synthesis, respectively, whereas PDP is used as the isoprenoid-derived tail in the synthesis of tocopherols, phylloquinones, and chlorophylls (Threlfall and Whistance, 1971; Schulze-Siebert et al., 1987; Oster et al., 1997). Thus, polyprenyltransferases act at biosynthetic branch points and are potential key regulatory enzymes for the synthesis of many essential compounds in photosynthetic organisms.
Figure 1.
Generalized overview of prenylquinone biosynthetic pathways in photosynthetic organisms. Prenylation steps and substrates in tocopherol and plastoquinone synthesis are shown in detail, whereas those for other prenyllipids are incomplete for clarity. Aromatic and prenyl-DP substrates are shared among the various polyprenyltransferases (see text for details). Enzymes are depicted as numbers in black circles: 1, homogentisate phytyltransferase (HPT); 2, homogentisate solanesyltransferase; 3, chlorophyll synthase; 4, 1,4-dihydroxy-2-naphthoate phytyltransferase; 5, GGDP reductase; 6, p-HPPD; and 7, SDP synthase. Compounds in parentheses indicate where GGDP may be used in place of PDP by HPT resulting in a tocotrienol product.
In plant chloroplasts, the synthesis of tocopherols and plastoquinones is closely related. Biochemical studies have shown that condensation of HGA with PDP or SDP yields 2-methyl-6-phytyl-1,4-benzoquinol (2-Me-6-Ph-1,4-BQ) and 2-demethylplastoquinol-9, the first prenylquinol intermediates in tocopherol and plastoquinone-9 (PQ-9) synthesis, respectively (Hutson and Threlfall, 1980; Soll et al., 1980; Marshall et al., 1985). Although these studies could not distinguish whether one or more polyprenyltransferases catalyzed these reactions, it was suggested that separate enzymes might be involved (Schulze-Siebert et al., 1987). In contrast, recent genetic data from Arabidopsis suggested involvement of a single polyprenyltransferase activity in tocopherol and PQ-9 synthesis. Two loci were identified, PDS1 and PDS2 (phytoene desaturation), which when mutated, decreased the levels of both tocopherols and plastoquinones below detection (Norris et al., 1995), consistent with the disruption of enzymes shared in their synthesis. The PDS1 locus has been cloned and encodes p-hydroxyphenylpyruvate dioxygenase (HPPD; Norris et al., 1998), which catalyzes formation of HGA. The pds2 mutation was proposed to disrupt another shared pathway enzyme, most likely a polyprenyltransferase, which could utilize either PDP or SDP as substrates for condensation with HGA (Norris et al., 1995). Unlike PDS1, the PDS2 locus has not yet been cloned.
As an alternative to purifying the membrane-bound PDS2 gene product or walking to the PDS2 locus, we attempted to clone an orthologous gene from the cyanobacterium Synechocystis sp. PCC 6803, which also synthesizes α-tocopherol. In this paper, we report the cloning and functional analysis of gene products from Synechocystis sp. PCC 6803 and Arabidopsis encoding polyprenyltransferases specific to tocopherol biosynthesis. We also present biochemical and physiological characterization of the corresponding Synechocystis sp. PCC 6803 polyprenyltransferase knockout mutant, which completely lacks tocopherols.
RESULTS
Identification and Disruption of a Polyprenyltransferase Involved in Tocopherol Biosynthesis in Synechocystis sp. PCC 6803
Due to the metabolic synteny observed for the prenyllipid biosynthetic pathways in photosynthetic organisms, we decided to utilize a genomics-based approach to identify the gene encoding the homogentisate polyprenyltransferase involved in tocopherol synthesis, first from cyanobacteria, and subsequently from plants. We hypothesized that this polyprenyltransferase would show some similarity to previously characterized polyprenyltransferases from cyanobacteria and plants that utilize similar prenyl-DPs as substrates.
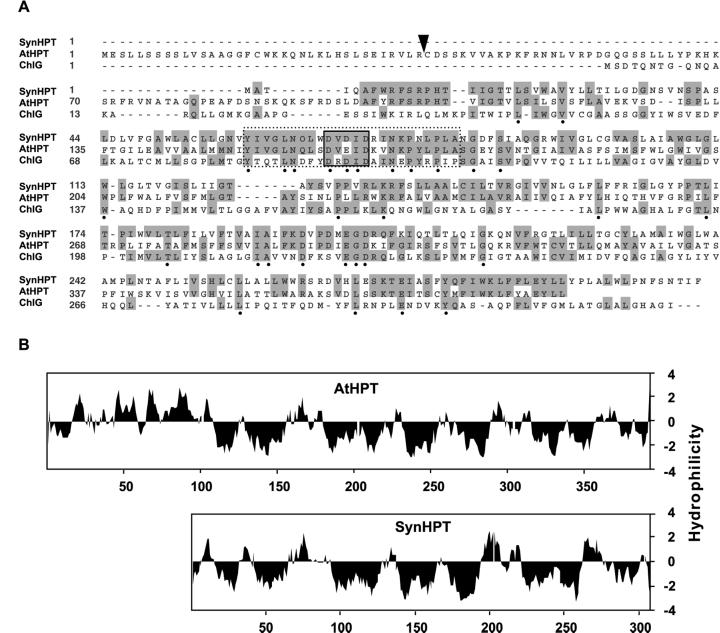
Chlorophyll synthase is a polyprenyltransferase that attaches PDP or GGDP to the tetrapyrrole core of chlorophyllide during chlorophyll biosynthesis (Lopez et al., 1996; Oster et al., 1997). The Synechocystis sp. PCC 6803 chlorophyll synthase open reading frame (ORF; ChlG, GenBank accession no. BAA10281) was used to query CyanoBase, which contains the complete Synechocystis sp. PCC 6803 genome sequence (Kaneko et al., 1996). Several ORFs showing varying degrees of similarity were identified and SLR1736 was selected as a putative HPT based on its protein identity to ChlG (21%) and the presence of prenyl-DP- and divalent cation-binding motifs characteristic of polyprenyltransferases (Lopez et al., 1996; Fig. 2A). SLR1736 is also a highly hydrophobic protein (Fig. 2B), as would be expected for a membrane-bound HPT (Soll et al., 1980, 1984).
Figure 2.
A, Alignment of Synechocystis sp. PCC 6803 and Arabidopsis polyprenyltransferases. HPTs from Synechocystis sp. PCC 6803 (SynHPT, GenBank accession no. S74813) and Arabidopsis (AtHPT, accession no. AF324344) share 41% protein identity, whereas SynHPT and ChlG (Synechocystis sp. PCC 6803 chlorophyll synthase, accession no. BAA10281) share 22% protein identity. Residues conserved in at least two of three sequences are shaded in gray, whereas residues identical in all three proteins are labeled by black dots. The conserved prenyl-DP and divalent cation binding domains are indicated by dashed and black boxes, respectively. The predicted AtHPT chloroplast-targeting domain cleavage site is indicated by a black arrow. B, Kyte/Doolittle hydrophillicity profiles of AtHPT and SynHPT. The two profiles are nearly identical. Negative values indicate hydrophobicity.
