Abstract
The reorganization of the cellulose-xyloglucan matrix is proposed to serve as an important mechanism in the control of strength and extensibility of the plant primary cell wall. One of the key enzymes associated with xyloglucan metabolism is xyloglucan endotransglycosylase (XET), which catalyzes the endocleavage and religation of xyloglucan molecules. As with other plant species, XETs are encoded by a gene family in tomato (Lycopersicon esculentum cv T5). In a previous study, we demonstrated that the tomato XET gene LeEXT was abundantly expressed in the rapidly expanding region of the etiolated hypocotyl and was induced to higher levels by auxin. Here, we report the identification of a new tomato XET gene, LeXET2, that shows a different spatial expression and diametrically opposite pattern of auxin regulation from LeEXT. LeXET2 was expressed more abundantly in the mature nonelongating regions of the hypocotyl, and its mRNA abundance decreased dramatically following auxin treatment of etiolated hypocotyl segments. Analysis of the effect of several plant hormones on LeXET2 expression revealed that the inhibition of LeXET2 mRNA accumulation also occurred with cytokinin treatment. LeXET2 mRNA levels increased significantly in hypocotyl segments treated with gibberellin, but this increase could be prevented by adding auxin or cytokinin to the incubation media. Recombinant LeXET2 protein obtained by heterologous expression in Pichia pastoris exhibited greater XET activity against xyloglucan from tomato than that from three other species. The opposite patterns of expression and differential auxin regulation of LeXET2 and LeEXT suggest that they encode XETs with distinct roles during plant growth and development.
The plant primary cell wall is a complex and dynamic structure that undergoes substantial reorganization during cell growth and differentiation, and plays a fundamental role in controlling cell morphology. A major component of the primary wall is a network of cellulose microfibrils embedded in a matrix of hemicellulosic and pectic polysaccharides (Carpita and Gibeaut, 1993). Xyloglucan, the principal hemicellulosic cell wall polysaccharide in dicotyledons, associates non-covalently with cellulose, coating and cross-linking adjacent cellulose microfibrils (Hayashi, 1989; McCann et al., 1990). The resulting extensive xyloglucan-cellulose network is thought to act as the major tension-bearing structure in the primary wall; accordingly, the rearrangement of xyloglucan cross-links that tether the microfibrils potentially serves as an important mechanism for controlling the strength and extensibility of the cell wall.
A class of enzymes known as xyloglucan endotransglycosylases (XETs) catalyze the endocleavage of xyloglucan polymers and the subsequent transfer of the newly generated reducing ends to other polymeric, or oligomeric, xyloglucan molecules (Fry et al., 1992; Nishitani and Tominaga, 1992). Therefore, XET action represents a potential means to achieve regulated wall loosening during turgor-driven expansion by rearranging load-bearing xyloglucan cross-links between cellulose microfibrils. XETs may also catalyze molecular grafting reactions required to integrate nascent xyloglucan polysaccharides into the existing cell wall, thus maintaining wall thickness and integrity.
XET activity has been extracted from a variety of plant tissues, and XET gene families have been identified in a wide range of species (Nishitani, 1997; Campbell and Braam, 1999b). Although for any given XET gene family typically only a few genes have been demonstrated to encode true XETs (Arrowsmith and de Silva, 1995; Purugganan et al., 1997; Schröder et al., 1998; Campbell and Braam, 1999a), the high homology among designated XET genes, together with the presence of conserved key motifs, strongly suggests that they encode proteins with XET activity.
In agreement with a proposed role in plant growth, XET activity levels are high in rapidly growing tissues (Nishitani and Tominaga, 1991; Fry et al., 1992; Pritchard et al., 1993), and gibberellic acid treatment, which induces the elongation of leaves and stems in several plant species, increases XET activity (Potter and Fry, 1994; Smith et al., 1996). Furthermore, specific XET genes have been shown to be up-regulated by the growth-promoting hormones auxin, gibberellins, and brassinosteroids (Zurek and Clouse, 1994; Xu et al., 1996; Catalá et al., 1997; Schünmann et al., 1997). However, XET activity does not always correlate with growth rate, and activity has also been detected in vegetative tissues that have ceased to elongate (Smith et al., 1996; Barrachina and Lorences, 1998) and in ripening fruit (Redgwell and Fry, 1993; Maclachlan and Brady, 1994), and divergent XET genes are associated with wall reorganization during cellular differentiation and fruit ripening (Arrowsmith and de Silva, 1995; Saab and Sachs, 1996; Schröder et al., 1998). Thus, the presence of XETs with different tissue-specific expression, hormonal regulation, and/or potentially different enzymatic properties seems to be necessary for the metabolism of xyloglucan during various different stages of plant growth and development. The characterization of individual XET genes, and their corresponding proteins, within a single species is essential to understand their specific role.
Within the tomato (Lycopersicon esculentum) XET gene family, the expression of only four members has been characterized to date: two highly homologous cDNA clones, tXET-B1 and tXET-B2, from ripe fruit (Arrowsmith and de Silva, 1995); a brassinosteroid-regulated XET gene, Le-BR1 (Koka et al., 2000); and LeEXT (Okazawa et al., 1993), also called EXT3 (Campbell and Braam, 1999b), which is abundantly expressed in the epidermis of the apical elongating region of the tomato hypocotyl (Catalá et al., 1997). LeEXT mRNA accumulation increases by auxin treatments that also induce the elongation of apical hypocotyl segments. Here, we report the identification of a new tomato XET gene, LeXET2, and we describe its hormonal regulation in etiolated hypocotyls. We show that LeXET2 exhibits the opposite pattern of expression and auxin regulation from LeEXT and that the LeXET2 protein, obtained by heterologous expression in Pichia pastoris, has XET activity against tomato xyloglucan.
RESULTS
Cloning and Phylogenetic Analysis of LeXET2
In an effort to characterize the expression of the XET gene family in tomato, reverse transcriptase (RT)-PCR was used to amplify XET cDNA fragments from tomato hypocotyl RNA using degenerate primers based on deduced amino acid domains conserved between XETs. The sequence of one of the resulting PCR products did not correspond to any previously reported tomato XET and was designated LeXET2. The PCR product was used to screen a tomato hypocotyl cDNA library and a 1.16-kb full-length clone was isolated that corresponded to the size of the LeXET2 mRNA transcript.
The deduced amino acid sequence of LeXET2 has 47% amino acid identity to tomato LeEXT, another XET identified in elongating tomato hypocotyls (Okazawa et al., 1993; Catalá et al., 1997), and a 58% and 60% identity, respectively, to tXETB1 and tXETB2, two tomato XETs isolated from ripe fruit (de Silva et al., 1994). Of the tomato XETs that have been cloned to date, LeXET2 shares the highest identity (73%) with LeBR1, a brassinosteroid-regulated XET (Koka et al., 2000).
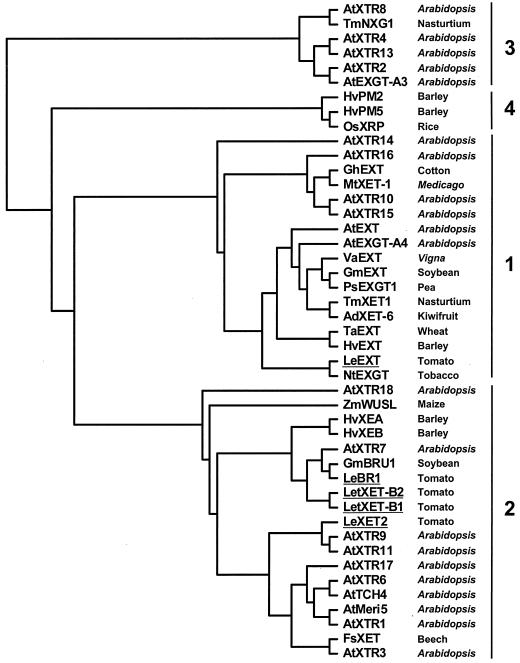
A phylogenetic tree generated from the alignment of the amino acid sequences of tomato LeXET2 and 44 published XET sequences revealed four distinct groups (Fig. 1), as has been reported by other authors (Nishitani, 1997; Campbell and Braam, 1999b). Group 1 contains genes that share a high level of sequence identity between different species and that are expressed in young developing tissues (Catalá et al., 1997; Shimizu et al., 1997; Akamatsu et al., 1999; Takano et al., 1999). Group 3 represents a divergent group of XETs, including NXG1 from nasturtium (Tropaeolum majus) that can act as a xyloglucan hydrolase and transglycosylase (de Silva et al., 1993). LeXET2 showed the highest homology to sequences in Group 2, which comprise XET genes from several species showing diverse patterns of expression and responses to hormonal or mechanical stimuli, including touch-inducible, flooding-responsive, brassinosteroid-inducible, and fruit ripening-related XETs.
