Abstract
Peroxisomes are important organelles in plant metabolism, containing all the enzymes required for fatty acid β-oxidation. More than 20 proteins are required for peroxisomal biogenesis and maintenance. The Arabidopsis pxa1 mutant, originally isolated because it is resistant to the auxin indole-3-butyric acid (IBA), developmentally arrests when germinated without supplemental sucrose, suggesting defects in fatty acid β-oxidation. Because IBA is converted to the more abundant auxin, indole-3-acetic acid (IAA), in a mechanism that parallels β-oxidation, the mutant is likely to be IBA resistant because it cannot convert IBA to IAA. Adult pxa1 plants grow slowly compared with wild type, with smaller rosettes, fewer leaves, and shorter inflorescence stems, indicating that PXA1 is important throughout development. We identified the molecular defect in pxa1 using a map-based positional approach. PXA1 encodes a predicted peroxisomal ATP-binding cassette transporter that is 42% identical to the human adrenoleukodystrophy (ALD) protein, which is defective in patients with the demyelinating disorder X-linked ALD. Homology to ALD protein and other human and yeast peroxisomal transporters suggests that PXA1 imports coenzyme A esters of fatty acids and IBA into the peroxisome for β-oxidation. The pxa1 mutant makes fewer lateral roots than wild type, both in response to IBA and without exogenous hormones, suggesting that the IAA derived from IBA during seedling development promotes lateral root formation.
Peroxisomes are small, ubiquitous organelles encased in a single lipid bilayer that contain hydrogen peroxide-producing oxidases and catalases to inactivate reactive molecules (for review, see Gerhardt, 1992; Kindl, 1993; Olsen, 1998; Tabak et al., 1999). Arabidopsis and other oilseed plants β-oxidize long-chain fatty acids (LCFAs) in peroxisomes to provide energy during germination. Plant peroxisomes also contain enzymes that act in photorespiration (Olsen, 1998) and the catabolism of branched-chain amino acids (Gerhardt, 1992; Zolman et al., 2001). In addition, seedlings and senescing tissues contain specialized peroxisomes called glyoxysomes that convert acetyl-coenzyme A (CoA) to succinate, which is transported to the mitochondria where it fuels the tricarboxylic acid cycle (Gerhardt, 1992; Olsen, 1998).
Mammals metabolize fatty acids in both mitochondria and peroxisomes, and each organelle shortens a distinct subset of fatty acids (Lazarow, 1993; Tabak et al., 1999). In contrast, plants and yeast catabolize fatty acids exclusively in peroxisomes (Gerhardt, 1992; Kindl, 1993). Because peroxisomes lack DNA, proteins required for β-oxidation and other peroxisomal processes are translated in the cytoplasm and then imported (Olsen, 1998; Subramani, 1998; Tabak et al., 1999). Peroxisomal matrix proteins contain one of two peroxisomal targeting signals (PTSs). The PTS1 is made up of the amino acids “SKL” (or a conserved variant) at the extreme C termini of peroxisomal matrix-bound proteins (Gould et al., 1989). The PEX5 receptor binds PTS1-containing proteins in the cytoplasm and translocates them into the peroxisome (Olsen, 1998; Subramani, 1998; Tabak et al., 1999). PEX7 imports proteins that have the nine-residue N-terminal PTS2 sequence (Olsen, 1998; Subramani, 1998; Tabak et al., 1999). Both the PEX5 and PEX7 matrix protein receptors have been identified in plants (Brickner et al., 1998; Kragler et al., 1998; Wimmer et al., 1998; Schumann et al., 1999).
In addition to matrix enzymes, peroxisomes must import the substrates and cofactors required in peroxisomal processes, such as fatty acids destined for catabolism. Fatty acids are synthesized and metabolized in different subcellular locations, and how they are transported between organelles is just beginning to be understood. Yeast and humans apparently transport LCFAs into peroxisomes via ATP-binding cassette (ABC)-containing ATPases in the peroxisomal membrane (Shani and Valle, 1998; for review, see Dubois-Dalcq et al., 1999; Holland and Blight, 1999). The transporter required for fatty acid uptake into peroxisomes has not yet been characterized in any plant species.
Yeast mutants defective in peroxisomal function or β-oxidation utilize alternative carbon sources poorly; screens for mutants with reduced growth on oleic acid have uncovered more than 20 proteins required for peroxisomal biogenesis, maintenance, and the import of enzymes and metabolites (for review, see Erdmann and Kunau, 1992; Lazarow, 1993; Olsen, 1998; Subramani, 1998; Tabak et al., 1999). In humans, mutations in peroxisomal proteins cause several life-threatening diseases, including Zellweger syndrome, X-linked adrenoleukodystrophy (X-ALD), and Refsum disease (for review, see Fujiki, 1997; Gärtner, 2000; Gould and Valle, 2000).
We are studying the phytohormone auxin, which influences virtually every aspect of plant growth and development, including root elongation, lateral root initiation, organ identity, and tropic responses (Davies, 1995). Two naturally occurring auxins found in plants are indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA). Although these compounds differ only in the two additional carbon atoms on the IBA side chain, they have different potencies in bioassays (Ludwig-Müller, 2000; Zolman et al., 2000; Bartel et al., 2001).
Experiments with labeled auxin indicate that several plant species can convert IBA to IAA (Epstein and Ludwig-Müller, 1993; Ludwig-Müller, 2000; Bartel et al., 2001). Because the conversion shortens the IBA side chain by two carbons, this process has been proposed to occur similarly to fatty acid β-oxidation (Wain and Wightman, 1954; Fawcett et al., 1960). Previously, we described a collection of Arabidopsis mutants that are resistant to the inhibitory effects of IBA on root elongation but that respond normally to IAA (Zolman et al., 2000). A subset of these mutants is distinguished by developmental defects in the absence of exogenous Suc and inefficient metabolism of LCFAs during germination (Zolman et al., 2000), suggesting defects in peroxisomal β-oxidation. Therefore, these IBA-response mutants probably have defects in the β-oxidation of both fatty acids and IBA, causing Suc-dependent seedling development and IBA-resistant root elongation. Some of these mutants are defective in proteins acting directly in β-oxidation. For example, enzymes defective in mutants resistant to IBA or the IBA analog 2,4-dichlorophenoxybutyric acid (2,4-DB) include an acyl-CoA oxidase (acx3; Eastmond et al., 2000), a multifunctional protein (aim1; Richmond and Bleecker, 1999), and a thiolase (ped1; Hayashi et al., 1998) that act in β-oxidation. In addition, because fatty acid β-oxidation is strictly peroxisomal in plants, mutations in peroxisomal biogenesis or maintenance proteins can disrupt β-oxidation. For example, a mutant defective in PEX5, the PTS1 peroxisomal matrix protein importer, is IBA resistant (Zolman et al., 2000) and the 2,4-DB-resistant ped2 mutant is defective in the peroxisomal membrane protein PEX14 (Hayashi et al., 2000).
Here, we describe an IBA-response mutant that requires exogenous Suc for development and is resistant to IBA in both root elongation and lateral root initiation. We used a map-based positional approach to determine that the mutant is defective in PXA1, which encodes an ABC transporter similar to peroxisomal fatty acid transporters and the human protein disrupted in X-ALD. PXA1 appears to act in the peroxisomal import of fatty acids and IBA for β-oxidation.
