Abstract
Understanding biological systems and the roles of their constituents is facilitated by the ability to make quantitative, sensitive, and comprehensive measurements of how their proteome changes, e.g., in response to environmental perturbations. To this end, we have developed a high-throughput methodology to characterize an organism's dynamic proteome based on the combination of global enzymatic digestion, high-resolution liquid chromatographic separations, and analysis by Fourier transform ion cyclotron resonance mass spectrometry. The peptides produced serve as accurate mass tags for the proteins and have been used to identify with high confidence >61% of the predicted proteome for the ionizing radiation-resistant bacterium Deinococcus radiodurans. This fraction represents the broadest proteome coverage for any organism to date and includes 715 proteins previously annotated as either hypothetical or conserved hypothetical.
The era of whole-genome DNA sequencing has yielded a nearly completed human genome (1) and many complete and initially annotated genomes for lower organisms, with several hundred more anticipated over the next few years. Effective exploitation of this information requires technologies that provide more comprehensive, higher-throughput, and quantitative analyses of proteomes (i.e., the array of proteins expressed by a cell, tissue, or organism at a given time).
To address this need, we have developed a method for identifying peptides based on accurate mass tags (AMTs) for each protein expressed by a given organism. This approach provides greater sensitivity and dynamic range than previously achievable, more comprehensive coverage of expressed proteins, a basis for precise quantitation, and greater throughput for measurements of proteomes, because protein identification using AMTs circumvents the need for routine tandem MS (MS/MS).
Our initial study has focused on the extremely radiation-resistant bacterium Deinococcus radiodurans, but the general approach can be used for any organism whose genome has been sequenced. Two independent annotations are available for the sequenced genome of D. radiodurans (2, 3). This organism has been the target of genetic manipulation for a decade, a candidate for bioremediation of radioactive waste sites, and a subject for the study of DNA repair pathways (3, 4). D. radiodurans has an extraordinary ability to tolerate both acute and chronic exposure to high levels of ionizing radiation. Exponentially growing cultures of the Gram-positive, nonmotile, red-pigmented, nonpathogenic bacterium are able to withstand 50–100 times more ionizing radiation than Escherichia coli (5, 6). D. radiodurans can survive 5,000–15,000 Gy of acute ionizing radiation with no loss of viability (depending on the culture conditions) (7), can grow continuously under 60 Gy/hr (8), and has the ability to reduce contaminant metals and radionuclides including Cr, Tc, and U to less soluble species (9). Its resistance to radiation and to other DNA-damaging conditions (e.g., UV light, hydrogen peroxide) and desiccation (10) is likely because of its efficient DNA damage repair (6, 11). It has been suggested that the multiple copies of the D. radiodurans genome (4–10 genome equivalents) (11) may be organized to facilitate recombinational repair processes (3, 12). However, the set of predicted genes for D. radiodurans appears conventional and does not reveal the basis for its extreme radiation resistance (3). The number of annotated DNA repair enzymes (2, 3) is less than reported for E. coli.
Most likely the DNA damage-resistance phenotype is determined collectively by a complex array of interacting proteins (3) as well as by many more subtle structural peculiarities of proteins and DNA. Neither of these resistance motifs is readily inferred from the D. radiodurans sequence, underscoring the potential importance of global studies to obtain a better understanding of the interactions involved, such as determining protein expression patterns under stressed and nonstressed conditions (i.e., proteome-wide analyses).
Experimental Protocol
Cell Culture.