Figure 1.
Phylogenetic alignment of the tomato LeXET2-deduced amino acid sequence with other plant XETs. LeXET2 was aligned with 44 full-length deduced amino acid sequences using the ClustalX method, and a phylogenetic tree was generated using the neighbor-joining method and the TreeView program. Tomato genes are shown underlined. Details and GenBank accession numbers are described in “Materials and Methods.”
Genomic Analysis of LeXET2
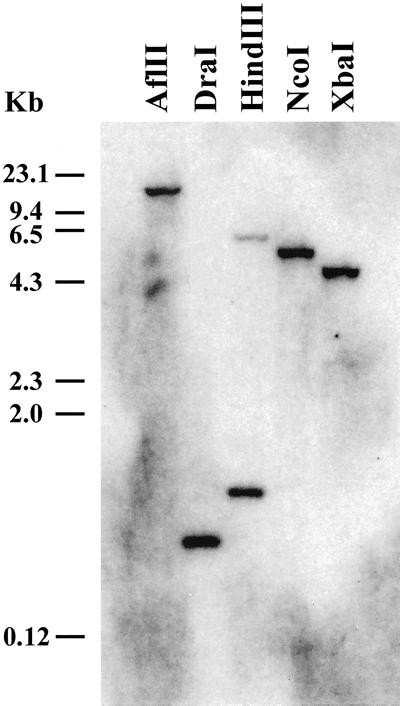
A gene-specific probe comprising the LeXET2 3′- end was used in genomic DNA gel-blot analysis at moderate stringency. The probe hybridized strongly to a single genomic fragment (Fig. 2), indicating the presence of a single gene. When the LeXET2 PCR fragment was used as a probe, weak hybridization was detected in some lanes to other fragments that may represent distantly related XET genes (data not shown). Both probes gave identical hybridization patterns when subsequently used to probe RNA gel blots (data not shown).
Figure 2.
Southern-blot analysis of LeXET2 gene in tomato. Genomic DNA (10 μg lane−1) was digested with the indicated restriction enzymes and the DNA gel blot hybridized with a radiolabeled LeXET2 3′-end cDNA as a probe. The blot was washed with 0.5× SSC and 0.5% (w/v) SDS at 65°C (14°C below the melting temperature).
Expression of LeXET2 in Different Tomato Tissues
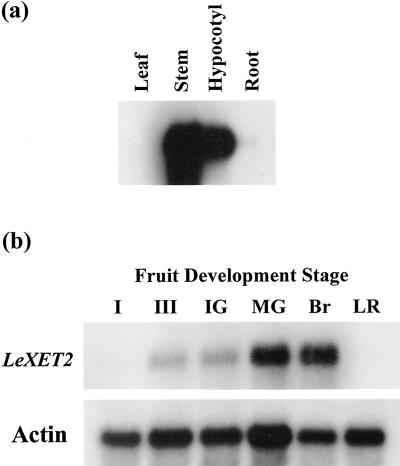
The pattern of LeXET2 expression was analyzed by RNA gel-blot analysis using poly(A+) RNA from different tomato vegetative tissues and from fruit at different developmental stages (Fig. 3). The strongest hybridization signal was detected in stems and hypocotyls, and only prolonged exposure showed some signal in roots and leaves (data not shown; Fig. 3a). LeXET2 was also expressed in fruit and although expression was barely detectable in the latest stages of fruit growth (stage III and immature green), mRNA levels increased in the mature green stage when growth had ceased and at the onset of ripening (breaker). However, LeXET2 expression was undetectable in ripe light-red fruit, a more advanced stage of ripening (Fig. 3b).
Figure 3.
Organ-specific expression pattern of tomato LeXET2. Poly(A+) RNA was isolated from different vegetative tissues (a) and tomato pericarp (b) at different stages of fruit development and was analyzed by northern blot (1 μg lane−1). Blots were hybridized with a LeEXT2 cDNA probe and washed in 0.5× SSC at 65°C. The fruit blot was subsequently hybridized with a tomato actin probe as a loading control. Exposure to photographic film was 30 times longer for the fruit tissue blot than for the vegetative tissue blot. I, 0.5- to 1-cm diameter fruit; III, 4- to 6-cm diameter fruit; IG, immature green; MG, mature green; Br, breaker; LR, light red.
LeXET2 and LeEXT Show Differential Spatial Localization and Regulation by Auxin in Etiolated Tomato Hypocotyls
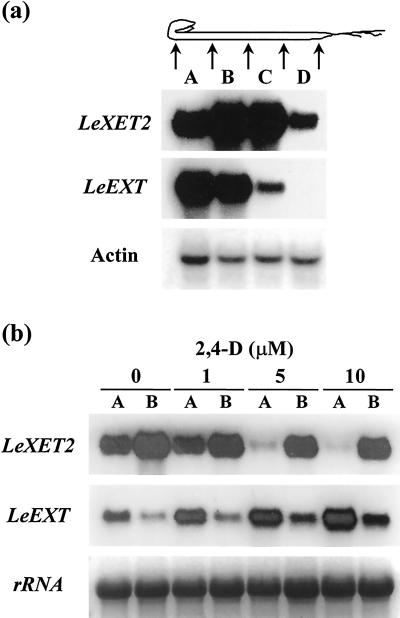
The expression of LeXET2 in different regions of etiolated hypocotyls was analyzed in comparison with a previously characterized XET gene, LeEXT (Catalá et al., 1997). A gradient in growth rate has been observed in etiolated tomato seedlings, with the maximum rate in the region immediately below the apical hook (Catalá et al., 2000). As previously described, LeEXT showed higher levels of expression in apical segments and mRNA levels decreased toward the base of the hypocotyl in good correlation with the growth rate. In contrast, LeXET2 mRNA abundance was lower in the apical region, increased in the middle zone of the hypocotyl (which has a much lower elongation rate), and diminished again in the basal region, where elongation had ceased (Fig. 4a).
Figure 4.
Differential expression and auxin regulation of the XET genes LeXET2 and LeEXT in etiolated tomato hypocotyls. a, RNA gel-blot analysis of LeXET2 and LeEXT expression along the tomato hypocotyl. Each lane contained 10 μg of total RNA isolated from consecutive 1-cm sections (A–D) of the hypocotyl. Gel blots were hybridized successively with the LeEXT2, LeEXT, and actin cDNA probes. b, Effect of auxin concentration on LeXET2 and LeEXT mRNA accumulation. Hypocotyl sections (1 cm) were cut immediately below the apical hook (A) or from a region 1.5 cm below the hook (B), incubated in the presence of the indicated concentration of 2,4-dichlorophenoxyacetic acid (2,4-d) for 24 h, and RNA was isolated. Total RNA (15 μg lane−1) gel blots were hybridized with the LeXET2 and LeEXT cDNA probe. Ethidium bromide staining of ribosomal RNA is shown as a loading control.
We had previously shown that LeEXT mRNA accumulation was induced by auxin during the elongation of etiolated hypocotyl segments (Catalá et al., 1997). To test whether auxin had a differential effect on the expression of different XET genes, mRNA levels of LeXET2 and LeEXT were examined in hypocotyl segments treated with buffer alone or with buffer plus the synthetic auxin 2,4-d (Fig. 4b). The presence of 2,4-d caused an increase in LeEXT expression in apical and basal hypocotyl segments; however, LeXET2 mRNA levels decreased in apical segments incubated with increasing auxin concentrations. Only a slight decrease was observed in basal segments treated with auxin.