RESULTS
pxa1 Has Altered Responses to IBA
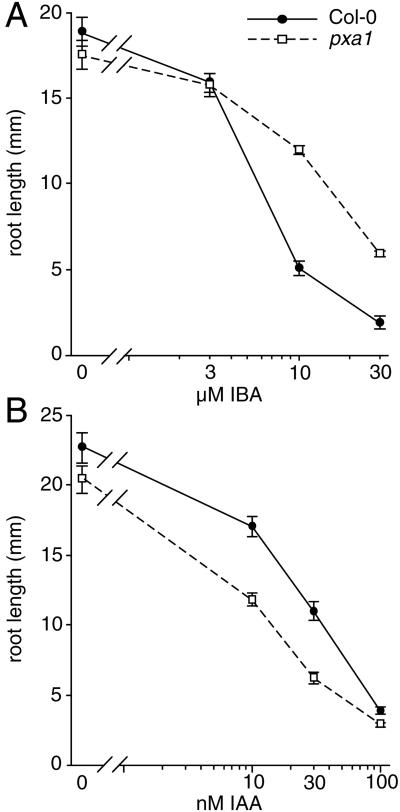
We previously described the isolation of 14 Arabidopsis IBA-response mutants (Zolman et al., 2000). Here, we report the characterization and cloning of the PXA1 gene, which is defective in one of the IBA-response mutants (see below). As shown in Figure 1, high levels of exogenous auxin inhibit wild-type root elongation. The pxa1 mutant is resistant to the inhibition of root elongation by IBA over a range of concentrations (Fig. 1A) but remains sensitive to inhibition by IAA (Fig. 1B). pxa1 also is resistant to the inhibitory effects of 2,4-DB but is sensitive to the synthetic auxins 2,4-dichlorophenoxyacetic acid (2,4-D; Zolman et al., 2000) and naphthalene-1-acetic acid (data not shown).
Figure 1.
pxa1 is IBA resistant. A, Root elongation on IBA. Eight-day-old wild-type and mutant seedlings grown on the indicated concentrations of IBA under yellow-filtered light were removed from the agar, and the length of the primary root was measured. Error bars indicate the se values (n > 9). B, Root elongation on IAA. Primary root length on IAA was measured as described above. Error bars indicate the se values (n > 12).
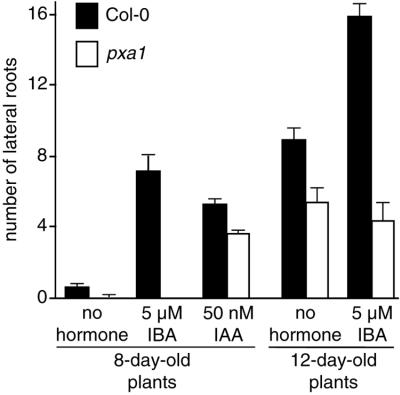
In addition, pxa1 has defects in lateral root initiation. We examined root initiation by growing plants on unsupplemented medium for 4 d, transferring seedlings to IBA, IAA, or unsupplemented medium, and counting lateral roots after an additional 4 d. On unsupplemented medium, wild-type plants form few lateral roots (approximately one per plant; Fig. 2), but more lateral roots are induced when plants are transferred to either IBA (approximately seven per plant) or IAA (approximately five per plant; Fig. 2). pxa1 has fewer lateral roots than wild type on unsupplemented medium and is completely unresponsive to IBA, making no lateral roots after 8 d (Fig. 2). However, the mutant does respond to the stimulatory effects of IAA (Fig. 2). To better quantify the defect, we extended the growth period for lateral root initiation to 6 d before and 6 d after transfer. Again, addition of IBA strongly initiates lateral roots in wild type (approximately 16 per plant) compared with plants on unsupplemented medium (approximately nine per plant; Fig. 2). Similar to the short experiment, pxa1 has fewer lateral roots than wild type after 12 d and does not initiate additional lateral roots with IBA treatment (Fig. 2).
Figure 2.
pxa1 is defective in lateral root formation. Seedlings were grown for 4 or 6 d on hormone-free medium and transferred to medium with no hormone, 50 nm IAA, or 5 μm IBA. After 8 or 12 d, plants were removed from the agar, and the number of lateral roots was counted. Error bars indicate se values (n > 16).
pxa1 Has Defects in Growth and Development
Mutants defective in peroxisomal β-oxidation have growth defects on minimal medium because oilseed plants (like Arabidopsis) use LCFAs as an energy source before photosynthesis begins (Hayashi et al., 1998). Plants that cannot catabolize fatty acids consequently cannot develop unless exogenous Suc is provided. pxa1 and certain other IBA-response mutants catabolize seed storage fatty acids slowly and have growth defects on Suc-free medium, suggesting β-oxidation defects (Zolman et al., 2000). In fact, pxa1 is among our most severe Suc-dependent mutants. To quantify the mutant defects, we examined germination (radicle emergence from the seed coat) and establishment (cotyledon expansion in light-grown plants; hypocotyl elongation in dark-grown plants) of wild-type and mutant seeds. Seeds were plated on medium either with or without Suc and grown in the light or in the dark. As shown in Table I, most (>80%) wild-type and mutant seeds germinated regardless of the growth conditions. Whereas Suc-grown pxa1 seedlings developed normally, mutant seedlings grown in the absence of Suc arrested after germination, even in the light. This phenotype is consistent with a strong defect in peroxisomal β-oxidation. Germinated mutant seeds could reinitiate normal development when transferred to medium containing Suc (data not shown), indicating that they were developmentally arrested rather than dead.
Table I.
Seed germination and development
| Genotype | Exogenous Suc | Light
|
Dark
|
||
|---|---|---|---|---|---|
| Germinateda | Establishedb | Germinated | Establishedc | ||
| mm | % | ||||
| pxa1 | 0 | 100 | 0 | 86 | 0 |
| pxa1 | 15 | 100 | 95 | 88 | 88 |
| Col-0 | 0 | 100 | 100 | 94 | 89 |
| Col-0 | 15 | 100 | 93 | 82 | 82 |
Wild-type (Col-0) and pxa1 mutant seeds were grown on medium in either the presence or absence of Suc for 6 d in the light or 1 d in the light and 5 d in the dark. At least 14 seeds were examined for each condition.
Radicle emergence from the seed coat.
Expansion of cotyledons.
Hypocotyl elongation.
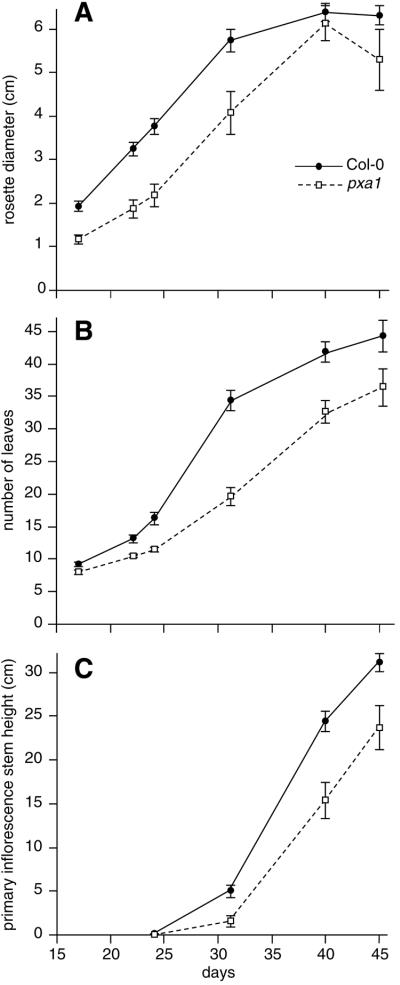
In addition to germination defects, pxa1 mutants are smaller than wild type. pxa1 has a slightly shorter root than wild type on unsupplemented medium, as seen at the “no hormone” data points in Figure 1. To further investigate the mutant growth defects, we examined wild-type and pxa1 plants over time. The mutant has a smaller rosette (Fig. 3A) and fewer leaves (Fig. 3B) than wild type throughout adult development. The primary inflorescence also is consistently shorter in pxa1 than wild type (Fig. 3C). Each of these characteristics suggests that pxa1 plants grow more slowly than wild type. pxa1 has a delayed time to flowering when measured by the number of days but flowers with a similar number of leaves as wild type (data not shown). These data indicate that pxa1 is developmentally delayed, but the defect in this mutant does not affect flowering time pathways. Other than decreased size, mutant plants are morphologically similar to wild type (data not shown).