All cell cultures were inoculated with 10 ml of starter culture in either defined (13) or rich media incubated overnight to confluence. The specific culture conditions are as follows: Defined medium mid-logarithm (log) phase: cells were cultured at 32°C; OD600 0.3–0.4. Defined medium late-log phase: cells were cultured at 32°C; OD600 0.6. Defined medium stationary phase: cells were cultured at 32°C; OD600 0.9. Tryptone/glucose/yeast (TGY) medium mid-log phase: cells were cultured at 32°C; OD600 0.3–0.4. TGY medium late-log phase: cells were cultured at 32°C; OD600 0.6. TGY medium stationary phase: cells were cultured at 32°C; OD600 0.9. For specific stress conditions, all cells were grown to mid-log phase (OD600 0.3–0.4) at 32°C in TGY media before stress unless otherwise noted. Heat shock: incubation temperature was raised to 42°C and further incubated for 1 hr before harvest. Cold shock: incubation temperature was lowered to 0°C and further incubated for 1 hr before harvest. Hydrogen peroxide: H2O2 was added to a final concentration of 60 μM and further incubated for 2 hr before harvest. One-week starvation: poststationary phase (OD600 0.9) D. radiodurans culture was incubated at 32°C for 1 wk without the addition of fresh medium. Four-week starvation: poststationary phase (OD600 0.9) D. radiodurans culture was incubated at 32°C for 4 wk without the addition of fresh medium. Chemical shock: 0.05% (vol/vol) trichloroethylene or xylene was added to the culture and incubated for 2 hr before harvest. Alkaline shock: the pH of culture was raised from 6.5 to 8.5 with 1 N NaOH and incubated for 1 hr before harvest. For experiments with irradiated cells, D. radiodurans was cultured to stationary phase in rich media, diluted, and irradiated without change in broth on ice at 10 kGy/hr [60Co Gammacell irradiation unit, J. L. Sheperd and Associates (San Fernando, CA) Shepard, Model JL 109]. After irradiation, cells were transferred to fresh media to recover before aliquots were taken at 10 time intervals: 0, 0.5, 1, 3, 5, 7, 9, 12, 24, and 36 hr. Cells were harvested by centrifugation at 10,000 × g at 4°C, washed three times with PBS, aliquoted, and quick frozen for storage at −80°C.
Cell Lysis and Tryptic Digestion.
Cell lysis was achieved by bead beating using three 90-sec cycles at 4,500 rpm in a Biospec (Bartlesville, OK) Minibeadbeater, with a 5-min cool down on ice between cycles. Lysates were immediately placed on ice to inhibit proteolysis. Protein concentration was determined by the BCA assay kit (Pierce). Before liquid chromatography (LC) MS analysis, the protein samples were denatured and reduced by the addition of guanidine hydrochloride (Gdn⋅HCl) (6 M) and DTT (1 mM) and boiled for 5 min. On reducing the Gdn⋅HCl concentration to below 2 M with 100 mM NH4HCO3 and 5 mM EDTA (pH 8.4), protein samples were digested by using bovine pancreas sequencing grade modified trypsin (Promega) (trypsin/protein, 1:50, wt/wt) at 37°C for 16 hr. Protein lysates were ultracentrifuged for 30 min at 356,000 × g, and clear supernatant was dialyzed against 50 mM Tris⋅HCl at 4°C with 500 molecular weight cutoff cellulose ester membrane. Lysates were subsequently aliquoted and quick frozen in nitrogen and stored in −80°C freezer until analyzed.
Capillary LC Separations.
The capillary LC system consisted of a pair of syringe pumps (100-ml ISCO model 100DM) and controller (series D ISCO) and an in-house manufactured mixer, capillary column selector, and sample loop for manual injections. Separations were achieved with 5,000 psi reversed-phase packed capillaries (150 μm i.d. × 360 μm o.d.; Polymicro Technologies, Phoenix) (14) by using two mobile-phase solvents consisting of 0.2% acetic acid and 0.05% trifluoroacetic acid (TFA) in water (A) and 0.1% TFA in 90% acetonitrile/10% water (B). The mobile-phase selection valve was switched from position A to B 10 min after injection, creating an exponential gradient as mobile phase B displaced A in the mixer. Flow through the capillary HPLC column was ≈1.8 μl/min when equilibrated to 100% mobile-phase A.
Initial Mass Tag Development.
Eluant from the HPLC was infused into a conventional ion trap MS (LCQ, ThermoFinnigan, San Jose, CA) operating in a data-dependent MS/MS mode over a series of segmented m/z ranges and also with prior fractionation by using ion exchange chromatography. For each cycle, the three most abundant ions from MS analysis were selected for MS/MS analysis by using a collision energy setting of 35%. Dynamic exclusion was used to discriminate against previously analyzed ions. The collision induced dissociation spectra from the conventional ion trap mass spectrometer were analyzed using sequest (15) and the genome sequence of D. radiodurans. Preliminary identifications were based on a minimum crosscorrelation score of 2, and these peptides were used as potential mass tags (PMTs) for validation by Fourier transform ion cyclotron resonance (FTICR) measurements. These analyses identified large numbers of polypeptide PMTs, of which ≈70% were then validated as AMTs based on the detection of a species having the predicted mass for the PMT to <1 ppm at the corresponding elution time in the LC-FTICR analysis. The methodology for PMT generation is described in detail elsewhere (16).