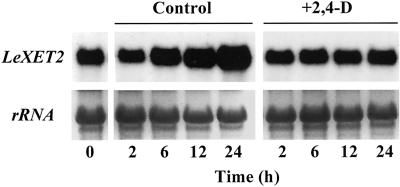
Time Course Analysis of LeXET2 mRNA Accumulation in Tomato Hypocotyl Segments
Previous studies have shown that etiolated hypocotyl segment elongation is stimulated 3-fold over a 12-h time period in the presence of 2,4-d and that no additional increase in segment length occurs between 12 and 24 h (Catalá et al., 1997). A time course analysis of LeXET2 mRNA accumulation in hypocotyl segments showed that during the incubation of segments with buffer alone, there was an increase in the levels of LeXET2 mRNA (Fig. 5). However, the addition of auxin to the incubation buffer prevented this increase and resulted in constant expression levels throughout the 24-h incubation period (Fig. 5). Therefore, Figure 4, which shows the effect of auxin at a single time point (24 h), reflects the lack of mRNA accumulation in the presence of varying concentrations of auxin.
Figure 5.
Time course analysis of LeXET2 mRNA accumulation in tomato hypocotyl segments. Apical segments were incubated for the indicated times in buffer alone (control) or buffer plus 5 μm 2,4-d and RNA was isolated. Total RNA (15 μg lane−1) gel blots were hybridized with the LeXET2 cDNA probe. Ethidium bromide staining of ribosomal RNA is shown as a loading control.
To check the potential effect of wounding on LeXET2 expression after segment excision, hypocotyl segments were cut and stored, without buffer solution, in a humidified chamber. LeXET2 mRNA abundance in samples collected 24 h after excision showed no changes compared with the control at 0 h (data not shown).
Hormonal Regulation of LeXET2
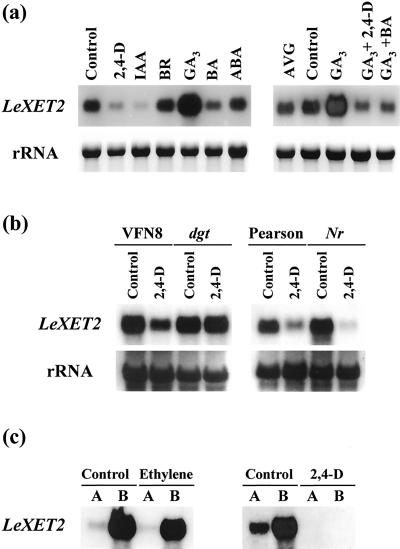
To further characterize the hormonal regulation of LeXET2 expression, the effect of several plant hormones on LeXET2 mRNA abundance was examined (Fig. 6). Apical segments were incubated with 2,4-d, the natural auxin, IAA, GA3, the cytokinin BA, BR, and ABA. After 20 h of incubation, LeXET2 mRNA levels were lower in the segments treated with auxin, as expected, but also in those treated with cytokinin. Brassinosteroid and ABA had no effect; however, LeXET2 mRNA accumulation was enhanced substantially following treatment with GA3 (Fig. 6a). Of these treatments, only 2,4-d, IAA, and BR induced segment elongation (data not shown), and segments did not elongate with GA3. When segments were incubated with GA3 plus auxin or GA3 plus BA, the increase in mRNA levels caused by GA3 was abolished. To test a potential effect of wound-induced ethylene, hypocotyl segments were incubated with aminoethoxyvinyl Gly, an inhibitor of ethylene biosynthesis. Aminoethoxyvinyl Gly showed no effect, and LeXET2 mRNA accumulated to the same extent as in the buffer control (Fig. 6a).
Figure 6.
Hormonal regulation of LeXET2 mRNA levels in tomato hypocotyl segments. a, Effect of several plant hormones on LeXET2 gene expression. Apical segments were incubated for 20 h with buffer, with buffer plus 5 μm 2,4-d, 10 μm indole-3-acetic acid (IAA), 1 μm brassinolide (BR), 10 μm GA3, 10 μm benzyladenine (BA), 10 μm abscisic acid (ABA), or 1 μm aminoethoxyvinyl Gly (AVG), or a combination of GA3 plus 2,4-d or BA. b, Auxin regulation of LeXET2 mRNA accumulation in Nr and dgt hypocotyl segments. Apical segments from the ethylene-insensitive mutant Nr and the corresponding wild type (Pearson), or from the auxin-insensitive mutant dgt and its corresponding wild type (VFN8) were incubated in buffer alone or in buffer plus 5 μm 2,4-d for 20 h. c, Intact etiolated seedlings were treated with a continuous flow of air, air containing 10 μL L−1 of ethylene, or were sprayed with a 1-mm solution of 2,4-d, and apical elongating (A) or mature (B) hypocotyl sections were cut after 48 h for RNA isolation. Total RNA (15 μg lane−1) gel blots were hybridized with the LeXET2 cDNA probe. Ethidium bromide staining of ribosomal RNA is shown as a loading control.
Auxin treatment of plant tissues including hypocotyl segments can lead to endogenous ethylene production. To discriminate between the possible regulatory roles of auxin compared with auxin-induced ethylene, the expression of LeXET2 mRNA was examined in two tomato mutants, dgt and Nr, defective in sensitivity to auxin (Kelly and Bradford, 1986) and ethylene (Lanahan et al., 1994), respectively. Figure 6b shows the mRNA levels in apical segments from both mutants and their corresponding wild types after incubation in buffer with or without auxin. Following the auxin treatment, LeXET2 mRNA abundance was reduced in the segments from VFN8 wild type but did not change in dgt segments. However, reduction in LeXET2 mRNA accumulation was similar in segments from the Nr mutant and wild type.
Regulation of LeXET2 expression by ethylene was further examined by treating intact etiolated seedlings with air containing 10 μL L−1 of ethylene (Fig. 6c). LeXET2 mRNA abundance in apical and mature regions of the hypocotyl did not change significantly with the ethylene treatment. However, spraying entire seedlings with 2,4-d, a treatment that caused similar thickening and shortening of the hypocotyls to the ethylene treatment, had a dramatic effect on LeXET2 mRNA abundance, which decreased to undetectable levels in elongating and mature regions.
Production and Activity of Recombinant LeXET2
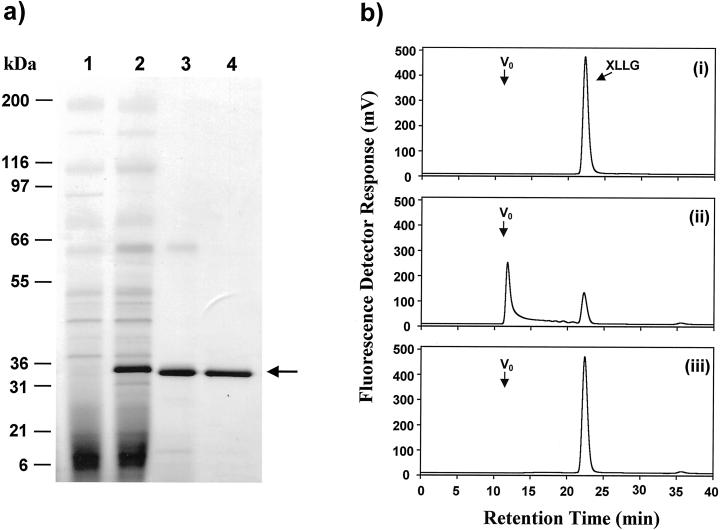
Recombinant LeXET2 was produced using the yeast P. pastoris heterologous expression system. An expression vector designed to produce the LeXET2 protein, including the putative N-terminal signal sequence, was used to transform P. pastoris, and the culture medium of transformed yeast was examined for the presence of LeXET2 protein and XET activity (Fig. 7). SDS-PAGE electrophoresis analysis showed the presence of a 32- to 33-kD protein in the culture medium of yeast transformed with the LeXET2 expression vector (Fig. 7a, lane 2) but not in the medium from control yeast transformed with the empty vector (Fig. 7a, lane 1). XET activity was assayed as the ability to generate a fluorescent high-Mr xyloglucan after transglycosylation between tamarind (Tamarindus indica) seed xyloglucan and an oligosaccharide (XLLG) tagged with a fluorescent group (Fig. 7b). XET activity was detected in the culture medium of yeast expressing LeXET2, indicating that the LeXET2 signal peptide is processed in yeast cells and the protein is secreted in an active form. Segments that void the exclusion volume of the Superdex-75 column were transferred from polymeric xyloglucan to the 1.4-kD oligosaccharide covalently linked to the fluorophor 9-aminopyrene-1,4,6-trisulfonate (APTS) (Fig. 7b, ii). No XET activity was present in the medium from control-transformed cultures (Fig. 7b, iii).