Figure 3.
pxa1 grows more slowly than wild type. Ten-day-old wild-type and pxa1 mutant plants were transferred to soil and examined weekly. A, Rosette diameter at widest point. B, Number of rosette leaves. C, Height of the primary inflorescence stem. Error bars indicate se values (n > 13).
Positional Cloning Reveals a Defect in an Apparent ABC Transporter
The IBA-resistant root elongation in pxa1 is dominant. As shown in Figure 4A, heterozygote plants can elongate roots similarly to the homozygous mutant on inhibitory concentrations of IBA. However, pxa1 mutant development on Suc-free medium is recessive because the PXA1/pxa1 heterozygote develops normally without Suc (Fig. 4B).
Figure 4.
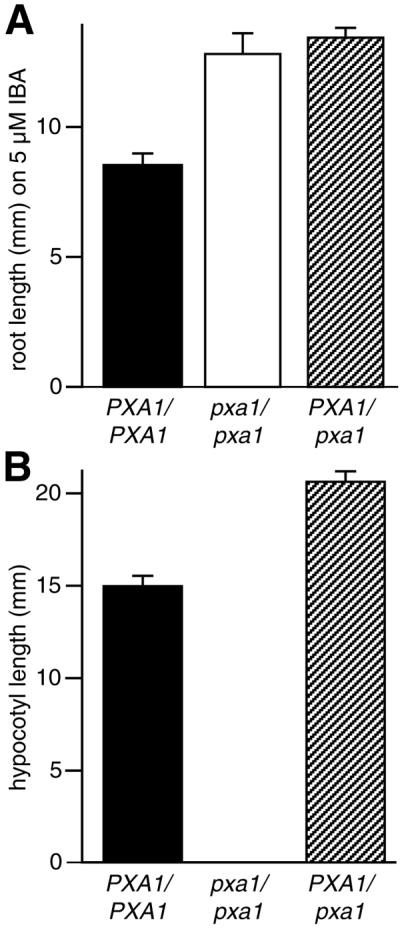
Genetic analysis of pxa1 phenotypes. A, pxa1 IBA-resistant root elongation is dominant. Root elongation on 5 μm IBA was measured as described in the legend to Figure 1. Error bars indicate se values (n > 13). B, pxa1 Suc dependence is recessive. Seedlings were grown on Suc-free medium for 1 d in the light and 5 d in the dark, and hypocotyl length was measured. Error bars indicate se values (n > 16). For A and B, PXA1/pxa1 seeds are F1 progeny from a backcross of pxa1 to wild-type Columbia (Col-0).
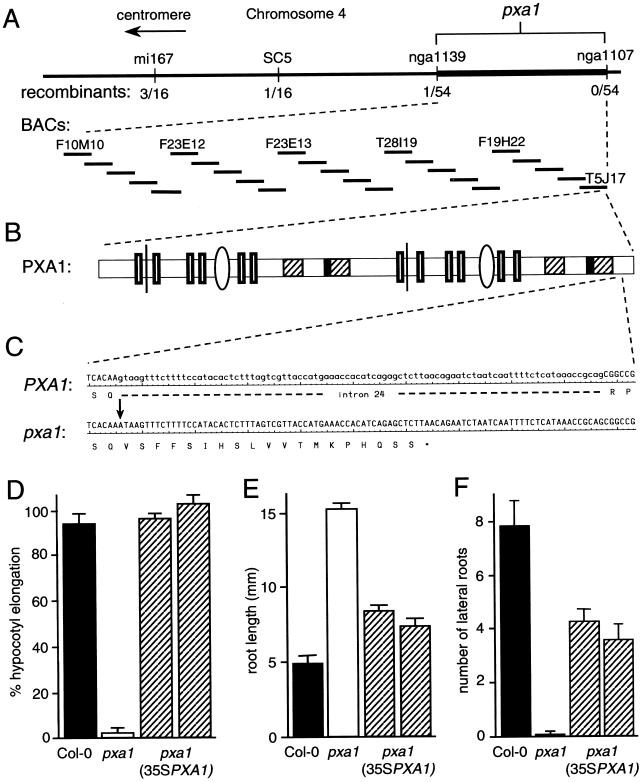
We used positional information to identify the molecular defect in the pxa1 mutant. We outcrossed a mutant plant (Col-0 background) to Wassilewskija (Ws) and selected IBA-resistant F2 progeny. Because the pxa1 IBA-resistant root elongation phenotype is dominant (Fig. 4A), we tested the progeny from IBA-resistant F2 plants on Suc-free medium to identify lines that were homozygous at the mutant locus. We then used PCR-based polymorphic markers to localize the mutant defect to the bottom of chromosome 4, south of nga1139 (Fig. 5A; see “Materials and Methods”). Examination of the genes between this marker and the telomere identified a candidate gene on the T5J17 bacterial artificial chromosome (GenBank accession no. AL035708). This bacterial artificial chromosome contains a gene (At4g39850) predicted to encode a protein resembling a subset of ABC-ATPase transporters. In yeast and humans, similar ATPases are required to import LCFAs into the peroxisome for catabolism (Shani and Valle, 1998; Dubois-Dalcq et al., 1999; Holland and Blight, 1999). We hypothesized that a defect in this protein would disrupt β-oxidation, resulting in an IBA-resistant mutant with developmental defects. We sequenced this gene using pxa1 mutant DNA (see “Materials and Methods”) and identified a G-to-A mutation at position 5,559 (where position 1 is the A of the initiator ATG; Fig. 5C).
Figure 5.
Positional cloning of PXA1. A, Recombination mapping of pxa1. Mapping with PCR-based markers mi167 (see “Materials and Methods”), SC5 (http://Arabidopsis.org/maps/CAPS_Chr4.html), nga1139, and nga1107 (Bell and Ecker, 1994) localized the defect between nga1139 and the telomere. Examination of the sequenced BACs (thick lines) in this region revealed the PXA1 gene (At4g39850) on T5J17. B, PXA1 structure. PXA1 contains several conserved domains in its predicted protein structure, including 12 transmembrane (TM) domains (hollow boxes), two loop 1 conserved regions (thin lines), two EAA-like domains (ovals), Walker A and B ATP-binding domains (hatched rectangles), and two ABC signature motifs, or C sequences (black rectangles). C, pxa1 has a G-to-A mutation at position 5559 (where 1 is the A of the initiator ATG) that destroys the 5′-splice site of the last intron. Reverse transcription-PCR analysis indicates that the last intron is not removed from the protein, and thus translation continues into the intron sequence. Nucleotides 7,349 through 7,452 are shown, corresponding to the end of the 24th exon and beginning of the 25th exon in wild type and the 24th exon in the mutant, which continues coding through the intron to a premature termination codon. D through F, 35SPXA1 rescues the pxa1 mutant phenotype. D, Seeds from wild-type (Col-0), pxa1-1, and two independent homozygous lines from pxa1-1 plants transformed with the 35SPXA1 construct were analyzed for hypocotyl elongation as described in the legend of Figure 4B. Error bars indicate se values (n > 12). E, Root elongation inhibition by 10 μm IBA was measured for wild-type, pxa1, and transformed plants as described in the legend of Figure 1A. Error bars indicate se values (n > 13). F, Lateral root initiation by wild-type, pxa1, and transformed plants was examined in response to 5 μm IBA after 8 d as described in the legend of Figure 2. Error bars indicate se values (n > 13).
To confirm that the nucleotide change in pxa1 causes the mutant phenotypes, we complemented the mutant with a wild-type copy of PXA1. We inserted a full-length PXA1 cDNA in the 35SpBARN plant transformation vector (LeClere and Bartel, 2001) behind the constitutive 35S cauliflower mosaic virus promoter. This construct (35SPXA1) was transformed into pxa1 mutant plants, and transformants were selected using gluphosinate ammonium (Basta, Crescent Chemical Co., Hauppauge, NY) herbicide. Mutant plants transformed with this construct developed normally on medium without Suc (Fig. 5D), suggesting a restored ability to β-oxidize LCFAs. In addition, transformants became sensitive to root elongation inhibition on IBA (Fig. 5E) and regained the ability to make lateral roots both in response to IBA (Fig. 5F) and without induction (data not shown). This phenotypic rescue indicates that we have identified the mutation causing the defect in pxa1. Therefore, we named this gene PXA1 (peroxisomal ABC transporter 1).