Validation of Mass Tags.
The 11.4-tesla FTICR mass spectrometer developed at our laboratory uses an electrospray ionization (ESI) interface to an electrodynamic ion funnel assembly coupled to a radio frequency quadrupole for collisional ion focusing and highly efficient ion accumulation and transport to the cylindrical FTICR cell for analysis (17). Mass spectra were acquired with ≈105 resolution. To obtain the desired <1-ppm mass measurement accuracy (MMA), a program that uses the multiple charge states (e.g., 2+, 3+) produced by ESI for many protonated polypeptides (18) is applied, followed by the use of “lock masses” (i.e., confidently known species that serve as effective internal calibrants) for each spectrum derived from commonly occurring polypeptides that are identified with high confidence from one capillary LC MS/MS analysis (for each organism) obtained by using FTICR with accurate mass measurements (16). Our calculations show ≈50% of the peptides predicted from in silico tryptic digestion of D. radiodurans have unique masses at MMA ≤1 ppm, and these peptides can be used to identify 99.5% of the predicted proteins potentially expressed by the organism. The fraction of peptides useful as AMTs is actually greater than 50% due to our use of elution time data, which serves to distinguish previously identified peptides having otherwise indistinguishable masses.
Results
Strategy for AMT-Based Measurements.
The approach utilizes instrumentation that combines high-pressure reversed-phase capillary LC with high magnetic field FTICR-MS (16, 19–21) and MS/MS. In our initial strategy, proteins extracted from cells grown under a variety of culture conditions are digested to yield a complex mixture of polypeptides that are analyzed to create a set of AMTs that serve as biomarkers for their parent proteins (16, 22). Our approach for defining AMTs provides high confidence for protein identification because of their initial identification by using MS/MS with standard database searching/identification (15) combined with validation by using high MMA and elution time (16). Although conventional capillary LC-MS/MS was used for most of the initial identifications, MS/MS with FTICR instrumentation was used in a limited number of cases to identify peptides present in concentrations too low to be analyzed effectively by conventional MS. The AMT-based protein identification allows subsequent high-throughput proteome measurements because only the distinctive masses and LC elution time are needed (16).
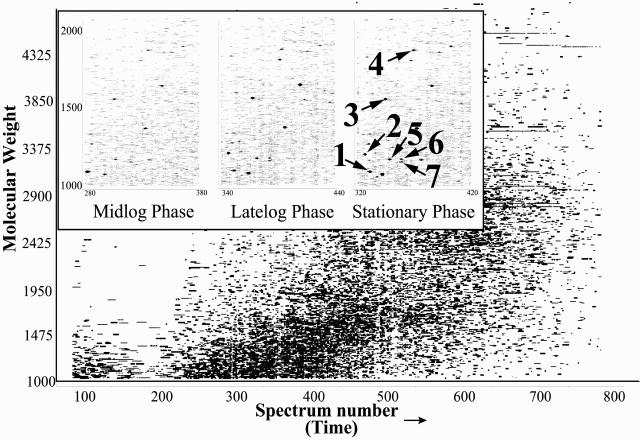
The D. radiodurans strain R1 genome consists of two chromosomes, one megaplasmid and one plasmid (3). Our analysis used the 3,116 protein-encoding ORFs predicted by the TIGR annotation (ftp://ftp.tigr.org/pub/data/d_radiodurans/GDR.pep) (2) (we exclude from this analysis 71 ORFs predicted to contain frame shifts). The proteomic measurements provide a physical validation that predicted ORFs actually encode a protein. A two-dimensional visualization of a portion of one analysis (Fig. 1) illustrates the ability to identify proteins from the peptides detected for D. radiodurans [grown in a defined minimal medium (13) and harvested at mid-log phase].
Fig 1.