Figure 7.
Purification of recombinant LeXET2 from P. pastoris culture medium and activity of the recombinant protein. a, Proteins were separated on a SDS-polyacrylamide gel and were stained with Coomassie Blue. Proteins secreted from yeast transformed with pPIC3.5K vector only (1); proteins secreted from yeast transformed with LeXET2 in pPIC3.5K (2); eluate after SP-Sepharose chromatography of the culture media from yeast expressing LeXET2 (3); and purified LeXET2 after gel permeation chromatography (4). Mr markers are shown. The arrow indicates the band corresponding to LeXET2. b, Gel permeation chromatography showing transglycosylation between high-Mr tamarind xyloglucan and a xyloglucan oligosaccharide (XLLG) labeled with APTS, a fluorescent tag. Ten microliters of P. pastoris culture medium was incubated with 40 μg of tamarind xyloglucan and 282 pmol of XLLG-APTS in a total volume of 40 μL for 1 h at 25°C. The reaction mixtures were then analyzed by gel permeation chromatography using a Superdex-75 HR 10/30 column, as described in “Materials and Methods,” and fluorescent transglycosylation products were detected in the eluate with a fluorescent detector. XLLG-APTS alone (i); reaction containing high-Mr tamarind xyloglucan, XLLG-APTS, and culture medium from P. pastoris transformed with LeXET2 in pPIC3.5K (ii); and reaction containing high-Mr tamarind xyloglucan, XLLG-APTS, and culture medium from P. pastoris transformed with empty pPIC3.5K (iii). Vo, Void volume.
The LeXET2 protein was purified from the P. pastoris culture medium and was used to compare its activity against xyloglucans from different plant species (Table I) to determine whether substrate structural specificity was apparent. LeXET2 was active against tamarind seed xyloglucan, which has a β-1→4-d glucan backbone bearing α-d-xylosyl or β-d-galactosyl-(1→2)-α-d-xylosyl side chains at O-6 of 75% of the backbone glucosyl residues (Vincken et al., 1997). In addition to tamarind xyloglucan, LeXET2 was active against xyloglucan from bean (Phaseolus vulgaris) and sycamore (Acer pseudoplatanus) suspension-cultured cells, both of which contain α-l-fucosyl-(1→2)-β-d-galactosyl-(1→2)-α-d-xylosyl side chains. However, LeXET2 showed the highest activity with xyloglucans from tomato suspension-cultured cells as donor substrates (Table I). Tomato xyloglucans are unusual in that the glucan backbone is less substituted and the side chains do not contain Fuc, but have arabinosyl residues linked directly to the xylosyl residues (York et al., 1996).
Table I.
Effect of different xyloglucan donor substrates on LeXET2 activity
| Donor Substrate | Fucose | XET Activity |
|---|---|---|
| % | ||
| Tamarind seed xyloglucan | No | 16 |
| Bean suspension-cultured cells xyloglucan | Yes | 14 |
| Sycamore suspension-cultured cells xyloglucan | Yes | 16 |
| Tomato suspension-cultured cells xyloglucan | No | 66 |
Reactions containing 100 μg of donor substrate, 282 pmol XLLG-APTS, and 50 ng of recombinant LeXET2 protein in a total volume of 40 μL were incubated for 1 h at 25°C and were analyzed by gel permeation chromatography on a Superdex-75 column as described in “Materials and Methods.” Fluorescent transglycosylation products were detected in the eluate with a fluorescent detector. XET activity was expressed as a percentage of the peak area of the high-Mr fluorescent transglycosylation products relative to the total peak area of all fluorescent derivatives.
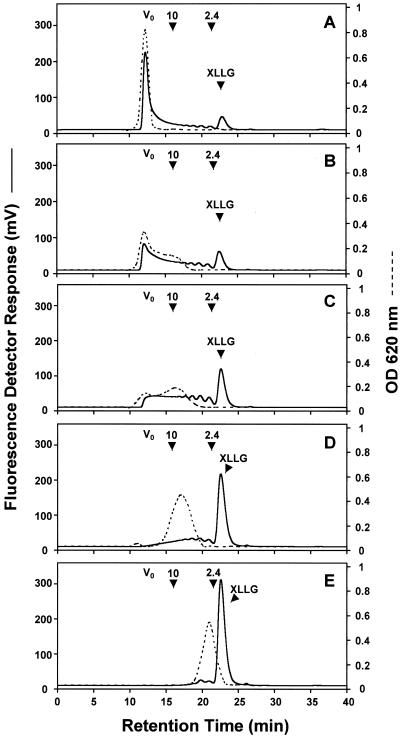
The activity of LeXET2 was further investigated using unlabeled tomato xyloglucans of different molecular sizes and fluorescently labeled XLLG as the donor and acceptor substrates, respectively (Fig. 8). Tomato xyloglucan isolated from the extracellular polysaccharide of suspension-cultured cells is a heterodisperse polymer comprising molecules of a wide range of molecular sizes. Separation of this polymer on a Sephacryl-200 column yielded fractions with molecular masses ranging from 70 to 10 kD (data not shown), which were used as donor substrates (Fig. 8, A–C). Smaller molecular mass xyloglucan fractions (<10 kD) and XLLGs, obtained by treatment of the tomato xyloglucan polymer with a fungal xyloglucanase (Pauly et al., 1999), were also tested as donor substrates (Fig. 8, D and E). Figure 8 shows the profiles of the xyloglucan donor substrates on a Superdex-75 column prior to the XET reaction. The presence of fluorescent peaks eluting from the column before the fluorescently labeled XLLG acceptor reflects transglycosylation between the acceptor and another xyloglucan molecule (Fig. 8). LeXET2 exhibited higher activity when donor substrates enriched in high-Mr xyloglucans were used (Fig. 8, A–D). Transglycosylation was barely detectable when an XLLG fraction containing mainly a trimer and dimer of an XXXG (for nomenclature, see Fry et al., 1993) repeating unit were used as donor substrates (Fig. 8E).
Figure 8.
Effect of the molecular size of the xyloglucan donor substrate on LeXET2 activity. Xyloglucan fractions obtained after separation of arabinoxyloglucans (AXGs) from tomato suspension-cultured cells were used as a substrate in a reaction containing 100 μg of donor substrate, 282 pmol of XLLG-APTS, and 50 ng of enzyme in a total volume of 40 μL. Reactions were incubated for 1 h at 25°C and were analyzed by gel permeation chromatography on a Superdex-75 column as described in “Materials and Methods.” Prior to the reaction, the profiles of the xyloglucan donor substrates were analyzed on the same column measuring the total sugar content in the eluate by the anthrone assay (OD = 620 nm, dotted lines). Fluorescent transglycosylation products were detected in the eluate with a fluorescent detector (solid lines). A, Donor substrate with a molecular mass of 40 to 70 kD; B, donor substrate with a molecular mass of 10 to 40 kD; C, donor substrate with a molecular mass of 10 to 30 kD; D, donor substrate with a molecular mass around 10 kD; E, donor substrate with a molecular mass of 2.4 to 10 kD. Vo, Void volume. The elution time of a 10-kD average molecular mass Dextran and of a 2.4-kD xyloglucan oligosaccharide is indicated by an arrow.