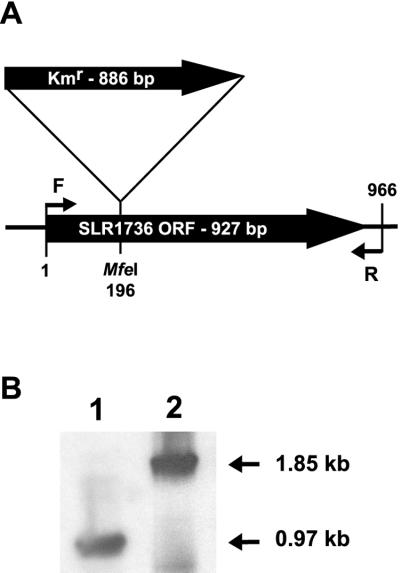
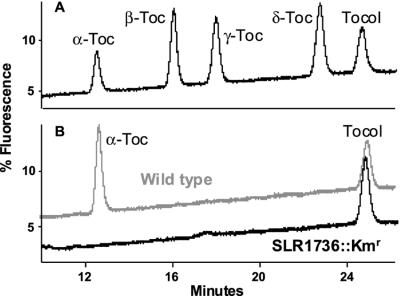
To test the hypothesis that SLR1736 is involved in tocopherol biosynthesis, a disruption mutant (SLR1736::Kmr) was generated by homologous recombination of the kanamycin cassette-disrupted SLR1736 gene into the wild-type SLR1736 locus (Fig. 3). If the SLR1736::Kmr mutation disrupted HPT activity, one would expect a complete absence of tocopherols and their prenylquinol intermediates. HPLC analysis shows that wild-type Synechocystis sp. PCC 6803 lipid extracts contain predominantly α-tocopherol (Fig. 4, Table I). In contrast, α-tocopherol and its prenylchromanol and quinol precursors are absent from SLR1736::Kmr lipid extracts (Fig. 4, Table I, and data not shown), consistent with the hypothesis that SLR1736 encodes a polyprenyltransferase involved in tocopherol synthesis.
Figure 3.
Construction of the Synechocystis sp. PCC 6803 SLR1736::Kmr mutant. A, Simplified scheme of the wild-type SLR1736 ORF in the Synechocystis sp. PCC 6803 genome. Insertion of a kanamycin (Kmr) cassette into MfeI site of the SLR1736 ORF and the SLR1736F and R PCR primers (F and R, small arrows) are indicated. B, Autoradiograph of the PCR products amplified from wild type (lane 1) and the SLR1736::Kmr mutant (lane 2) genomic DNA. No wild-type copies of SLR1736 were detected in the SLR1736::Kmr mutant.
Figure 4.
HPLC traces of tocopherol standards and lipids extracted from wild-type Synechocystis sp. PCC 6803 and SLR1736::Kmr. Equivalent weights of fresh cells were extracted for the analysis shown. Tocopherol analysis was performed on a normal phase column using 8% (v/v) di-isopropyl ether in hexane as a solvent. A, Separation of α-, β-, γ-, and δ-tocopherol (Toc, tocopherol) and tocol standards. B, Wild-type cells accumulate predominantly α-tocopherol (gray trace). No tocopherols were detected in the SLR1736::Kmr mutant (black trace). Tocol was used as an internal standard.
Table I.
Prenyllipid contents of photoautotrophically grown wild-type Synechocystis sp. PCC 6803 and the SLR1736::Kmr mutant
| Prenyllipid | Wild Type | SLR1736::Kmr |
|---|---|---|
| α-Tocopherola | 18.7 ± 1.4 | Not detected |
| Plastoquinone-9a | 31.6 ± 4.3 | 28.4 ± 1.7 |
| Phylloquinonea | 13.8 ± 1.1 | 12.5 ± 1.2 |
| Chlorophyll ab | 4.3 ± 0.3 | 4.6 ± 0.4 |
Other than the absence of tocopherols in the SLR1736::Kmr mutant, no significant differences were observed between wild-type and mutant cells for the presented parameters. Each value is the mean ± sd of at least five separate measurements per experiment. Each experiment was repeated at least three times.
mmol mol−1 chlorophyll a.
mmol mg−1 cells.
As shown in Figure 1, the various polyprenyltransferases in photosynthetic organisms utilize many of the same aromatic and prenyl-DP substrates. HGA is the aromatic precursor in both tocopherol and plastoquinone synthesis and PDP is a substrate for tocopherol, phylloquinone, and chlorophyll polyprenyltransferases (Threlfall and Whistance, 1971; Schulze-Siebert et al., 1987; Oster et al., 1997). Given this biosynthetic relationship, disrupting SLR1736 activity could directly or indirectly affect the synthesis of other prenylated compounds in pathways that also utilize these substrates. To determine the effect of the SLR1736 gene disruption on the synthesis of other prenylated compounds, we analyzed plastoquinone, phylloquinone, and chlorophyll levels in the SLR1736::Kmr mutant relative to wild type. No significant differences were observed in the levels of these compounds (Table I).
Biochemical Characterization of the SLR1736 Gene Product
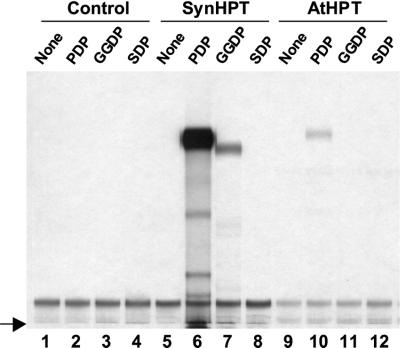
The SLR1736::Kmr phenotype strongly suggests that SLR1736 encodes a polyprenyltransferase specific to tocopherol synthesis. To determine the activity and substrate specificity of the SLR1736 gene product, HGA polyprenyltransferase assays were performed using SLR1736 protein expressed in Escherichia coli. These assays are based on thin-layer chromatography (TLC) separation and subsequent autoradiography or HPLC separation of prenylated quinones formed from radioactive HGA and various unlabeled prenyl-DPs in the presence of a putative polyprenyltransferase.
When various prenyl-DPs at the same molar concentrations were tested as potential substrates for the SLR1736 protein, PDP was used most efficiently, though GGDP could also be utilized (Figs. 5 and 6, B and D). The amount of geranylgeranylated benzoquinone product formed was approximately 18% that of the phytylated product. No products were observed when SDP, the prenyl-DP substrate for PQ-9 synthesis, was used (Fig. 5). In the case of PDP, the main reaction product comigrated with 2′-trans-2-Me-6-Ph-1,4-BQ in both TLC and HPLC analyses (Figs. 5 and 6B).
Figure 5.