To examine the protein sequence and the intron/exon junctions of the gene, we obtained and sequenced an apparently full-length cDNA (see “Materials and Methods”), which revealed a predicted coding sequence of 1,338 amino acids (GenBank accession no. AF378120). Comparing the cDNA and genomic sequences revealed that PXA1 is interrupted by 24 introns. The G-to-A change in the pxa1 mutant is at the exon/intron junction following the 24th exon. It is interesting that sequencing the PXA1 cDNA also revealed that the protein predicted by the sequencing project (T5J17.20, GenBank accession no. AL035708) was incorrectly spliced at 15 (of 48) intron/exon junctions, including three exons that were missing completely (data not shown). These discrepancies reinforce the continued importance of cDNA analysis in protein predictions.
The consensus 5′-splice site for wild-type exon/intron junctions is AG/gt, and previous work has established that the intronic GT bases are absolutely required for splicing (Brown et al., 1996). The pxa1 mutation alters an essential G in the 5′-exon/intron splice site (Fig. 5C), converting the sequence AA/gt to AAAT. To determine how the splicing mutation affects the mutant protein, we made RNA from 5-d-old wild-type and mutant plants. We reverse transcribed the RNA to make the PXA1 and pxa1 cDNAs and amplified these templates using primers spanning the mutation (see “Materials and Methods”). We initially expected that a second AAGT immediately following the original splice site would reinitiate splicing, removing the intron but shifting the remainder of the protein out of frame. Sequencing the pxa1 cDNA, however, revealed that the entire intron was present in the mutant cDNA, which would cause translation through the intron and the coding of a premature stop codon (Fig. 5C). In the resultant pxa1 protein, 19 amino acids (encoded by intron 24) replace the final 32 amino acids (encoded by exon 25) of PXA1.
DISCUSSION
pxa1 originally was identified as an IBA-response mutant (Zolman et al., 2000). It is resistant to the inhibitory effects of IBA on root elongation (Fig. 1A) but remains sensitive to IAA (Fig. 1B) and the synthetic auxin naphthalene-1-acetic acid (data not shown). The mutant also is resistant to the IBA analog 2,4-DB (Zolman et al., 2000), which is converted to the synthetic auxin 2,4-D in a mechanism similar to the β-oxidation of IBA (Wain and Wightman, 1954; Hayashi et al., 1998). In addition, pxa1 is defective in lateral root initiation, making fewer lateral roots than wild type in the absence of hormone and particularly in response to IBA induction (Fig. 2).
The pxa1 mutant also has developmental defects. Although mutant seeds germinate normally, mutant plants do not develop beyond germination unless provided with exogenous Suc (Table I). This phenotype suggests severe peroxisomal defects, because peroxisomal β-oxidation mutants cannot catabolize stored fatty acids for energy before photosynthesis begins (Hayashi et al., 1998). Throughout development, pxa1 mutant plants grow more slowly than wild-type plants, with smaller rosettes, fewer leaves, and shorter inflorescence stems (Fig. 3). pxa1 also has a slightly shorter root than wild type (Fig. 1).
The IBA resistance of the pxa1 mutant is dominant; heterozygous PXA1/pxa1 plants elongate roots on IBA similarly to homozygous pxa1/pxa1 plants (Fig. 4A). In contrast, the Suc dependence of mutant seedling establishment appears fully recessive (Fig. 4B). Also, the ped1 mutant, which is defective in a thiolase acting in fatty acid β-oxidation (Hayashi et al., 1998), the ped2 mutant, which is defective in the peroxisomal membrane protein PEX14 (Hayashi et al., 2000), and the ped3 mutant are dominant for resistance to the IBA analog 2,4-DB but recessive for Suc dependence during germination (Hayashi et al., 1998). These results suggest that haplo-insufficiency, rather than a gain-of-function in the pxa1 protein, causes the dominance of pxa1 IBA resistance. Because heterozygous plants apparently metabolize sufficient fatty acids for normal development, but do not β-oxidize enough IBA to inhibit root elongation, IBA resistance appears to provide a more sensitive assay for β-oxidation defects than does Suc dependence. The defect in ped3 has not been reported, but ped3 maps to the same interval as pxa1 on the bottom of chromosome 4 and also is Suc dependent for seedling establishment (Hayashi et al., 1998), suggesting that it may also have a defect in PXA1.
The gene defective in pxa1 encodes a protein homologous to members of the ABC-ATPase superfamily, which are ATP-driven pumps or channels transporting substrates ranging from small ions to large polypeptides across membranes (for review, see Holland and Blight, 1999; Davies and Coleman, 2000). More than 100 putative ABC transporters have been identified in Arabidopsis (Davies and Coleman, 2000; Sánchez-Fernández et al., 2001). As shown in Figure 6, the Arabidopsis PXA1 protein resembles two yeast peroxisomal ABC transporters (Pxa1p/Pat2p/Pal1p and Pxa2p/Pat1p; Shani et al., 1995; Hettema et al., 1996; Shani and Valle, 1996; Swartzman et al., 1996) and four human transporters (PMP70/PXMP1, P70R, ALDP, and ALDRP; Gärtner et al., 1992, 1998; Kamijo et al., 1992; Mosser et al., 1993; Lombard-Platet et al., 1996; Holzinger et al., 1999). Several of these proteins have been localized to the peroxisomal membrane (Kamijo et al., 1992; Contreras et al., 1994; Imanaka et al., 1996, 1999; Swartzman et al., 1996; Holland and Blight, 1999), and experiments using semi-intact yeast cell systems show that oleic acid enters the peroxisome as a CoA ester using PXA2 in an ATP-dependent manner (Verleur et al., 1997). Because Arabidopsis PXA1 closely resembles these proteins and the pxa1 mutant has β-oxidation defects, we predict that this protein is an ABC transporter acting in peroxisomal import. To our knowledge, this is the first characterization of a potential peroxisomal fatty acid transporter in plants.
Figure 6.