No oligomers were produced when LeXET2 was incubated with xyloglucan alone, which suggests that LeXET2 does not function as a xyloglucan hydrolase (data not shown).
DISCUSSION
Tomato XETs constitute a gene family, which suggests that individual XETs may exhibit distinct patterns of expression and/or hormonal regulation, together with specific enzymatic properties conferring on them unique functions in cell wall modification. Therefore, a careful analysis of the individual tomato genes, and the corresponding proteins, is necessary to understand the functional significance of specific members of the XET gene family. We have identified an additional member of the tomato XET gene family, LeXET2, which is expressed in tomato stems and hypocotyls and, much less abundantly, in fruit. In etiolated hypocotyls and during fruit development, LeXET2 mRNA accumulates after rapid elongation has ceased (Figs. 3 and 4). This contrasts with the pattern of expression of a previously identified tomato XET gene, LeEXT, which is primarily expressed in the apical elongating zone of etiolated hypocotyls (Fig. 4a) and in the stages of rapid expansion during fruit development (Catalá et al., 2000). Furthermore, LeXET2 and LeEXT expression in hypocotyl segments shows diametrically opposite patterns of regulation by auxin, a growth-promoting hormone. Although LeXET2 mRNA abundance is negatively regulated by auxin, LeEXT mRNA levels increase substantially with auxin treatment (Fig. 4b). A time course analysis revealed that auxin suppresses the increase in LeXET2 mRNA levels that occurs during the incubation of hypocotyl segments in buffer alone. Catalá et al. (1997) have shown that the time dependence of LeEXT mRNA accumulation during the incubation of hypocotyl segments in buffer follows the opposite trend: LeEXT mRNA levels decrease to undetectable levels after 24 h, coinciding with a drop in the rate of elongation. Incubation with auxin maintains the rate of elongation and the levels of LeEXT mRNA.
Together, these data suggest that although LeEXT is likely to play a role in cell wall metabolism during rapid cell expansion, LeXET2 may be involved in cell wall restructuring after expansion has occurred, such as in the trimming and reorganization of non-cellulose-bound domains of newly deposited xyloglucan (Thompson and Fry, 1997). Another possibility is that LeXET2 participates in incorporating xyloglucan to reinforce the walls of cells that have recently completed expansion. In addition to showing strikingly opposite patterns of expression, LeXET2 and LeEXT belong to divergent phylogenetic groups (Fig. 1). LeEXT belongs to group 1, which comprises closely related XETs from different plant species that are expressed in rapidly expanding tissues (Catalá et al., 1997; Shimizu et al., 1997; Akamatsu et al., 1999; Takano et al., 1999), whereas LeXET2 is more closely related to XETs in group 2, which contains genes regulated by a diverse range of environmental and hormonal stimuli not always associated with growth (Xu et al., 1995; Saab and Sachs, 1996). However, a detailed study of the expression of XET genes included in those diverse phylogenetic groups is far from complete. It is likely that the divergence between groups 1 and 2 also reflects distinctly different biological functions, but it still needs to be addressed whether there are differences in enzymatic properties between members within the two groups.
The increase in LeXET2 mRNA levels during incubation of segments in buffer alone (Fig. 5a) is intriguing. It is possible that segment excision causes wound-induced LeXET2 expression. However, we could not detect induction of LeXET2 gene expression upon wounding of hypocotyls or leaves that were not incubated in buffer (data not shown). Induction by wound-induced ethylene seems unlikely due to the inability of an inhibitor of the ethylene biosynthesis to prevent the increase in LeXET2 mRNA levels during the incubation of segments in buffer (Fig. 6a). Our hypothesis is that during the incubation in buffer alone, the segments are depleted of a factor that represses LeXET2 transcription or enhances its mRNA degradation. It is likely that this factor is auxin; this would explain why the addition of exogenous 2,4-d or IAA prevented the increase in LeXET2 mRNA levels (Figs. 5 and 6a). This is further supported by the observation that exogenous auxin has no effect on LeXET2 mRNA accumulation in segments of the auxin-insensitive tomato mutant dgt (Fig. 6b). Transcriptional down-regulation of gene expression by auxin has been demonstrated in several instances, although it has not been established whether down- and up-regulated auxin responses share similar mechanisms (Sitbon and Perrot-Rechenman, 1997). The earliest detectable decrease in LeXET2 mRNA levels, compared with the control, occurs after 2 h of incubation. Thus, LeXET2 does not belong to a class of early auxin-regulated genes (Sitbon and Perrot-Rechenman, 1997), but rather to a class of secondary response genes associated with long-term responses to auxin. Auxin treatment of tomato hypocotyl segments can lead to ethylene production (Kelly and Bradford, 1986); however, the auxin inhibition of LeXET2 mRNA accumulation was unaffected by a mutation in the ethylene response pathway (Fig. 6b). In addition, exogenous ethylene did not alter LeXET2 expression in intact etiolated seedlings, whereas spraying with 2,4-d caused a dramatic reduction in LeXET2 mRNA levels (Fig. 6c). Together, these results indicate that auxin inhibition of LeXET2 mRNA accumulation is likely due to changes mediated by auxin and not to an indirect effect of auxin-induced ethylene production. The effect of auxin on LeXET2 expression in hypocotyls segments was mimicked by a treatment with the cytokinin BA. Incubation of segments with BA resulted in lower LeXET2 mRNA levels similar to the 2,4-d or IAA treatments (Fig. 6a). Interactions between auxin and cytokinin signaling pathways, as well as cytokinin-induced increases in free auxin, have been reported in other plant systems (Coenen and Lomax, 1997), and both could explain the similar effects of auxin and BA on regulating LeXET2 gene expression.
A significant increase in LeXET2 mRNA accumulation was observed after incubation of hypocotyls segments with the gibberellin GA3 (Fig. 6a). Gibberellins regulate tissue elongation in several plants, and an effect of gibberellins on wall extensibility, including promotion of wall loosening reactions, has been proposed (Cosgrove and Sovonick-Dunford, 1989). GA3 modulation of XET activity during elongation has also been described (Potter and Fry, 1993), and induction of XET gene expression, concomitant with organ or tissue elongation, has been shown in rice (Oryza sativa) and barley (Hordeum vulgare; Schünmann et al., 1997; Uozu et al., 2000). Although GA3 stimulates elongation in intact plants, it is less effective when applied to excised segments. The significance of the GA3-induced increase in LeXET2 mRNA levels in our system is unclear given that GA3 has no detectable effect on the growth of tomato-etiolated hypocotyl segments. Auxin and BA were able to counteract the increase in LeXET2 mRNA accumulation caused by incubation of segments with GA3 (Fig. 6a), indicating that the effect of auxin and BA predominates over GA3 up-regulation of LeXET2 gene expression.
We have shown that LeXET2 encodes a protein with XET activity. When LeXET2 containing its own putative signal peptide is expressed in P. pastoris, the protein is secreted to the culture media in an active form. The LeXET2 mature protein has a predicted molecular mass of 28.87 kD. The experimental molecular mass of the heterologous protein is 30.7 kD, as indicated by mass spectrometry (data not shown). This suggests that the LeXET2 polypeptide is glycosylated in P. pastoris. LeXET2 contains a N-glycosylation consensus site that is conserved among most XETs (Campbell and Braam, 1999b).