Homogentisate polyprenyltransferase assays. Individual reactions contained the indicated prenyl-DP and protein extracts from E. coli expressing empty vector or the indicated phytyltransferases. Radiolabeled prenylquinol reaction products were extracted, oxidized to corresponding quinones, separated by TLC, and subjected to autoradiography. SynHPT can utilize both PDP and GGDP as prenyl-DP substrates (lanes 6 and 7), whereas AtHPT can only use PDP (lane 10). Neither enzyme could catalyze condensation of HGA and SDP (lanes 8 and 12). No prenylquinone products were detected in control reactions (lanes 1–5 and 9). The arrow indicates the origin.
Figure 6.
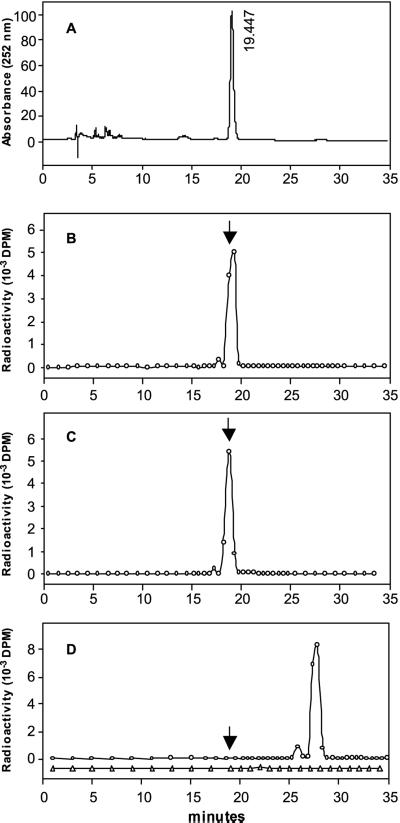
Normal phase HPLC separation of radiolabeled prenylquinones produced from HGA and prenyl-DP substrates by HPTs. Homogentisate polyprenyltransferase reactions were performed in a total volume of 0.5 mL for SynHPT with PDP (B) and 5 mL for SynHPT with GGDP, and AtHPT with PDP or GGDP (C–E, respectively) as described in “Materials and Methods.” Elution of the internal standard 2′-trans-2-Me-6-Ph-1,4-BQ was monitored at 252 nm, whereas that of the prenylquinones formed during the assay was monitored by scintillation counting of collected fractions. The UV traces are not shown for B through D for clarity, but when aligned with the radioelution profiles shown, the major radiolabeled phytylated products co-chromatographed with authentic 2′-trans-2-Me-6-Ph-1,4-BQ standard (indicated by arrows). A, Elution of 2′-trans-2-Me-6-Ph-1,4-BQ; B and C, SynHPT and AtHPT catalyzed formation of 2′-trans-2-Me-6-Ph-1,4-BQ from HGA and PDP; D, SynHPT catalyzed formation of 2-Me-6-GG-1,4-BQ from HGA and GGDP (circles). AtHPT did not produce a product with HGA and GGDP as substrates (triangles).
We also observed a couple of minor products on TLC and HPLC (Figs. 5 and 6). On TLC, a band (Fig. 5, lane 6, RF approximately 0.3) likely corresponds to the quinol form of 2-Me-6-Ph-1,4-BQ because intensity of this band increases when the samples are not oxidized with AgO prior to TLC (data not shown). A small radioactive peak eluting before the major 2′-trans-2-Me-6-Ph-1,4-BQ peak was also observed in HPLC analysis (Fig. 6B). This peak probably corresponds to 2′-cis-2-Me-6-Ph-1,4-BQ formed by isomerization of the trans-isomer as previously reported (Hutson and Threlfall, 1980; Henry et al., 1987). It is unlikely that this peak represents the corresponding quinol because quinols are eluted much later than quinones in the HPLC system used. Due to their low abundance, further analyses of these minor peaks could not be performed.
When GGDP was used as a substrate the SLR1736 enzyme also produced a major and minor product. It is unfortunate that 2-methyl-6-geranylgeranyl-1,4-benzoquinone (2-Me-6-GG-1,4-BQ), the expected product of HGA and GGDP condensation, was not available. However, indirect evidence suggests that the major GGDP reaction product is 2-Me-6-GG-1,4-BQ. First, consistent with previous reports (Soll and Schultz, 1979; Hutson and Threlfall, 1980), this product migrates slightly slower than its phytylated counterpart in the TLC system used (Compare lane 6 with lane 7, Fig. 5). Moreover, the chromatographic properties of the major product in normal-phase HPLC are consistent with those previously published for 2-Me-6-GG-1,4-BQ (Hutson and Threlfall, 1980). To be specific, it elutes 9 min after the internal control 2′-trans-2-Me-6-Ph-1,4-BQ (Fig. 6C), which is in good agreement with the previously reported relative elution difference between 2-Me-6-Ph-1,4-BQ and 2-Me-6-GG-1,4-BQ (Hutson and Threlfall, 1980). As with PDP, an unknown minor GGDP product migrating prior to the major peak is also present in HPLC analysis. It is unfortunate that neither GGPD products are produced in sufficient quantity to allow further analytical characterization. Based on the combined results of these polyprenyltransferase assays and the tocopherol-specific phenotype of the SLR1736::Kmr mutant, the protein encoded by the SLR1736 gene was named SynHPT, for Synechocystis sp. PCC 6803 HPT.
Identification and Characterization of an HPT Homolog from Arabidopsis
To identify an HPT homolog from plants, we used the SynHPT protein sequence as a database query for BLAST searches (Altschul et al., 1990). A single predicted Arabidopsis gene on chromosome 2 (bacteria artificial chromosome clone F19F24) containing regions of significant similarity to SynHPT was identified. The corresponding cDNA subsequently was isolated from an Arabidopsis seed cDNA library and fully sequenced. The predicted protein encoded by this cDNA (GenBank accession no. AF324344) shares 41% identity with SynHPT. In addition, both proteins have remarkably similar hydrophobicity profiles and contain prenyl-DP and divalent cation binding motifs conserved in both location and sequence (Fig. 2). The Arabidopsis protein also contains an additional 95-amino acid N-terminal extension that is not present in SynHPT. The first 36 amino acids of this domain exhibit features of a chloroplast targeting sequence (Emanuelsson et al., 1999), consistent with the reported chloroplast envelope localization of HPT activity in plants (Soll et al., 1980, 1984). The Arabidopsis protein was tentatively named AtHPT for Arabidopsis HPT.
To determine the activity and substrate specificity of the putative AtHPT and compare it with SynHPT, AtHPT was expressed in E. coli and HGA polyprenyltransferase assays were performed. Like SynHPT, AtHPT catalyzed condensation of HGA and PDP to form 2′-trans-2-Me-6-Ph-1,4-BQ as a major product and was not active with the substrates HGA and SDP. Unlike SynHPT, no products were observed when HGA and GGDP were used as substrates (Figs. 5 and 6D). To test whether this difference between the two enzymes was due to the presence of chloroplast-targeting sequences in AtHPT, we also tested two truncated versions of the protein. One truncation removed the predicted 36-amino acid chloroplast transit peptide, whereas the second removed 95 N-terminal amino acids not present in SynHPT. Neither truncation altered the specific activity or substrate specificity of AtHPT (results not shown).