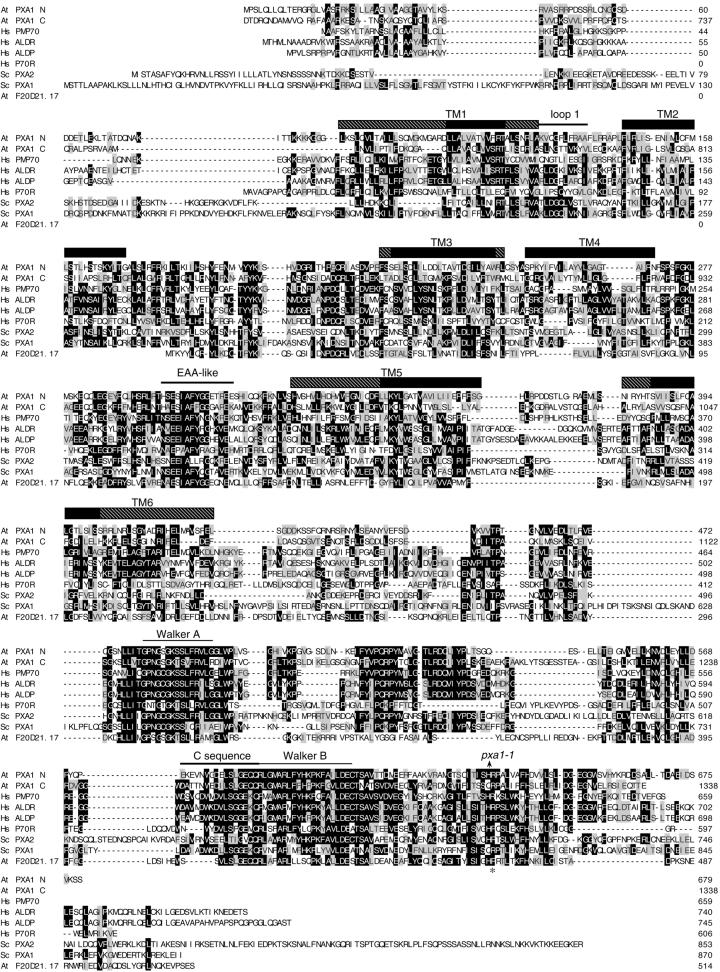
Alignment of PXA1 and its homologs. Alignment of PXA1 with the PMP70 (GenBank accession no. XP_010507, 45% identical to Arabidopsis PXA1), ALDR (NP_005155, 42% identical), ALDP (XP_010174, 42% identical), and P70R (NP_064731, 36% identical) proteins from human (Hs); the Pxa2p (NP_012733, 30% identical) and Pxa1p (NP_015178, 24% identical) proteins from yeast (Sc); and the F20D21.17 (AAD25615, 21% identical) protein from Arabidopsis (At). The PXA1 protein is divided into halves to show its homology with the hemitransporters; PXA1 N is the N terminus of the protein from amino acids 1 to 679; PXA C is the C terminus of the protein from amino acids 680 to 1,338. Sequences were aligned with the MegAlign program (DNASTAR, Inc., Madison, WI) using the ClustalW method. Amino acid residues identical in at least three of the sequences are boxed in black and similar amino acids are boxed in gray. Hyphens indicate gaps introduced to maximize alignment. The arrow above the alignment marks the position of the pxa1 splicing defect and conserved domains are indicated above the sequence. The 12 TM domains were predicted using homology to the human (Shani and Valle, 1998; Dubois-Dalcq et al., 1999) and yeast proteins (Shani and Valle, 1996; Swartzman et al., 1996) and by TM prediction programs TMAP (Perrson and Argos, 1994, 1996), SMART; (Schultz et al., 1998, 2000), and TMpred (Hofmann and Stoffel, 1993). TM domains are indicated by rectangles above the sequence; regions of high certainty are indicated by black boxes; regions of lower certainty are indicated by hatched boxes. An asterisk marks an Arg residue in ALDP that is a site of recurrent mutations in X-ALD patients (Dubois-Dalcq et al., 1999), which coincides with the first amino acid affected in the pxa1 mutant.
ABC transporters are composed of two homologous halves, which can be encoded either by one gene as a single polypeptide or by separate genes that each encode a hemitransporter (Dubois-Dalcq et al., 1999; Holland and Blight, 1999; Davies and Coleman, 2000). Each half has four to six TM domains, a cytoplasmic ATP-binding and hydrolysis region containing the highly conserved Walker A and B domains (GX2GXGKS/T; Figs. 5B, hatched boxes, and 6), and a 19-amino acid C sequence (Figs. 5B, black boxes, and 6; Dubois-Dalcq et al., 1999; Holland and Blight, 1999; Davies and Coleman, 2000).
The yeast and human peroxisomal ABC transporters are composed of two hemitransporters (Dubois-Dalcq et al., 1999; Holland and Blight, 1999; Liu et al., 1999). In contrast, the Arabidopsis PXA1 protein appears to be a complete transporter with two homologous halves: amino acids 1 through 679 make up the first half of the transporter, whereas amino acids 680 through 1,338 make up the second half (Figs. 5B and 6). These peroxisomal ABC transporters contain two additional highly conserved motifs. The loop following the TM domain 1 (loop 1) is in the peroxisomal matrix and probably controls membrane insertion (Figs. 5B, black lines, and 6; Dubois-Dalcq et al., 1999). The EAA-like domain (Figs. 5B, ovals, and 6) is a second conserved region on the cytoplasmic face of the protein between TM domains 4 and 5 and is thought to control substrate specificity by binding fatty acids (Shani et al., 1995; Shani and Valle, 1996). The mechanism of fatty acid transport by these peroxisomal proteins has not been elucidated. Some evidence indicates that the human ABC protein P-glycoprotein acts as a flippase that transports phospholipids between leaflets of the plasma membrane (Romsicki and Sharom, 2001), although other mechanisms also have been suggested for protein P-glycoprotein and other ABC transporters (van Veen and Konings, 1997; van Veen, 2001).
Yeast PXA1 and PXA2 deletion mutants have morphologically intact peroxisomes but have reduced (approximately 20%–50%) LCFA β-oxidation and consequently cannot use oleic acid as a carbon source (Shani et al., 1995; Hettema et al., 1996; Shani and Valle, 1996). The β-oxidation of short-chain fatty acids is unaffected, indicating that β-oxidation enzymes and matrix protein import remain intact (Hettema et al., 1996). The phenotype of the yeast pxa1/pxa2 double mutant is comparable with the single mutants and Pxa1p and Pxa2p interact in yeast two-hybrid assays and co-immunoprecipitation experiments, suggesting that the two proteins function together (Shani et al., 1995; Hettema et al., 1996; Shani and Valle, 1996).
In humans, disruption of the PXA1 homolog ALDP is lethal. Patients with X-ALD accumulate very LCFAs (VLCFAs) in serum and tissues, resulting in adrenal insufficiency and myelin destruction in the central nervous system (Hettema et al., 1996; Dubois-Dalcq et al., 1999; Gärtner, 2000). Fibroblasts from X-ALD patients have decreased LCFA β-oxidation, causing the fatty acid accumulation (Lazo et al., 1989; Braiterman et al., 1999). The reduced β-oxidation and consequent accumulation of LCFAs in yeast and humans with defects in these ABC-type transporters suggests that these proteins facilitate the peroxisomal import of LCFA CoA esters. Consistent with this hypothesis, β-oxidation of long-chain CoA esters requires Pxa2p in a semi-intact yeast system (Verleur et al., 1997).
However, other data suggest that VLCFA transport across the peroxisomal membranes is normal in X-ALD fibroblasts (Singh et al., 1992). In addition, X-ALD fibroblasts can β-oxidize long-chain CoA esters but not LCFAs (Hashmi et al., 1986; Lazo et al., 1988). The VLCFA synthetase responsible for activating the fatty acids to the CoA ester has reduced activity in X-ALD fibroblasts (Lazo et al., 1988, 1989; Wanders et al., 1988; Singh et al., 1992), leading to a second hypothesis that ALDP acts in the activation of VLCFAs or the stabilization of VLCFA synthetase (Smith et al., 2000). Further characterization of fatty acid transport and enzyme activity in the Arabidopsis pxa1 mutant may allow determination of how PXA1 functions and its role in fatty acid β-oxidation in plants.
Examination of the virtually complete Arabidopsis genome sequence reveals only one PXA1 homolog (F20D21.17/At1g54350, Fig. 6). Unlike PXA1, this protein is a hemitransporter, containing one TM domain region followed by a single nucleotide-binding fold. The identity between the two Arabidopsis proteins (21%) is less than the identity between PXA1 and the yeast (24%–30%) and mammalian (36%–45%) proteins. Furthermore, the human and yeast proteins are more similar to Arabidopsis PXA1 than to F20D21.17, with the single exception of P70R. It is interesting that the predicted F20D21.17 protein apparently contains only four TM domains (Fig. 6), whereas each of the homologous hemitransporters has six TM domains and PXA1 has 12 predicted TM domains. Because humans and yeast have multiple proteins that dimerize to form the functional peroxisomal transporter, F20D21.17 may homodimerize and form an Arabidopsis transporter. It remains to be determined whether both the full transporter (PXA1) and the hemitransporter (F20D21.17) play similar roles in plant peroxisomal β-oxidation, or whether the proteins have different expression patterns or substrate specificities making their roles unique.