A number of structural features of xyloglucan appear to be important for its suitability as a substrate for XETs, including the pattern of side chain substitution (Nishitani, 1997). Xyloglucans from most dicotyledonous plants are highly branched polysaccharides with approximately 75% of the β-1→4 glucosyl residues in the backbone carrying an α-d-xylosyl residue at O-6 (Vincken et al., 1997). The presence of β-d-galactosyl or α-l-fucosyl-(1→2)-β-d-galactosyl residues linked to the xylosyl side chains is also a characteristic of most dicotyledonous plant xyloglucan, although it is not a requirement for XET activity. Several previous reports have described XETs that show greater affinity for non-fucosylated xyloglucan (Rose et al., 1996; Purugganan et al., 1997; Campbell and Braam, 1999a). In contrast, although LeXET2 was active against fucosylated xyloglucan from two different plant species and non-fucosylated xyloglucans from tamarind seeds, it showed a substantially greater activity against tomato xyloglucan (Table I). Xyloglucan from tomato and other solanaceous species is distinct from xyloglucan that has so far been characterized from other plant families because only 40% of the backbone glucosyl residues are substituted and the side chains do not contain α-l-fucosyl-(1→2)-β-d-galactosyl moieties. Instead, up to 60% of the α-d-xylosyl residues are substituted with α-l-arabinosyl residues (York et al., 1996), and therefore, these xyloglucans have been called AXGs. The higher activity of LeXET2 against tomato AXG could result from the lower degree of backbone substitution in this polysaccharide, and/or from the different structure and composition of its side chains. We are currently studying the importance of specific structural features of AXG for its suitability as donor and acceptor substrate for tomato XETs.
LeXET2 showed a higher activity with xyloglucan polymers than with xyloglucan oligosaccharides as the donor substrates (Fig. 8). These results are similar to those obtained by Nishitani and Tominaga (1992) with a Vigna angularis XET that exhibits donor substrate specificity for xyloglucans with a molecular mass larger than 10 kD, but differ from the reported activity of a divergent nasturtium XET that is able to catalyze endotransglycosylation between xyloglucan oligomers (Fanutti et al., 1993). Xyloglucans undergo substantial changes in Mr following synthesis and deposition in the cell wall, during cell expansion (Talbott and Ray, 1992; Thompson and Fry, 1997). The presence of a family of XETs that are active against donor substrates of different sizes might contribute to the variation in the Mr distribution of xyloglucans during cell growth.
In conclusion, the up-regulation of LeXET2 mRNA accumulation by GA3 and the potential for shared signal transduction elements between auxin and BA in repressing LeXET2 mRNA accumulation imply that XET expression is regulated through a complex interacting network of plant hormones. The diametrically opposite patterns of expression and auxin regulation of two tomato XETs, LeXET2 and LeEXT, suggests that they possess distinct functions within the cell wall and makes them an excellent system in which to study the divergent roles of individual XETs.
MATERIALS AND METHODS
Plant Materials
Tomato (Lycopersicon esculentum cv T5) seeds were sown in moist vermiculite and etiolated seedlings were grown in the dark for 6 to 7 d at 25°C. Mutant Nr seeds were in a Pearson genetic background and mutant dgt seeds were in a VFN8 background. Fruit and vegetative tissues were harvested from greenhouse-grown tomatoes. Fruit were staged as described in Catalá et al. (2000).
Hormone Treatments
Tomato hypocotyl sections (6 mm) were cut directly below the apical hook or from basal regions and were incubated in 25 mm potassium phosphate buffer (pH 6) and 2% (w/v) Suc for 2 to 3 h. Buffer was replaced with fresh buffer or buffer containing a particular hormone concentration and segments were incubated at 25°C in the dark with gentle agitation.
Intact seedlings were exposed to a continuous flow of air containing 10 μL L−1 of ethylene in the dark, or were sprayed with a solution of 1 mm 2,4-d and incubated for 48 h. After incubation, apical and basal segments were excised and frozen. All experiments were repeated at least twice and representative data are shown.
PCR Amplification and cDNA Library Screening
Degenerate primers were designed from known conserved deduced amino acid domains of XETs (Okazawa et al., 1993). Primers (5′) GARCAYGAYGARATHGAYTTYG and (3′) TCNGTRCARTARTTRTADATNG were used to amplify an XET cDNA fragment from tomato hypocotyl RNA, as described in Rose et al. (1996), with an annealing temperature of 40°C. The resulting 485-bp cDNA fragment corresponding to amino acids 98 through 259 of the full-length clone was cloned into PCRII (Invitrogen, San Diego) and sequenced using universal primers and the Sequenase version 2.0 sequencing kit (USB, Cleveland), according to the manufacturer's instructions.
The PCR fragment was used to screen a tomato hypocotyl cDNA library in the pARC7 vector (O'Neill et al., 1990), a generous gift of Prof. Sharman O'Neill (University of California, Davis). Twenty-four independent inserts were isolated, and three of the largest clones were subcloned into pBluescript II and DNA sequence determined with universal and specific internal primers (Genset Corporation, La Jolla, CA), using an ABI 377 (Perkin Elmer, Foster City, CA) utilizing dye terminator chemistry with AmpliTaq DNA polymerase, FS (Taq; FS; Perkin Elmer). The longest clone was designated LeXET2 (accession no. AF176776).
The full deduced amino acid sequence of tomato LeXET2 was aligned with the corresponding sequences of other XETs using the ClustalX method (Thompson et al., 1997), and a tree was generated using the neighbor-joining method and the TreeView program (Page, 1996). The GenBank accession numbers are: Arabidopsis AtEXT, D164454; EXGT-A3, D63509; AtEXGT-A4, AF163822; AtMeri5, D63508; TCH4, AF051338; AtXTR1, AC004512; AtXTR2, U43487; AtXTR3, U43485; AtXTR4, U43486; AtXTR6, U43488; AtXTR7, U43489; AtXTR8, X92975; AtXTR9, AF093672; AtXTR10, AL021684; AtXTR11, AB011482; AtXTR13, AL021711; AtXTR14, AAC5398; AtXTR15, AL035709; AtXTR16, AC005275; AtXTR17, AC005724; AtXTR18, AL035353; azuki bean (Vigna angularis) VaEXT, D16458; barley (Hordeum vulgare) HvEXT, X91659; HvXEA, X93174; HvXEB, X93175; HvPM2, X91660; HvPM5, X93173; beech (Fagus spp.) FsXET, AJ130885; cotton (Gossypium hirsutum) GhEXT, D88413; kiwifruit (Actinidia deliciosa) AdXET-5, L46792; Medicago truncatula MtXET-1, AF093507; nasturtium (Tropaeolum majus) TmNXG1, X68254; TmXET1, L43094; maize (Zea mays) WUSL, U15781; pea (Pisum sativum) PsEXGT1, AB015428; rice (Oryza sativa) OsXRP, JE0156; soybean (Glycine max) GmBRU1, L22162; GmEXT, D16455; tobacco (Nicotiana tabacum) NtEXGT, D86730; tomato LeEXT, D16456; LeXET2, AF176776; LetXET-B1, X82685; LetXET-B2, X82684; LeBR1, AF205069; and wheat (Triticum aestivum) TaEXT, D16457.
Genomic DNA Isolation and Analysis
Genomic DNA was isolated from tomato leaves as described in Murray and Thompson (1980). Genomic DNA samples (10 μg) were digested with the indicated restriction enzymes, fractionated by electrophoresis on 0.8% (w/v) agarose gels, and transferred to Hybond-N membrane (Amersham, Arlington Heights, IL). Blots were hybridized with a PflMI/HindIII cDNA fragment from the full-length clone or with the 485-bp LeXET2 PCR product, radiolabeled by random hexamer priming using [α-32P] dATP (3,000 Ci mmol−1, DuPont-NEN, Boston) and Klenow DNA polymerase (New England Biolabs, Beverly, MA). Hybridization was performed at 42°C in 50% (w/v) formamide, 6× SSPE, 0.5% (w/v) SDS, 5× Denhardt's solution, and 100 mg mL−1 of sheared salmon sperm DNA. The blot was washed three times in 5× SSC, 1% (w/v) SDS at 42°C for 15 min, followed by three washes in 0.5× SSC at 65°C for 20 min (14°C below the melting temperature).