The specific activity of AtHPT expressed in E. coli was approximately 3% that of SynHPT expressed from the same vector. Several explanations are plausible for this difference, including decreased protein stability, poor protein expression in E. coli due to codon bias, or an improper lipid environment relative to that of chloroplasts. Neither AtHPT nor SynHPT could be visualized on Coomassie Blue-stained gels following induction, indicating both are expressed at low levels in E. coli. Addition of lipids extracted from Arabidopsis leaves or seeds to reactions had no discernible effect on AtHPT activity (data not shown). Finally, addition of Tween 80 or CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid} to final concentrations of 0.2%, 0.5%, 1%, or 2% (w/v) moderately stimulated both AtHPT and SynHPT activities without appreciably changing their specific activities relative to each other (data not shown). It appears that a combination of lower expression and/or lower stability of AtHPT relative to SynHPT may be the cause of limited AtHPT activity in E. coli.
Physiological Consequences of Tocopherol Deficiency in Synechocystis sp. PCC 6803
Given the importance of tocopherols in free radical scavenging, a photosynthetic organism lacking tocopherols might be expected to be compromised in growth or exhibit increased sensitivity to high light stress. To address this question, we compared growth of the tocopherol deficient SLR1736::Kmr mutant and wild-type Synechocystis sp. PCC 6803 under low-light or high-light conditions (approximately 30 and 110 μE m−2 s−1, respectively). It is surprising that the doubling times of both strains under photoautotrophic or heterotrophic conditions in both low and high light were comparable (Table II and data not shown). Whole-chain oxygen evolution measured at 0.75, 2, and 5 mE m−2 s−1 was also found to be similar in both strains, indicating that the initial rates of photosynthesis in SLR1736::Kmr and wild type are comparable (Table II).
Table II.
Growth and O2 evolution rates of wild-type Synechocystis sp. PCC 6803 and SLR1736::Kmr
| Parameter | Wild Type | SLR1736::Kmr |
|---|---|---|
| Growth, doubling time (h)a | ||
| Photoautotrophic | 22 ± 3 | 19 ± 3 |
| Photoheterotrophic | 12 ± 1 | 13 ± 1 |
| Oxygen evolution ratesb | ||
| Whole chain | 156 ± 6 | 163 ± 8 |
Doubling times and photosynthetic activity of wild-type and mutant cells are similar. Each value is the mean ± sd of three independent measurements in a representative experiment. Each experiment was repeated at least three times.
The cells were grown at high light (105–110 μE m−2 s−1).
The cells were grown photoautotrophically; O2 evolution rates are in mmol O2 mg−1 chlorophyll h−1 and measured at 5 mE m−2 s−1.
DISCUSSION
Due to the conserved evolution of photosynthetic organisms, many biosynthetic pathways in cyanobacteria and plants are often quite similar (Whistance and Threlfall, 1970; Marechal et al., 1997). The rapid growth of expressed sequence tag and genome databases in a wide variety of organisms allows plant biochemists to utilize such inter-kingdom conservation in their research more effectively. In particular, the availability of the fully sequenced Synechocystis sp. PCC 6803 genome in a searchable online database, CyanoBase (Kaneko et al., 1996), coupled with straightforward gene disruption methods for analysis of gene function in this organism (Williams, 1988) makes Synechocystis sp. PCC 6803 an attractive system to complement studies of tocopherol synthesis in plants. Two tocopherol biosynthetic enzymes, GGDP reductase (Addlesee et al., 1996; Keller et al., 1998) and γ-tocopherol methyltransferase (Shintani and DellaPenna, 1998), have already been cloned and characterized from Synechocystis sp. PCC 6803 and used successfully as database probes to identify orthologs from Arabidopsis. The γ-tocopherol methyltransferases from both organisms were shown to have nearly identical activities (Shintani and DellaPenna, 1998). We have employed a similar genomics-based approach to identify and characterize a third enzyme of the tocopherol pathway from Synechocystis sp. PCC 6803 and Arabidopsis, HGA phytyltransferase, and assess whether this step in tocopherol synthesis is also conserved between cyanobacteria and plants.
In photosynthetic organisms, condensation of HGA with either a 20- or 45-carbon prenyl-DP is the branch point in tocopherol and plastoquinone synthesis, respectively. Early biochemical studies established that the tocopherol and plastoquinone pathways are remarkably similar in oxygenic cyanobacteria, algae, and plants (Whistance and Threlfall, 1970). Although these studies could not distinguish separate polyprenyltransferase activities for tocopherol and plastoquinone synthesis, it was suggested that separate prenylation enzymes might be involved (Schulze-Siebert et al., 1987). Genetic analysis of the pathways in Arabidopsis more recently identified two loci whose mutant phenotypes are consistent with the disruption of enzymes shared in the synthesis of tocopherols and plastoquinones (Norris et al., 1995). This was found to be the case for the PDS1 locus, which encodes HPPD, the enzyme that produces the aromatic head group HGA in both the plastoquinone and tocopherol pathways (Norris et al., 1998). The PDS2 locus was suggested to encode a similarly shared polyprenyltransferase that could utilize either PDP or SDP for tocopherol and plastoquinone synthesis, respectively (Norris et al., 1995). The cloning of plant and cyanobacterial HPTs now allows us to directly address the nature of polyprenyltransferases involved in tocopherol and plastoquinone synthesis in oxygenic photosynthetic organisms.
The Synechocystis sp. PCC 6803 SLR1736::Kmr mutant lacks tocopherols but accumulates wild-type levels of PQ-9, consistent with the existence of separate HGA polyprenyltransferases in tocopherol and plastoquinone synthesis. This conclusion is also supported by enzymatic studies showing that SynHPT can utilize the 20-carbon tocopherol substrates, PDP or GGDP, but is inactive with the 45-carbon PQ-9 substrate, SDP. Thus, it appears that Synechocystis sp. PCC 6803 contains a single polyprenyltransferase specific to tocopherol synthesis, SynHPT, and a separate, yet-to-be characterized polyprenyltransferase specific to plastoquinone synthesis.
In Arabidopsis, the prenyltransferase reaction involving HGA and PDP substrates appears to be nearly identical to that in Synechocystis sp. PCC 6803. The AtHPT and SynHPT enzymes share 61% protein similarity and both enzymes use PDP as their preferred prenyl-DP substrate in vitro to form 2′-trans-2-Me-6-Ph-1,4-BQ as a major product. This is in agreement with previous biochemical studies of HPT activity in isolated spinach, lettuce, and pea chloroplasts where 2-Me-6-Ph-1,4-BQ was the only product detected (Hutson and Threlfall, 1980; Soll et al., 1980; Marshall et al., 1985). Like SynHPT, AtHPT did not generate detectable prenylquinone products with SDP as a substrate, suggesting Arabidopsis likely contains separate polyprenyltransferases for tocopherol and plastoquinone synthesis. An Arabidopsis HPT knockout mutant is needed to rigorously address this question.