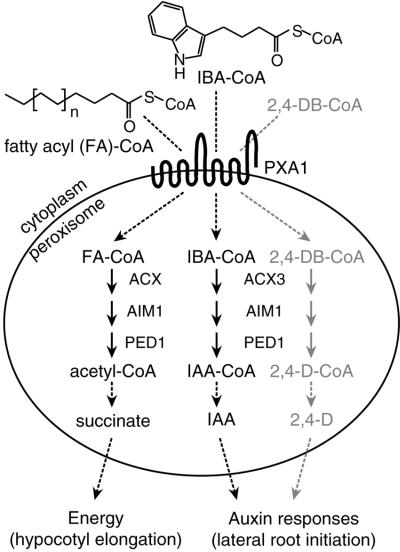
The use of IBA resistance coupled with Suc-dependent seedling development as a screen to identify Arabidopsis peroxisomal β-oxidation mutants, such as pxa1, is providing an unbiased approach to explore the specifics of both IBA and fatty acid metabolism in plants (Bartel et al., 2001). The identification of PXA1 as the gene defective in an IBA-response mutant supports the hypothesis that peroxisomes convert IBA to IAA using a pathway resembling fatty acid β-oxidation. A model of this process, based on IBA- and 2,4-DB-resistant mutants, is shown in Figure 7. PXA1 likely transports fatty acyl-CoA esters into the peroxisome, where they are β-oxidized to acetyl-CoA, which is metabolized to succinate via the glyoxylate cycle (Gerhardt, 1992; Olsen, 1998). Because pxa1 is resistant to IBA and the IBA analog 2,4-DB (Zolman et al., 2000), PXA1 probably also imports IBA-CoA and 2,4-DB-CoA into peroxisomes for oxidation to IAA-CoA and 2,4-D-CoA, respectively. These compounds presumably are hydrolyzed and exit the peroxisome to elicit specific phenotypic effects, including root elongation inhibition and lateral root initiation.
Figure 7.
Proposed model for PXA1 function in Arabidopsis. Based on its homology to human and yeast proteins, PXA1 is predicted to be a transporter localized in the peroxisomal membrane. Because of the mutant phenotype, PXA1 is likely to import fatty acyl (FA)-CoA esters, IBA-CoA, and 2,4-DB-CoA into the peroxisome, where they are catabolized to succinate, IAA, and 2,4-D, respectively. 2,4-DB-resistant mutants defective in an acyl-CoA oxidase (acx3, Eastmond et al., 2000), a multifunctional protein (aim1, Richmond and Bleecker, 1999), and a thiolase (ped1, Hayashi et al., 1998) also are IBA resistant (Zolman et al., 2000; B. Zolman and B. Bartel, unpublished data), suggesting that these isozymes act directly in IBA and 2,4-DB β-oxidation. See text for details.
IBA is used widely to propagate plant cuttings because it efficiently induces lateral and adventitious roots (Hartmann et al., 1990). One hypothesis explaining the high rooting ability of IBA is that IBA acts as a “slow-release” form of IAA, similar to certain auxin conjugates (Hangarter and Good, 1981). Because IBA is β-oxidized to IAA and because numerous mutants defective in β-oxidation do not form lateral roots in response to IBA (Zolman et al., 2000), it is likely that the enhanced lateral root production by wild-type plants in response to IBA is based on its slow conversion to IAA. Furthermore, the observation that pxa1 makes fewer lateral roots on unsupplemented medium suggests that the β-oxidation of endogenous IBA to IAA may be important for the development of lateral roots in wild-type seedlings.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis accessions Col-0 and Ws were used. The pxa1 mutant was previously described as B40, an ethyl methanesulfonate-induced IBA-response mutant in the Col-0 background (Zolman et al., 2000). Plants were grown in soil (Metromix 200, Scotts, Marysville, OH) at 22°C to 25°C under continuous illumination by Cool White fluorescent bulbs (Sylvania, Danvers, MA). Plants grown aseptically were plated on PNS (plant nutrient medium with 0.5% [w/v] Suc; Haughn and Somerville, 1986) solidified with 0.6% (w/v) agar, either alone or supplemented with hormones (from 0.1, 1, or 100 mm stocks in ethanol) or Basta (from a 50-mg mL−1 stock in 25% [v/v] ethanol). Plates were wrapped with gas-permeable surgical tape (LecTec Corp., Minnetonka, MN) and grown at 22°C under continuous light. Plates containing auxin were incubated under yellow filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter, 1990).
Phenotypic Analyses
The pxa1 mutant was backcrossed at least once prior to analyses, and all assays were conducted at least twice with similar results. Seeds were surface sterilized (Last and Fink, 1988) and plated on PNS with the indicated hormone concentration. In root elongation assays, seedlings were grown for 8 d and removed from the agar, and the length of the primary root was measured (Figs. 1, 4A, and 5E). In lateral root assays (Figs. 2 and 5F), seeds were grown on PNS for either 4 or 6 d, transferred to medium containing IBA, IAA, or no hormone, and grown for an additional 4 or 6 d. The number of lateral roots was counted under a dissecting microscope. In the seed germination assay (Table I), seeds were plated on either PNS or PN (plant nutrient medium without Suc) and grown in the light for 6 d or in the light for 1 d to induce germination, followed by 5 d in the dark. Germination, defined as radicle emergence from the seed coat, was scored using a dissecting microscope. Establishment was defined as seedling emergence and cotyledon expansion in the light or hypocotyl elongation in the dark. Seeds that germinated but did not establish were transferred to PNS and scored again after an additional 4 d of growth under white light. For hypocotyl elongation assays (Figs. 4B and 5D), seeds were plated on PN (without Suc) or PNS and incubated for 24 h under white light before being transferred to the dark. The length of the hypocotyl was measured after an additional 5 d.
In the adult growth studies, wild-type and mutant plants were grown on PNS for 10 d under white light before being transferred to soil. Rosette diameter was measured at the widest point of the plant without disturbing any leaves (Fig. 3A). For flowering time determination, plants were examined two to three times per week and the number of leaves was counted at the appearance of the first bud in the shoot apex.
Genetic Analysis and Mutant Complementation
The mutant was outcrossed to Ws for mapping, and DNA was isolated (Celenza et al., 1995) from IBA-resistant F2 plants that had 100% Suc-dependent progeny. The mutation was mapped using published simple sequence length polymorphisms (Bell and Ecker, 1994) and cleaved amplified polymorphic sequences (Konieczny and Ausubel, 1993). For the marker mi167, PCR amplification with the primers 5′-CACTAGATCTTCAAGCGCTCGATG-3′ and 5′-GACATATCCATAGAGTAACTTCAC-3′ yield a 390-bp product with two RsaI sites in Col-0 and no sites in Ws.
A candidate gene (PXA1) within the mapping interval was examined for defects in the mutant. Genomic DNA extracted from mutant plants was amplified using seven pairs of oligonucleotides (5′-CTTCAGGTGTTTTGGACACTTGTTGTCAAG-3′ and 5′-CATCCAGTATAAGATCGCTCAACTCTGAGG-3′, 5′-ATATCACACGTGGATGGTCGGATTACGC-3′ and 5′-CAGAAGATTAGACCCTTGCTC-AACTCG-3′, 5′-GTTACTCCAACCGGAAATGTTTTGGT-GG-3′ and 5′-CCATCCTCCTTCACCGTCTAATGACAG-AAC-3′, 5′-GCACAAGTGCTGTCACAACTGATATGG-3′ and 5′-GAGCAATTTAGTAAGTCGTGACAAGGTG-3′, 5′-GGACCACTGTGAAGTATGTCTTGGAGCAAG-3′ and 5′-CAATTCCAAGTTCACTTCCGGTAGCTTTG-3′, 5′-CTTAATTGCCCTAGCTATAGCTGCTGG-3′ and 5′-CCATTCCC-TGACCCAAGTTCTTTGATATCC-3′, and 5′-GAAATAG-TTTCAGGGAAAAGCCTGCTCGTC-3′ and 5′-ATTCTCTT-CCACTCCTTGCGATCGAGGAAG-3′) with a program of 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 3 min. The resulting overlapping fragments were approximately 1200 bp each and covered the gene from 90 bp upstream of the putative translation start site to 140 bp downstream of the stop codon. Amplification products were purified by sequential ethanol, polyethylene glycol, and ethanol precipitations (Ausubel et al., 1999) and sequenced directly using an automated DNA sequencer (Rice University Sequencing Facility, Houston) with the primers used for amplification.