RNA Isolation and Analysis
Total RNA was isolated from etiolated tomato hypocotyls with the RNeasy Plant Total RNA kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. RNA was extracted from frozen tomato fruit pericarp and vegetative tissues by the method of Wan and Wilkins (1994). poly(A+) RNA was isolated with the Oligotex mRNA kit (QIAGEN). Total RNA (15 μg per lane) was subjected to electrophoresis on 1.2% (w/v) agarose and 10% (w/v) formaldehyde gels, visualized with ethidium bromide to confirm equal sample loading, and transferred to Hybond-N membrane (Amersham). Blots were hybridized with the LeXET2 PCR product or with a PflMI/HindIII cDNA fragment from the full-length clone as described above, and washed three times in 5× SSC and 1% (w/v) SDS at 42°C for 15 min, followed by three washes in 0.5× SSC at 65°C for 20 min. Blots were stripped and reprobed with a LeEXT PCR product corresponding to amino acids 104 through 279 of the full-length clone. A 200-bp tomato actin cDNA fragment was used as a loading control for a gene expressed constitutively throughout fruit development and along the tomato hypocotyl.
Construction of the Expression Vector
Recombinant LeXET2 protein was produced using the Pichia pastoris expression system (Invitrogen, Carlsbad, CA). The entire LeXET2 sequence, including the putative signal peptide and the native stop codon, was amplified by PCR using the primers 5′-AAAACAGGATCCAAACA-ACATGAT-3′ and 5′-GATATTTTAACTCTAGAACTTAT-TAA-3′ to introduce BamHI and XbaI restriction sites at the 5′ and 3′ ends, respectively. The LeXET2 cDNA was used as a template. Denaturation, annealing, and extension temperatures of 94°C (1 min), 45°C (1 min), and 72°C (2 min) were used. After digestion with BamHI and XbaI, the PCR product was cloned into the pPIC3.5K P. pastoris expression vector (Invitrogen), and the LeXET2-coding region in the resulting plasmid was sequenced to verify that no sequence errors were introduced.
Recombinant Protein Production and Purification
The LeXET2 expression vector and the pPIC3.5K empty vector, used as a negative control, were linearized with SacI and used for electroporation transformation of P. pastoris (strain KM71) following the protocol in the P. pastoris Expression System Manual (Invitrogen). Recombinant yeast colonies were used to inoculate 10 mL of buffered glycerol complex medium (Invitrogen) in 50-mL plastic tubes. After overnight growth at 30°C, 1 mL of culture was used to inoculate 100 mL of the same media in 1-L flasks and the cultures were shaken at 30°C until a culture OD600 = 2 to 6 was reached. The yeast cells and culture media were separated by centrifugation at 2,500g for 5 min, and the collected cells were resuspended in 12.5 mL of buffered methanol complex medium (Invitrogen) in 125-mL flasks and shaken at 30°C for 2 d. Methanol was added to the cultures every 24 h to give a final concentration of 0.5% (w/v).
The LeXET2 protein was purified from the supernatant recovered from 100 mL of culture medium of P. pastoris cells expressing LeXET2. The culture medium was centrifuged at 3,000g for 10 min and the supernatant was dialyzed four times against 4 L of 25 mm MES (2-[N-morpholino]ethanesulfonic acid) buffer, pH 6. The dialyzed sample was loaded onto a 5-mL SP-Sepharose Hi-Trap column (Amersham Pharmacia, Piscataway, NJ) equilibrated with 50 mm MES buffer, pH 6, and after washing the column with 25 mL of the same buffer, the LeXET2 protein was eluted with a 0 to 0.4 m NaCl gradient in 50 mm MES buffer, pH 6. Fractions containing XET2 protein were pooled, concentrated using a Centriprep-10 microconcentrator (Millipore, Bedford, MA), applied to a Superdex 75 (HR 10/30) gel filtration column (Amersham Pharmacia), and eluted with 0.25 m sodium acetate buffer, pH 5.7, containing 0.25 m NaCl at 0.2 mL min−1. Protein purification was monitored by electrophoresis of column fractions in SDS 4% to 12% acrylamide (w/v) NuPAGE gels (Invitrogen) and staining with Coomassie Blue. Protein concentration was determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA).
Preparation of Xyloglucan Substrates
Xyloglucan from tamarind seeds (Tamarindus indica) and from the media of bean (Phaseolus vulgaris) and sycamore (Acer pseudoplatanus) suspension-cultured cells was obtained as described in York et al. (1990) and in Wilder and Albersheim (1973), respectively. Tomato xyloglucan was isolated from the medium of suspension-cultured tomato cv Bonnie Best cells, as described in York et al. (1996). To separate xyloglucans with different molecular masses, 50 mg of tomato xyloglucan was dissolved in water, fractionated on a Sephacryl-200 (Amersham Pharmacia) column (3.5 × 98 cm), and eluted with 20 mm ammonium formate at a flow rate of 1 mL min−1. Fractions of 8 mL were collected and assayed for carbohydrate content by the anthrone assay (Dische, 1962). The fractions corresponding to major peaks were pooled, rechromatographed on a Superdex-200 HR 10/30 column (Amersham Pharmacia) using the same eluent as above at a flow rate of 0.6 mL min−1, and lyophilized. To obtain tomato xyloglucan donor substrates of lower molecular mass, 100 mg of tomato xyloglucan was partially hydrolyzed using 1 unit of a xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus (Pauly et al., 1999) at 25°C for 1 h and fractionated in a Sephacryl-200 column as above. This treatment caused a shift in the elution profile toward smaller polysaccharides (<10 kD) and some low-molecular mass oligosaccharides were also produced (1.4–3 kD). Fractions corresponding to carbohydrate peaks with an elution volume equal to or smaller than a 10-kD Dextran marker were pooled, rechromatographed on a Superdex-75 HR 10/30 column (Amersham Pharmacia) for further purification, and lyophilized. Average molecular masses of xyloglucan molecules were estimated by comparison of elution volumes with those of Dextran's with known average Mrs (Amersham Pharmacia).
Preparation of a Labeled Xyloglucan Oligomer
A xyloglucan nonasaccharide (XLLG; for nomenclature, see Fry et al., 1993) obtained from tamarind xyloglucan as described in York et al. (1990) was labeled covalently at its reducing end with the fluorophore APTS (Molecular Probes, Eugene, OR) by reductive amination following the procedure described in Evangelista et al. (1995). The APTS-derivatized XLLG was purified by elution in water from a column of Sephadex G-10 (25 × 1 cm).
Assay of XET Activity
XET activity was assayed by measuring the transfer of non-labeled xyloglucan donor molecules to a fluorescently labeled XLLG-APTS acceptor molecule. The reaction mixture contained 1 mg mL−1 of xyloglucan, 282 pmol of XLLG, and 2 to 25 μL of enzyme preparation in a total volume of 40 μL of 25 mm sodium acetate buffer, pH 5.2. Reactions were incubated at 25°C for 1 h and were stopped by boiling for 10 min. Reaction mixtures were subsequently analyzed by gel filtration chromatography on a Superdex-75 HR 10/30 column (Amersham Pharmacia) eluted with 25 mm sodium acetate buffer, pH 5.2, at a flow rate of 0.5 mL min−1. Fluorescent xyloglucan-APTS derivatives were detected in the eluate using a spectrofluorometer (excitation, 424 nm; emission, 504 nm). In separate experiments carried out in the absence of XET and acceptor substrate, carbohydrates in the eluate were detected by the anthrone assay (Dische, 1962). XET activity was quantified as described in Nishitani and Tominaga (1992) by measuring the peak area of high-Mr fluorescent product(s) formed.
ACKNOWLEDGMENTS
We thank Qiang Qin, Carl Bergmann, and Zhonghua Jia for technical assistance and advice.
Footnotes
This work was supported in part by the U.S. Department of Energy (grant nos. DE–FG02–96ER20220 and DE–FG05–93ER20097).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010481.