In considering the nature of homogentisate polyprenyltransferase reactions in plants, it is important to note that our original goal of cloning the Arabidopsis PDS2 locus has not been achieved. Tocopherol and plastoquinone levels are reduced below detection in pds2, leading to the hypothesis that PDS2 encodes a polyprenyltransferase shared in tocopherol and plastoquinone synthesis (Norris et al., 1995). However, given that AtHPT is a phytyltransferase encoded by a single-copy gene on chromosome 2, whereas PDS2 maps to chromosome 3, this proposal now seems unlikely. If PDS2 is not a polyprenyltransferase shared in tocopherol and plastoquinone synthesis, what does it encode? One explanation is that PDS2 encodes an enzyme specific to plastoquinone synthesis (i.e. HGA solanesyltransferase or SDP synthase) and that the absence of tocopherols in pds2 is a pleiotropic effect of this mutation. In this scenario, the absence of plastoquinone, the main lipid soluble electron carrier in plastids, results in such high levels of oxidative stress in pds2 that any tocopherols produced are rapidly oxidized and degraded, and hence undetectable. In an alternate manner, plastoquinone may be a cofactor required for the synthesis of tocopherols and its absence arrests tocopherol synthesis. Regardless of mechanism, it appears likely that the tocopherol deficiency in pds2 is an indirect, rather than a direct effect of the pds2 mutation.
Though SynHPT and AtHPT are similar in their substrate specificities, there is one notable exception: SynHPT can use both PDP and GGDP as substrates, whereas AtHPT only uses PDP. The utilization of both PDP and GGDP as substrates by a polyprenyltransferase is not unprecedented. An analogous reaction occurs in chlorophyll biosynthesis where chlorophyll synthase can attach either PDP or GGDP to the tetrapyrrole moiety, and in cyanobacteria, PDP is the preferred substrate (Oster et al., 1997). We observed a similarly strong preference of SynHPT for PDP over GGDP. The use of GGDP by SynHPT in vivo would yield tocotrienol intermediates and end products that only differ from their tocopherol counterparts in having an unsaturated rather than saturated hydrophobic tail. This would necessitate subsequent enzymes in the pathway being active toward geranylgeranylated substrates. At least one other tocopherol biosynthetic enzyme from cyanobacteria has been shown to utilize both phytylated and geranylgeranylated intermediates in vitro, tocopherol cyclase from Anabaena variabilis (Stocker et al., 1996). However, Synechocystis sp. PCC 6803 does not accumulate tocotrienols, suggesting that any geranylgeranylated intermediate produced by SynHPT is either efficiently reduced (most likely by GGDP reductase), or that GGDP is not a substrate in vivo. Additional work is required to delineate the in vivo substrate(s) and product(s) of SynHPT.
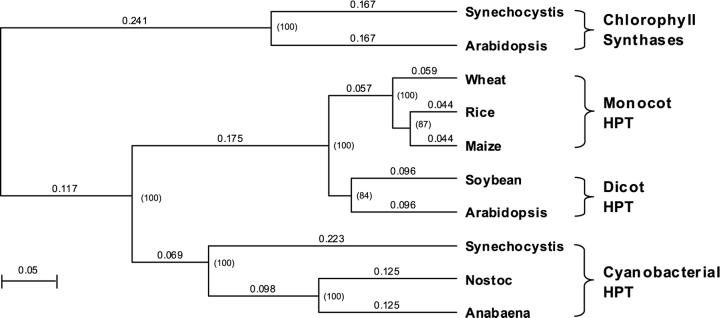
Within the limits of our assay sensitivity (approximately 3% of PDP product levels), AtHPT did not utilize GGDP as a substrate. Other researchers also failed to demonstrate condensation of HGA and GGDP using isolated spinach, lettuce, and pea chloroplasts (Hutson and Threlfall, 1980; Soll et al., 1980). These data are consistent with the general observation that dicots do not produce tocotrienols (Piironen et al., 1986; Franzen and Haas, 1991). However, many monocots and gymnosperms do produce both tocopherols and tocotrienols (Piironen et al., 1986; Franzen and Haas, 1991; Franzen et al., 1991) and we speculate that HPTs from such organisms would utilize GGDP and PDP as substrates, analogous to SynHPT. Phylogenetic analysis of polyprenyltransferases from various photosynthetic organisms shows that HPTs from cyanobacteria, monocots, and dicots form separate groups (Fig. 7), which probably represents taxonomic differences, although it may in part reflect differences in the substrate specificities of these enzymes. As with Synechocystis sp. PCC 6803, we would also anticipate that pathway enzymes after HPT in monocots would be active toward both geranylgeranylated and phytylated intermediates. The substrate specificity of tocopherol biosynthetic enzymes from monocots has not been characterized; however, in spinach (a dicot), where enzymology of the pathway has been best studied, later methyltransferases of the pathway are active toward various geranylgeranyl intermediates (Soll and Schultz, 1979). It appears that at some point in evolution, dicots, like monocots, could likely produce tocotrienols but have lost this ability as their HPTs have evolved substrate specificity for PDP over GGDP.
Figure 7.
Phylogenetic analysis of various prenyllipid polyprenyltransferases. Sequence alignment and phylogenetic analysis were performed using MacVector software (Genetic Computer Group, Madison, WI). Numbers indicate distances between protein sequences estimated by the uncorrected p distance method. Numbers in parentheses indicate percentages of the tree confidence calculated by bootstrapping. Chlorophyll synthases form a separate clade from HPTs. Within HPTs, cyanobacterial, monocot, and dicot HPTs also form distinct subgroups.
Numerous studies suggest that tocopherols are important antioxidants involved in photoprotection of plants. Tocopherol levels correlate well with the degree of oxidative stress in numerous plant species grown under various stress conditions, including high light, drought, and low temperatures (Wildi and Lutz, 1996; Streb et al., 1998; Bartoli et al., 1999; Havaux et al., 2000; Munne-Bosch and Alegre, 2000). Given this suggestive role of tocopherols in antioxidant and photoprotective function, a mutation that eliminates tocopherol synthesis in a photosynthetic organism would be anticipated to increase sensitivity to oxidative stress, reducing growth or viability of this organism under stressful conditions. Various mutants and transgenic plants with decreased tocopherol levels have been reported to exhibit photobleaching phenotypes and compromised growth (Henry et al., 1986; Norris et al., 1995; Tanaka et al., 1999). However, in all these cases, other prenyllipids such as phylloquinone, chlorophylls, carotenoids, or plastoquinone were affected in addition to tocopherols (Henry et al., 1986; Norris et al., 1995; Tanaka et al., 1999). Therefore, it was impossible to specifically attribute these phenotypes to tocopherol deficiency. The tocopherol-specific phenotype of SLR1736::Kmr provides a unique tool to begin to specifically address the question of tocopherol function in photosynthetic organisms.