An apparently full-length PXA1 cDNA in pBluescript II SK(−) (H1A6T7) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus; Kieber et al., 1993). This cDNA was sequenced using vector-derived and internal primers (GenBank accession no. AF378120). To remove an open reading frame upstream of the PXA1 open reading frame, we linearized the plasmid with AseI and removed the 3′ overhang with T4 DNA polymerase. The cDNA was excised by digestion with EcoRI and ligated into pBluescript II KS(+) cut with SmaI and EcoRI, forming pKS-PXA1c. The cDNA was excised by digestion with NotI and subcloned in the sense orientation behind the constitutive 35S cauliflower mosaic virus promoter in the 35SpBARN plant transformation vector (LeClere and Bartel, 2001). This plasmid (35SPXA1) was electroporated (Ausubel et al., 1999) into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1992), which was used to transform pxa1 mutant plants using the floral dip method (Clough and Bent, 1998). Transformants were identified on PNS plates supplemented with 7.5 μg mL−1 Basta after 10 d under white light. Rescue assays were done using seeds from homozygous progeny of Basta-resistant transformants (Fig. 5, D–F).
RNA Analyses
Wild-type and pxa1 mutant plants were grown for 5 d under white light on PNS plates covered with filter paper. Tissue was harvested by immersion in liquid nitrogen, and RNA was isolated as described before (Nagy et al., 1988). Reverse transcription of the RNA was done using the Retroscript reverse transcription-PCR kit (Ambion, Austin, TX) according to the manufacturer's instructions. PCR amplification of the cDNA was performed using primers spanning the pxa1 mutation, and the resulting products were purified and sequenced with the primers used for amplification (see above).
ACKNOWLEDGMENTS
We thank Sherry LeClere for the 35SpBARN vector, Beth Thompson for initial mapping attempts, and the Arabidopsis Biological Resource Center at Ohio State University for the cDNA clone. We are grateful to Raquel Adham, Sherry LeClere, Seiichi Matsuda, Melanie Monroe-Augustus, Rebekah Rampey, Luise Rogg, and Andy Woodward for critical comments on the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9982611), by the Robert A. Welch Foundation (grant no. C–1309), and by the American Society of Plant Biologists (Undergraduate Research Fellowship to I.D.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010550.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley-Interscience; 1999. [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regul (in press)
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Braiterman LT, Watkins PA, Moser AB, Smith KD. Peroxisomal very long chain fatty acid β-oxidation activity is determined by the level of adrenoleukodystrophy protein (ALDP) expression. Mol Genet Metab. 1999;66:91–99. doi: 10.1006/mgme.1998.2789. [DOI] [PubMed] [Google Scholar]
- Brickner DG, Brickner JH, Olsen LJ. Sequence analysis of a cDNA encoding Pex5p, a peroxisomal targeting signal type 1 receptor from Arabidopsis thaliana (PGR 98-154) Plant Physiol. 1998;118:330. [Google Scholar]
- Brown JWS, Smith P, Simpson CG. Arabidopsisconsensus intron sequences. Plant Mol Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Contreras M, Mosser J, Mandel JL, Aubourg P, Singh I. The protein coded by the X-adrenoleukodystrophy gene is a peroxisomal integral membrane protein. FEBS Lett. 1994;344:211–215. doi: 10.1016/0014-5793(94)00400-5. [DOI] [PubMed] [Google Scholar]
- Davies PJ. Plant Hormones. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–38. [Google Scholar]
- Davies TGE, Coleman JOD. The Arabidopsis thalianaATP-binding cassette proteins: an emerging superfamily. Plant Cell Environ. 2000;23:431–443. [Google Scholar]
- Dubois-Dalcq M, Feigenbaum V, Aubourg P. The neurobiology of X-linked adrenoleukodystrophy, a demyelinating peroxisomal disorder. Trends Neurosci. 1999;22:4–12. doi: 10.1016/s0166-2236(98)01319-8. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA. Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsisseed germination. J Biol Chem. 2000;275:34375–34381. doi: 10.1074/jbc.M004945200. [DOI] [PubMed] [Google Scholar]
- Epstein E, Ludwig-Müller J. Indole-3-butyric acid in plants: occurrence, synthesis, metabolism, and transport. Physiol Plant. 1993;88:382–389. [Google Scholar]
- Erdmann R, Kunau W-H. A genetic approach to the biogenesis of peroxisomes in the yeast Saccharomyces cerevisiae. Cell Biochem Funct. 1992;10:167–174. doi: 10.1002/cbf.290100306. [DOI] [PubMed] [Google Scholar]
- Fawcett CH, Wain RL, Wightman F. The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc R Soc Lond B Biol Sci. 1960;152:231–254. doi: 10.1098/rspb.1960.0035. [DOI] [PubMed] [Google Scholar]
- Fujiki Y. Molecular defects in genetic diseases of peroxisomes. Biochim Biophys Acta. 1997;1361:235–250. doi: 10.1016/s0925-4439(97)00051-3. [DOI] [PubMed] [Google Scholar]
- Gärtner J. Disorders related to peroxisomal membranes. J Inherit Metab Dis. 2000;23:264–272. doi: 10.1023/a:1005636113499. [DOI] [PubMed] [Google Scholar]
- Gärtner J, Jimenez-Sanchez G, Roerig P, Valle D. Genomic organization of the 70-kDa peroxisomal membrane protein gene (PXMP1) Genomics. 1998;48:203–208. doi: 10.1006/geno.1997.5177. [DOI] [PubMed] [Google Scholar]
- Gärtner J, Moser H, Valle D. Mutations in the 70K peroxisomal membrane protein gene in Zellweger syndrome. Nat Genet. 1992;1:16–23. doi: 10.1038/ng0492-16. [DOI] [PubMed] [Google Scholar]
- Gerhardt B. Fatty acid degradation in plants. Prog Lipid Res. 1992;31:417–446. doi: 10.1016/0163-7827(92)90004-3. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Valle D. Peroxisome biogenesis disorders. Trends Genet. 2000;16:340–344. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- Hangarter RP, Good NE. Evidence that IAA conjugates are slow-release sources of IAA in plant tissues. Plant Physiol. 1981;68:1424–1427. doi: 10.1104/pp.68.6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann HT, Kester DE, Davies FT. Plant Propagation: Principles and Practices. Englewood Cliffs, NJ: Prentice-Hall; 1990. pp. 199–245. [Google Scholar]
- Hashmi M, Stanley W, Singh I. Lignoceroyl-CoASH ligase: enzyme defect in fatty acid β-oxidation system in X-linked childhood adrenoleukodystrophy. FEBS Lett. 1986;196:247–250. doi: 10.1016/0014-5793(86)80256-3. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 2000;19:5701–5710. doi: 10.1093/emboj/19.21.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, van Roermund CWT, Distel B, van den Berg M, Vilela C, Wanders RJA, Tabak HF. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996;15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- Holzinger A, Mayerhofer P, Berger J, Lichtner P, Kammerer S, Roscher AA. Full length cDNA cloning, promoter sequence, and genomic organization of the human adrenoleukodystrophy related (ALDR) gene functionally redundant to the gene responsible for X-linked adrenoleukodystrophy. Biochem Biophys Res Comm. 1999;258:436–442. doi: 10.1006/bbrc.1999.0535. [DOI] [PubMed] [Google Scholar]
- Imanaka T, Aihara K, Takano T, Yamashita A, Sato R, Suzuki Y, Yokota S, Osumi T. Characterization of the 70-kDa peroxisomal membrane protein, an ATP binding cassette transporter. J Biol Chem. 1999;274:11968–11976. doi: 10.1074/jbc.274.17.11968. [DOI] [PubMed] [Google Scholar]