LITERATURE CITED
- Akamatsu T, Hanzawa H, Ohtake Y, Takahashi T, Nishitani K, Komeda Y. Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiol. 1999;121:715–721. doi: 10.1104/pp.121.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith DA, de Silva J. Characterization of two tomato fruit-expressed cDNAs encoding xyloglucan endotransglycosylases. Plant Mol Biol. 1995;28:391–403. doi: 10.1007/BF00020389. [DOI] [PubMed] [Google Scholar]
- Barrachina C, Lorences EP. Xyloglucan endotransglycosylase activity in pine hypocotyls: intracellular localization and relationship with endogenous growth. Physiol Plant. 1998;102:55–60. doi: 10.1034/j.1399-3054.1998.1020108.x. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 1999a;18:371–382. doi: 10.1046/j.1365-313x.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999b;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. Auxin regulation and spatial localization of an endo-1,4-β-d-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J. 1997;12:417–426. doi: 10.1046/j.1365-313x.1997.12020417.x. [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. Auxin-regulated genes encoding cell wall modifying proteins are expressed during early tomato fruit growth. Plant Physiol. 2000;122:527–534. doi: 10.1104/pp.122.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenen C, Lomax TL. Auxin-cytokinin interactions in higher plants: old problems and new tools. Trends Plant Sci. 1997;2:351–356. doi: 10.1016/S1360-1385(97)84623-7. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Sovonick-Dunford SA. Mechanism of gibberellin-dependent stem elongation in peas. Plant Physiol. 1989;89:184–191. doi: 10.1104/pp.89.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva J, Arrowsmith DA, Whiteman S, Robinson S. Xyloglucan endotransglycosylase and plant growth. J Exp Bot. 1994;45:1693–1701. [Google Scholar]
- de Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Chengappa S, Sidebottom C, Reid JS. Molecular characterization of a xyloglucan-specific endo-(1→4)-β-d-glucanase (xyloglucan endotransglycosylase) from nasturtium seeds. Plant J. 1993;3:701–711. [PubMed] [Google Scholar]
- Dische Z. Color reactions of carbohydrates. In: Whistler RL, Wolfrom ML, editors. Methods in Carbohydrate Chemistry. Vol. 1. New York: Academic Press; 1962. pp. 478–481. [Google Scholar]
- Evangelista RA, Liu M, Chen FA. Characterization of 9-aminopyrene-1,4,6-trisulfonate-derivatized sugars by capillary electrophoresis with laser-induced fluorescence detection. Anal Chem. 1995;67:2239–2245. doi: 10.1006/abio.1995.1474. [DOI] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1→4)-β-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J. 1993;3:691–700. doi: 10.1046/j.1365-313x.1993.03050691.x. [DOI] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill AG, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Plant Physiol. 1993;89:1–3. [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- Kelly MO, Bradford KJ. Insensitivity of the diageotropica tomato mutant to auxin. Plant Physiol. 1986;82:713–717. doi: 10.1104/pp.82.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000;122:85–98. doi: 10.1104/pp.122.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Yen H, Giovannoni JJ, Klee HJ. The never ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclachlan G, Brady C. Endo-1,4-β-glucanase, xyloglucanase and xyloglucan endotransglycosylase activities versus potential substrates in ripening tomatoes. Plant Physiol. 1994;105:965–974. doi: 10.1104/pp.105.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Wells B, Roberts K. Direct visualization of cross-links in the primary cell wall. J Cell Sci. 1990;96:323–334. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cells. Int Rev Cytol. 1997;173:157–205. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. In vitro molecular weight increase in xyloglucan by an apoplastic enzyme preparation from epicotyls of Vigna angularis. Physiol Plant. 1991;82:490–497. [Google Scholar]
- Nishitani K, Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates the molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem. 1993;268:25364–25368. [PubMed] [Google Scholar]
- O'Neill SD, Tong Y, Sporlein B, Forkmann G, Yoder JI. Molecular genetic analysis of chalcone synthase in Lycopersicon esculentum and an anthocyanin-deficient mutant. Mol Gen Genet. 1990;224:279–288. doi: 10.1007/BF00271562. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinnen S, Kofod LV, York WS, Albersheim P, Darvill AG. A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology. 1999;9:93–100. doi: 10.1093/glycob/9.1.93. [DOI] [PubMed] [Google Scholar]
- Potter I, Fry SC. Xyloglucan endotranglycosylase activity in pea internodes. Plant Physiol. 1993;103:235–241. doi: 10.1104/pp.103.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter I, Fry SC. Changes in xyloglucan endotransglycosylase (XET) activity during hormone-induced growth in lettuce and cucumber hypocotyls and spinach cell suspension cultures. J Exp Bot. 1994;45:1703–1710. [Google Scholar]
- Pritchard J, Hetherington PR, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and profiles of cell wall properties along growing regions of maize roots. J Exp Bot. 1993;44:1281–1289. [Google Scholar]
- Purugganan MM, Braam J, Fry SC. The Arabidopsis TCH4 xyloglucan endotransglycosylase. Plant Physiol. 1997;115:181–190. doi: 10.1104/pp.115.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgwell RJ, Fry SC. Xyloglucan endotransglycosylase activity increases during kiwifruit (Actinidia deliciosa) ripening. Plant Physiol. 1993;100:1318–1325. doi: 10.1104/pp.103.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Brummell DA, Bennett AB. Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in nasturtium. Plant Physiol. 1996;110:493–499. doi: 10.1104/pp.110.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab I, Sachs M. A flooding-induced xyloglucan endotransglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol. 1996;112:385–391. doi: 10.1104/pp.112.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder R, Atkinson RG, Langenkämper G, Redgwell RJ. Biochemical and molecular characterization of xyloglucan endotransglycosylase from ripe kiwifruit. Planta. 1998;204:242–251. doi: 10.1007/s004250050253. [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Smith RC, Lång V, Matthews PR, Chandler PM. Expression of XET-related genes and its relation to elongation in leaves of barley (Hordeum vulgare L.) Plant Cell Environ. 1997;20:1439–1450. [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T. Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol. 1997;38:375–378. doi: 10.1093/oxfordjournals.pcp.a029178. [DOI] [PubMed] [Google Scholar]
- Sitbon F, Perrot-Rechenman C. Expression of auxin-regulated genes. Physiol Plant. 1997;100:443–455. [Google Scholar]
- Smith RC, Matthews PR, Schünmann PHD, Chandler PM. The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in “Himalaya” barley (Hordeum vulgare L.) J Exp Bot. 1996;47:1395–1404. [Google Scholar]
- Takano M, Fuji N, Higashitani A, Nishitani K, Hirasawa T, Takahashi H. Endoxyloglucan transferase cDNA isolated from pea roots and its fluctuating expression in hydrotropically responding roots. Plant Cell Physiol. 1999;40:135–142. doi: 10.1093/oxfordjournals.pcp.a029520. [DOI] [PubMed] [Google Scholar]
- Talbott LD, Ray PM. Changes in molecular size of previously deposited and newly synthesized pea cell wall matrix polysaccharides. Plant Physiol. 1992;98:369–379. doi: 10.1104/pp.98.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Fry SC. Trimming and solubilization of xyloglucan after deposition in the walls of cultured rose cells. J Exp Bot. 1997;48:297–305. [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M. Characterization of XET-related genes of rice. Plant Physiol. 2000;122:853–859. doi: 10.1104/pp.122.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, York WS, Beldman G, Voragen AG. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol. 1997;144:9–13. doi: 10.1104/pp.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton. Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Wilder BM, Albersheim P. The structure of plant cell walls: IV. A structural comparison of the wall hemicellulose of cell suspension cultures of sycamore (Acer pseudoplatanus) and of red kidney bean (Phaseolus vulgaris) Plant Physiol. 1973;51:889–893. doi: 10.1104/pp.51.5.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Kolli VSK, Orlando R, Albersheim P, Darvill AG. The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res. 1996;285:99–128. doi: 10.1016/s0008-6215(96)90176-7. [DOI] [PubMed] [Google Scholar]
- York WS, van Halbeek H, Darvill AG, Albersheim P. Structure of plant cell walls: XXIX. Structural analysis of xyloglucan oligosaccharides by 1H-NMR spectroscopy and fast atom bombardment mass spectrometry. Carbohydr Res. 1990;200:9–31. doi: 10.1016/0008-6215(90)84179-x. [DOI] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epycotyls. Plant Physiol. 1994;104:161–170. doi: 10.1104/pp.104.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]