It is surprising that SLR1736::Kmr growth rates under photoautotrophic and photoheterotrophic conditions in low and high light were indistinguishable from wild type. Mutant and wild-type whole-chain oxygen evolution rates were also similar. The observation that the absence of tocopherols did not appreciably affect growth, photosynthetic electron transport, and tolerance to high-light stress seemingly contradicts the concept that tocopherols are essential lipid soluble antioxidants. However, there are several possible explanations for these apparently incongruous results. First, α-tocopherol is not the only antioxidant present in photosynthetic membranes. Photosynthetic organisms have evolved multiple mechanisms for protection from oxidative stresses (carotenoids, ascorbate, superoxide dismutases, etc.) that if up-regulated, could partially or fully compensate for the absence of tocopherols in SLR1736::Kmr under certain conditions. In an alternate manner, tocopherols may protect from a specific type of lipid peroxidation or at a particular site. Finally, the limited oxidative treatments used in this report may not be sufficient to produce detectable differences between wild-type Synechocystis sp. PCC 6803 and SLR1736::Kmr at the level of culture growth rates. A detailed analysis of membrane lipids, lipid peroxidation products, and other component of oxidative stress compensation and adaptation are needed to discern any effects of tocopherol deficiency in SLR1736::Kmr.
Finally, one potential application of the described work would be to allow engineering of elevated tocopherol levels in food crops for nutritional purposes. Given the central position of HPT in tocopherol synthesis, it seems likely that the enzyme may be an important step for controling flux into the pathway. A crucial observation is that eliminating HPT activity in Synechocystis sp. PCC 6803 does not affect the levels of other biosynthetically related compounds (plastoquinone, phylloquinone, and chlorophylls). This suggests that altering HPT enzyme levels in plants may also be tocopherol specific and have little effect on the synthesis of other prenylquinone compounds in the plastid. Experiments are under way to positively modify AtHPT expression in Arabidopsis to test whether the activity is a target for engineering tocopherol levels in plants.
MATERIALS AND METHODS
Chemicals and Bacterial Strains
Prenyl-DPs were more than 99% pure. PDP was kindly provided by Dr. Stephanie Sen (Purdue University, Indianapolis), and GGDP and SDP were purchased from American Radiolabeled Chemicals (St. Louis). (U-14C)-HPP (0.6–1.5 μm) was prepared from (U-14C)-Tyr (specific activity 464 mCi mmol−1; Amersham, Arlington Heights, IL) as described by Schulz et al. (1993). Tocol was a gift from Eisai Company (Tokyo). A mixture of various cis- and trans-methyl-phytyl-1,4-benzoquinone isomers was synthesized by Dr. Daniel Liebler (University of Arizona, Tucson). 2′-trans-2-Me-6-Ph-1,4-BQ was purified from the mixture by a combination of TLC and HPLC (Henry et al., 1987). PQ-9 was extracted from Iris hollandica bulbs and purified by TLC (Pennock, 1985) and HPLC (Johnson et al., 2000). Wild-type Synechocystis sp. PCC 6803 was grown on BG-11 plates or liquid media (Williams, 1988) either photoheterotrophically (with 15 mm Glc) or photoautotrophically (without Glc) at 20 to 30 μE m−2 s−1 and 30°C unless otherwise stated. Synechocystis sp. PCC 6803 cells were subcultured at least three times in liquid media prior to growth experiments. Escherichia coli strains DH5α (Stratagene, La Jolla, CA) and BL-21 (DE3; Novagen, Milwaukee, WI) were used for conventional subcloning and protein expression, respectively.
Plasmids and Mutants
Primers 5′-TATTCATATGGCAACTATCCAAGCTTTTTG-3′ (SLR1736F) and 5′-GGATCCTAATTGAAGAAGATACTAAATAGTTC-3′ (SLR1736R) containing engineered NdeI and BamHI sites (underlined) and Vent DNA polymerase (New England Biolabs, Beverly, MA) were used to amplify the SLR1736 ORF (GenBank accession no. BBA17774) from Synechocystis sp. PCC 6803 genomic DNA. The amplified fragment was subcloned into the EcoRV site of pBluescript II KS (+) to generate pKS1736. pKS1736 was digested with MfeI and ligated with the EcoRI-digested kanamycin resistance cassette from pUC4K (Taylor and Rose, 1988). Two constructs with opposite orientation of the kanamycin resistance cassette relative to the SLR1736 ORF were used to transform wild-type Synechocystis sp. PCC 6803 and generate disruption mutants by homologous recombination (Williams, 1988). Transformants were subcultured on kanamycin containing media for several plating cycles and the absence of wild-type SLR1736 gene copies was confirmed by PCR using SLR1736F and R primers followed by Southern-blot analysis (Fig. 3, A and B). Because the two orientation disruption mutants were phenotypically indistinguishable (data not shown), that with the Kmr cassette in the same orientation as the SLR1736 ORF, referred to hereafter as the SLR1736::Kmr mutant (Fig. 3A), was used for further analyses. The NdeI-BamHI fragment from pKS1736 encoding the entire SLR1736 ORF was ligated into NdeI-BamHI-digested pET30b (Novagen) to create pSynHPT, which was transformed into BL-21 (DE3) cells for protein expression.
The SLR1736 protein sequence was used to search the Arabidopsis database and identify a single genomic clone, F19F24, containing a homologous sequence. Primers 5′-TTGTTTTCAGGCTGTTGTTGCAGCTCTC-3′ and 5′-CGTTTCTGACCCAGAGTTACAGAGAATG-3′ were used to amplify a 977-bp fragment from F19F24 for use as a probe to screen an Arabidopsis seed cDNA library (a gift of Dr. John Ohlrogge, Michigan State University, East Lansing). The longest of 15 positive clones was sequenced and shown to encode a protein similar to SLR1736 that was designated AtHPT. For protein expression purposes the full-length clone encoding AtHPT was amplified using primers (5′-CCATGGAGTCTCTGCTCTC-3′ and 5′-GGATCCCAAGCAGAGACTTCTTTACC-3′) and subcloned into NcoI-BamHI-digested pET3d vector (Novagen) to generate pAtHPT.