- Imanaka T, Shiina Y, Takano T, Hashimoto T, Osumi T. Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J Biol Chem. 1996;271:3706–3713. doi: 10.1074/jbc.271.7.3706. [DOI] [PubMed] [Google Scholar]
- Kamijo K, Kamijo T, Ueno I, Osumi T, Hashimoto T. Nucleotide sequence of the human 70 kDa peroxisomal membrane protein: a member of ATP-binding cassette transporters. Biochim Biophys Acta. 1992;1129:323–327. doi: 10.1016/0167-4781(92)90510-7. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kindl H. Fatty acid degradation in plant peroxisomes: function and biosynthesis of the enzymes involved. Biochimie. 1993;75:225–230. doi: 10.1016/0300-9084(93)90080-c. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. T-DNA transformation and insertion mutagenesis. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis Research. River Edge, NJ: World Scientific; 1992. pp. 224–273. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsismutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kragler F, Lametschwandtner G, Christmann J, Hartig A, Harada JJ. Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc Natl Acad Sci USA. 1998;95:13336–13341. doi: 10.1073/pnas.95.22.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Fink GR. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science. 1988;240:305–310. doi: 10.1126/science.240.4850.305. [DOI] [PubMed] [Google Scholar]
- Lazarow PB. Genetic approaches to studying peroxisome biogenesis. Trends Cell Biol. 1993;3:89–93. doi: 10.1016/0962-8924(93)90079-g. [DOI] [PubMed] [Google Scholar]
- Lazo O, Contreras M, Bhushan A, Stanley W, Singh I. Adrenoleukodystrophy: impaired oxidation of fatty acids due to peroxisomal lignoceroyl-CoA ligase deficiency. Arch Biochem Biophys. 1989;270:722–728. doi: 10.1016/0003-9861(89)90555-9. [DOI] [PubMed] [Google Scholar]
- Lazo O, Contreras M, Hashmi M, Stanley W, Irazu C, Singh I. Peroxisomal lignoceroyl-CoA ligase deficiency in childhood adrenoleukodystrophy and adrenomyeloneuropathy. Proc Natl Acad Sci USA. 1988;85:7647–7651. doi: 10.1073/pnas.85.20.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol. 2001;46:695–703. doi: 10.1023/a:1011699722052. [DOI] [PubMed] [Google Scholar]
- Liu LX, Janvier K, Berteaux-Lecellier V, Cartier N, Benarous R, Aubourg P. Homo- and heterodimerization of peroxisomal ATP-binding cassette half-transporters. J Biol Chem. 1999;274:32738–32743. doi: 10.1074/jbc.274.46.32738. [DOI] [PubMed] [Google Scholar]
- Lombard-Platet G, Savary S, Sarde CO, Mandel JL, Chimini G. A close relative of the adrenoleukodystrophy (ALD) gene codes for a peroxisomal protein with a specific expression pattern. Proc Natl Acad Sci USA. 1996;93:1265–1269. doi: 10.1073/pnas.93.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J. Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 2000;32:219–230. [Google Scholar]
- Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature. 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua NH. Analysis of gene expression in transgenic plants. In: Gelvin SB, Schilperourt RA, editors. Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. B4/1–B4/29. [Google Scholar]
- Olsen LJ. The surprising complexity of peroxisome biogenesis. Plant Mol Biol. 1998;38:163–189. [PubMed] [Google Scholar]
- Perrson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Perrson B, Argos P. Topology prediction of membrane proteins. Protein Sci. 1996;5:363–371. doi: 10.1002/pro.5560050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999;11:1911–1923. doi: 10.1105/tpc.11.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsicki Y, Sharom FJ. Phospholipid flippase activity of the reconstituted P-glycoprotein multidrug transporter. Biochemistry. 2001;40:6937–6947. doi: 10.1021/bi0024456. [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández R, Davies TGE, Coleman JOD, Rea PA. The Arabidopsis thalianaABC protein superfamily, a complete inventory. J Biol Chem. 2001;267:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Gietl C, Schmid M. Sequence analysis of a cDNA encoding Pex7p, a peroxisomal targeting signal 2 receptor from Arabidopsis thaliana (PGR 99-060) Plant Physiol. 1999;120:339. [Google Scholar]
- Shani N, Valle D. A Saccharomyces cerevisiaehomolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc Natl Acad Sci USA. 1996;93:11901–11906. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani N, Valle D. Peroxisomal ABC transporters. Methods Enzymol. 1998;292:753–776. doi: 10.1016/s0076-6879(98)92058-4. [DOI] [PubMed] [Google Scholar]
- Shani N, Watkins PA, Valle D. PXA1, a possible Saccharomyces cerevisiaeortholog of the human adrenoleukodystrophy gene. Proc Natl Acad Sci USA. 1995;92:6012–6016. doi: 10.1073/pnas.92.13.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Lazo O, Dhaunsi GS, Contreras M. Transport of fatty acids into human and rat peroxisomes. J Biol Chem. 1992;267:13306–13313. [PubMed] [Google Scholar]
- Smith BT, Sengupta TK, Singh I. Intraperoxisomal localization of very-long-chain fatty acyl-CoA synthetase: implication in X-adrenoleukodystrophy. Exp Cell Res. 2000;254:309–320. doi: 10.1006/excr.1999.4757. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Swartzman EE, Viswanathan MN, Thorner J. The PAL1 gene product is a peroxisomal ATP-binding cassette transporter in the yeast Saccharomyces cerevisiae. J Cell Biol. 1996;132:549–563. doi: 10.1083/jcb.132.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Braakman I, Distel B. Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol. 1999;9:447–453. doi: 10.1016/s0962-8924(99)01650-5. [DOI] [PubMed] [Google Scholar]
- van Veen HW. Towards the molecular mechanism of prokaryotic and eukaryotic multidrug transporters. Semin Cell Dev Biol. 2001;12:239–245. doi: 10.1006/scdb.2000.0249. [DOI] [PubMed] [Google Scholar]
- van Veen HW, Konings WN. Multidrug transporters from bacteria to man: similarities in structure and function. Semin Cancer Biol. 1997;8:183–191. doi: 10.1006/scbi.1997.0064. [DOI] [PubMed] [Google Scholar]
- Verleur N, Hettema EH, van Roermund CWT, Tabak HF, Wanders RJA. Transport of activated fatty acids by the peroxisomal ATP-binding-cassette transporter Pxa2 in a semi-intact yeast cell system. Eur J Biochem. 1997;249:657–661. doi: 10.1111/j.1432-1033.1997.00657.x. [DOI] [PubMed] [Google Scholar]
- Wain RL, Wightman F. The growth-regulating activity of certain ω-substituted alkyl carboxylic acids in relation to their β-oxidation within the plant. Proc R Soc Lond B Biol Sci. 1954;142:525–536. doi: 10.1098/rspb.1954.0041. [DOI] [PubMed] [Google Scholar]
- Wanders RJA, van Roermund CWT, van Wijland MJA, Schutgens RBH, van den Bosch H, Schram AW, Tager JM. Direct demonstration that the deficient oxidation of very long chain fatty acids in X-linked adrenoleukodystrophy is due to an impaired ability of peroxisomes to activate very long chain fatty acids. Biochem Biophys Res Commun. 1988;153:618–624. doi: 10.1016/s0006-291x(88)81140-9. [DOI] [PubMed] [Google Scholar]
- Wimmer C, Schmid M, Veenhuis M, Gietl C. The plant PTS1 receptor: similarities and differences to its human and yeast counterparts. Plant J. 1998;16:453–464. doi: 10.1046/j.1365-313x.1998.00320.x. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B. chy1, an Arabidopsismutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem. 2001;276:31037–31046. doi: 10.1074/jbc.M104679200. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thalianareveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]