Prenyllipid Analysis
Fifteen to 20 mg of 14-d-old plate-grown Synechocystis sp. PCC 6803 cells were harvested, their lipids extracted (Bligh and Dyer, 1959), and dissolved in 100 μL of hexane or ethyl acetate. Ten microliters of each sample was withdrawn for chlorophyll determination (Lichtenthaler, 1987), whereas 50 μL was subjected to HPLC (Hewlett-Packard 1100, Wilmington, DE) on a LiChrosorb 5 Si60A 4.6- × 250-mm normal phase column (Phenomenex, Torrance, CA) at 42°C as described by Syvaoja et al. (1986). Tocopherols were detected by fluorescence using 290 nm excitation and 325 nm emission. For plastoquinone and phylloquinone analysis, separation was achieved on a reverse phase column (Spherisorb 5 ODS2 4.6 × 250 mm, Waters, Marlborough, MA) as described by Johnson et al. (2000). PQ-9 and phylloquinone were detected at 250 and 275 nm, respectively.
Homogentisate Polyprenyltransferase Assay
Each 0.2-mL reaction contained freshly prepared (U-14C)-HPP (approximately 0.2 μm, specific activity 464 mCi mmol−1), 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.6, 4 mm MgCl2, 50 mm potassium ascorbate, 100 μm KF, 0.2% (w/v) CHAPS, and 0.1 mg of total protein extracted from E. coli expressing HPPD (Norris et al., 1995). Individual reactions contained 100 μm PDP, GGDP, or SDP and the insoluble protein fraction from E. coli expressing pSynHPT (0.03 mg of protein), pAtHPT (1 mg of protein), or the empty pET vector (0.03 or 1 mg of protein). Reactions were incubated for 1 h at room temperature, extracted with two volumes of methanol:chloroform (1:1, v/v), and any newly formed prenylquinols were oxidized with AgO (Pennock, 1985). The organic phase was transferred to a fresh tube, evaporated to dryness, dissolved in ethyl acetate, and subjected to TLC on silica gel (J.T. Baker, Phillisburg, NJ) developed with 20% (v/v) ethyl ether in petroleum ether (Pennock, 1985). Labeled prenylquinones were detected by autoradiography for 14 d.
For HPLC analyses, the polyprenyltransferase assays were performed as described above except that larger volumes were used for the individual reactions to ensure formation of sufficient products: 0.5 mL for SynHPT with PDP and 5 mL for SynHPT with GGDP, and AtHPT with PDP or GGDP. At the end of incubation, reactions were spiked with 2′-trans-2-Me-6-Ph-1,4-BQ and extracted prenyllipids resolved by TLC as above. The areas corresponding to prenylquinones (RF 0.36–0.67) were scraped from the TLC plates, eluted with ethyl ether, dried under nitrogen, and dissolved in hexane. Samples were then subjected to HPLC on a normal phase column (LiChrosorb 5 Si60A, 4.6 × 250 mm) using 0.1% (v/v) dioxane in hexane as a mobile phase to separate various methyl-phytyl benzoquinone isomers (Henry et al., 1987). For geranylgeranylated quinone products, a mobile phase consisting of 0.15% (v/v) dioxane in iso-octane was used (Hutson and Threlfall, 1980). Prenylquinones were detected at 252 nm. Eluents were collected at 30- to 60-s intervals and the associated radioactivity determined by liquid scintillation counting.
Growth Curves
Wild-type Synechocystis sp. PCC 6803 and the SLR1736::Kmr mutant were inoculated to a final optical density at 730 nm of 0.05 in 50 mL of liquid BG-11 medium and grown at 30°C with vigorous shaking in four possible combinations: with or without 15 mm Glc and at 20 to 30 (low light) or 105 to 110 (high light) μE m−2 s−1. The optical density at 730 nm was measured every 6 to 12 h and used to calculate cell density (Williams, 1988).
Oxygen Evolution
Liquid cultures of photoautotrophically grown wild-type and mutant Synechocystis sp. PCC 6803 cells were washed twice and resuspended in fresh BG-11 medium at a concentration of 3 mg chlorophyll mL−1. The cells were exposed to three different high-light intensities for 5 min (0.75, 2, and 5 mE m−2 s−1). Oxygen measurements were performed with a Clark-type electrode at 25°C using a Hansatech CB1-D3 recording unit with Minirec recording software (Hansatech Instruments, King's Lynn, England). The oxygen evolution rate was calculated from the slope within the linear region of the curves.
Phylogenetic Analysis
Sequence alignment (ClustalW alignment using the BLOSUM series matrix) and subsequent phylogenetic analysis were performed using MacVector software (Genetic Computer Group). The N-terminal 96-amino acid extension of AtHPT and the corresponding N termini of the other polyprenyltransferases were not included in the phylogenetic analysis. The following protein sequences were used: Synechocystis sp. PCC 6803 ChlG (accession no. BAA10281), Arabidopsis ChlG (accession no. S60222), Synechocystis sp. PCC 6803 HPT (accession no. S74813), Nostoc HPT (480–1,445 bp of contig 566), Anabaena HPT (6,672–7,625 bp of contig c295), rice HPT (accession no. AX046728), maize HPT (accession no. AX046716), Arabidopsis HPT (accession no. AF324344), soybean HPT (accession no. AX046734), and wheat HPT (accession no. BE471221, overlapped BE471221 and BG604641 corresponded to the C-terminal part of AtHPT starting at Asp-160). For phylogenetic analysis, distances between the amino acid sequences were estimated by using the uncorrected p distance method with gaps distributed proportionally. The best tree was constructed by the Unweighted Pair-Group Method with Arithmetic Mean with random tie breaking. Bootstrapping (10,000 repetitions) confirmed the confidence of the best tree structure.
ACKNOWLEDGMENTS
We would like to thank Dr. Heiko Lokstein for technical assistance with oxygen evolution measurements and Dr. Zigang Cheng for purification of individual methyl-phytyl benzoquinone isomers. We are very grateful to Dr. Dave Shintani and the members of the DellaPenna laboratory for reviewing the manuscript and great moral support.
Footnotes
This work was supported by a grant from Pioneer Hi-Bred, Inc.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010421.
LITERATURE CITED
- Addlesee HA, Gibson LCD, Jensen PE, Hunter CN. Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chlP, of Synechocystissp. PCC 6803. FEBS Lett. 1996;389:126–130. doi: 10.1016/0014-5793(96)00549-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Simontacchi M, Tambussi E, Beltrano J, Montaldi E, Puntarulo S. Drought and watering-dependent oxidative stress: effect on antioxidant content in Triticum aestivumL. leaves. J Exp Bot. 1999;50:375–383. [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R, Traber MC. Vitamin E: function and metabolism. FASEB J. 1999;13:1145–1155. [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Prot Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen J, Bausch J, Glatzle D, Wagner E. Distribution of vitamin E in spruce seedlings and mature tree organs, and within the genus. Phytochemistry. 1991;30:147–151. [Google Scholar]
- Franzen J, Haas MM. Vitamin E content during development of some seedlings. Phytochemistry. 1991;30:2911–2913. [Google Scholar]
- Fryer MJ. The antioxidant effects of thylakoid vitamin E (α-tocopherol) Plant Cell Environ. 1992;15:381–392. [Google Scholar]
- Fryer MJ. Evidence for the photoprotective effects of vitamin E. Photochem Photobiol. 1993;58:304–312. doi: 10.1111/j.1751-1097.1993.tb09566.x. [DOI] [PubMed] [Google Scholar]
- Havaux M, Bonfils J-P, Lutz C, Niyogi KK. Photodamage of the photosynthetic apparatus and its dependence on the leaf developmental stage in the npq1Arabidopsis mutant deficient in the xanthophyll cycle enzyme violaxanthin de-epoxidase. Plant Physiol. 2000;124:273–284. doi: 10.1104/pp.124.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Powls R, Pennock JF. Scenedesmus obliquusPS28: α tocopherol-free mutant which cannot form phytol. Biochem Soc Trans. 1986;14:958–959. [Google Scholar]
- Henry A, Powls R, Pennock JF. Intermediates of tocopherol synthesis in the unicellular alga Scenedesmus obliquus. Biochem J. 1987;242:367–373. doi: 10.1042/bj2420367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson KG, Threlfall DR. Synthesis of plastoquinone-9 and phytylplastoquinone from homogentisate in lettuce chloroplasts. Biochim Biophys Acta. 1980;632:630–648. doi: 10.1016/0304-4165(80)90339-6. [DOI] [PubMed] [Google Scholar]
- Johnson TW, Shen G, Zybailov B, Kolling D, Reategui R, Beauparlant S, Vassiliev IR, Bryant DA, Jones AD, Golbeck J. Recruitment of a foreign quinone into the A1site of photosystem I. J Biol Chem. 2000;275:8523–8530. doi: 10.1074/jbc.275.12.8523. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S. Sequence analysis of the genome of the unicellular Cyanobacterium Synechocystissp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Keller Y, Bouvier F, D'Harlingue A, Camara B. Metabolic compartmentation of plastid prenyllipid biosynthesis: evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem. 1998;251:413–417. doi: 10.1046/j.1432-1327.1998.2510413.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Liebler DC. Antioxidant chemistry of α-tocopherol in biological systems: roles of redox cycles and metabolism. Subcell Biochem. 1998;30:301–317. doi: 10.1007/978-1-4899-1789-8_13. [DOI] [PubMed] [Google Scholar]
- Lopez JC, Ryan S, Blankenship RE. Sequence of the bchG gene from Chloroflexus aurantiacus: relationship between chlorophyll synthase and other polyprenyltransferases. J Bacteriol. 1996;178:3369–3373. doi: 10.1128/jb.178.11.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal E, Block MA, Dorne AJ, Douce R, Joyard J. Lipid synthesis and metabolism in the plastid envelope. Physiol Plant. 1997;100:65–77. [Google Scholar]
- Marshall PS, Morris SR, Threlfall DR. Biosynthesis of tocopherols: a re-examination of the biosynthesis and metabolism of 2-demethyl-6-phytyl-1,4-benzoquinol. Phytochemistry. 1985;24:1705–1711. [Google Scholar]
- Munne-Bosch S, Alegre L. Changes in carotenoids, tocopherols and diterpenes during drought and recovery, and the biological significance of chlorophyll loss in Rosmarinus officinalisplants. Planta. 2000;210:925–931. doi: 10.1007/s004250050699. [DOI] [PubMed] [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic disection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Shen X, DellaPenna D. Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 1998;117:1317–1323. doi: 10.1104/pp.117.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U, Bauer CE, Rudiger W. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Biol Chem. 1997;272:9671–9676. doi: 10.1074/jbc.272.15.9671. [DOI] [PubMed] [Google Scholar]
- Pennock JF. Biosynthesis of plastoquinone. Methods Enzymol. 1985;110:313–319. [Google Scholar]
- Piironen V, Syvaoja E-L, Varo P, Salminen K, Koivistoinen P. Tocopherols and tocotrienols in finish foods: vegetables, fruits, and berries. J Agric Food Chem. 1986;34:742–746. [Google Scholar]
- Schulz A, Ort O, Beyer P, Kleinig H. SC-0051, a 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993;318:162–166. doi: 10.1016/0014-5793(93)80013-k. [DOI] [PubMed] [Google Scholar]
- Schulze-Siebert D, Homeyer U, Soll J, Schultz G. Synthesis of plastoquinone-9, α-tocopherol and phylloquinone (vitamin K1) and its integration in chloroplast carbon metabolism of higher plants. In: Stumpf P, Mudd J, Nes W, editors. The Metabolism, Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 29–36. [Google Scholar]
- Shintani D, DellaPenna D. Elevating the vitamin E content of plants through metabolic engineering. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- Soll J, Kemmerling M, Schultz G. Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch Biochem Biophys. 1980;204:544–550. doi: 10.1016/0003-9861(80)90066-1. [DOI] [PubMed] [Google Scholar]
- Soll J, Schultz G. Comparison of geranylgeranyl and phytyl substituted methylquinols in the tocopherol synthesis of spinach chloroplasts. Biochem Biophys Res Comm. 1979;91:715–720. doi: 10.1016/0006-291x(79)91939-9. [DOI] [PubMed] [Google Scholar]
- Soll J, Schultz G, Joyard J, Douce R, Block MA. Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. In: Siegenthaler P-A, Eichenberger W, editors. Structure, Function and Metabolism of Plant Lipids. Amsterdam: Elsevier Science Publishers B. V.; 1984. pp. 263–266. [Google Scholar]
- Stocker A, Fretz H, Frick H, Ruttimann A, Woggon W-D. The substrate specificity of tocopherol cyclase. Bioorg Med Chem. 1996;4:1129–1134. doi: 10.1016/0968-0896(96)00125-3. [DOI] [PubMed] [Google Scholar]
- Streb P, Shang W, Feierabend J, Bligny R. Divergent strategies of photoprotection in high-mountain plants. Planta. 1998;207:313–324. [Google Scholar]
- Syvaoja EL, Piironen V, Varo P, Koivistoinen P, Salminen K. Tocopherols and tocotrienols in finish foods: oils and fats. J Am Oil Chem Soc. 1986;63:328–329. [Google Scholar]
- Tanaka R, Oster U, Kruse E, Rudiger W, Grimm B. Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 1999;120:695–704. doi: 10.1104/pp.120.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LA, Rose RE. Correction in the nucleotide sequence of the TN903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall DR, Whistance GR. Biosynthesis of isoprenoid quinones and chromanols. In: Goodwin T, editor. Aspects of Terpenoid Chemistry and Biochemistry. Liverpool, UK: Academic Press; 1971. pp. 357–404. [Google Scholar]
- Whistance GR, Threlfall DR. Biosynthesis of phytoquinones: homogentisic acid: a precursor of plastoquinones, tocopherols and α-tocopherolquinone in higher plants, green algae and blue-green algae. Biochem J. 1970;117:593–600. doi: 10.1042/bj1170593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildi B, Lutz C. Antioxidant composition of selected high alpine plant species from different altitudes. Plant Cell Environ. 1996;19:138–146. [Google Scholar]
- Williams JGK. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]