Two-dimensional display of a capillary LC-FTICR analysis in which >50,000 putative polypeptides were detected from a tryptic digest of D. radiodurans proteins harvested in mid-log phase (OD600 0.3–0.4; 30°C). Inset shows portions of displays for peptides from D. radiodurans harvested in mid-log (Left), late-log (Center), and poststationary (Right) phases (spot size reflects relative abundance). Each individual spot corresponds to a peptide that can be identified along with the parent protein using AMTs. Spot identifications: 1: DR1314 (hypothetical protein); 2: DR1790 (homolog of yellow/royal jelly protein of insects); 3: DR1314 (hypothetical protein); 4: DR0309 (elongation factor Tu); 5: DR2577 (S-layer protein); 6: DR0989 (membrane protein); and 7: DR1124 (S-layer protein).
Overall Analysis Yields Identification of Over 60% of the Predicted Proteome.
The 200 LC-MS/MS analyses of peptides from collective culture conditions yielded PMT identifications for 9,159 peptides having a sequest score >2. Our measurements verified with high-confidence 6,997 AMTs, corresponding to 1,910 ORFs from D. radiodurans, representing 61% of the predicted ORFs (Table 1), the broadest proteome coverage for an organism achieved to date. Because the method uses two distinct MS measurements, matching accurate mass with MS/MS measurements, the use of the FTICR in these analyses lends higher confidence to those peptides identified compared to those detected (16). In a functional category breakdown (Table 2), coverages for categories associated with housekeeping functions were significantly higher than hypothetical proteins. In a single FTICR analysis, the masses for ≈1,500 AMTs are typically detected corresponding to ≈700 ORFs (depending on the culture condition) and 15–20% of the D. radiodurans proteome.
Table 1.
D. radiodurans proteome coverage
| Category | Size | Predict ORFs | Obs ORFs | % coverage |
|---|---|---|---|---|
| Total | 3.29 Mbp | 3,116 | 1,910 | 61 |
| Chromosome 1 | 2.65 Mbp | 2,633 | 1,586 | 60 |
| Chromosome 2 | 412 kbp | 369 | 34 | 63 |
| Mega plasmid | 177 kbp | 145 | 76 | 52 |
| Small plasmid | 46 kbp | 40 | 4 | 35 |
Table 2.
D. radiodurans proteome coverage by TIGR assigned functional category
| Category | Total | Seq cov | St dev | Obs | % Cat |
|---|---|---|---|---|---|
| Amino acid biosynthesis | 80 | 21.5 | 15.1 | 70 | 88 |
| Cofactor biosynthesis, etc. | 61 | 16.1 | 9.7 | 41 | 67 |
| Cell envelope | 77 | 17.7 | 19 | 62 | 81 |
| Cellular processes | 89 | 21.9 | 21.5 | 64 | 72 |
| Central metabolism | 154 | 14.7 | 10.5 | 111 | 72 |
| DNA metabolism | 81 | 13.2 | 10.1 | 55 | 68 |
| Energy metabolism | 199 | 24.7 | 20 | 152 | 76 |
| Fatty acid metabolism | 53 | 23.8 | 16.2 | 40 | 75 |
| Conserved hypothetical | 499 | 16.6 | 13.1 | 276 | 55 |
| Phage related/transposon | 47 | 11.7 | 3.7 | 9 | 19 |
| Protein fate | 86 | 22.2 | 20.6 | 65 | 76 |
| Protein synthesis | 114 | 38.8 | 24.2 | 100 | 88 |
| Nucleic acid synthesis | 53 | 19.2 | 14.7 | 42 | 79 |
| Regulatory functions | 126 | 15.5 | 10.8 | 76 | 60 |
| Transcription | 28 | 18.5 | 11.8 | 22 | 79 |
| Transport proteins | 191 | 16.5 | 15.5 | 138 | 72 |
| Unknown function | 176 | 15.4 | 12.4 | 107 | 61 |
| Hypothetical | 1,002 | 18.7 | 14.2 | 479 | 48 |
Number of ORFs assigned to each category by TIGR.
Percentage of protein sequence represented by AMTs averaged over all proteins in a single category.
Standard deviation of sequence coverage for the entire category.
Number of ORFs detected in each category.
Percentage of ORFs identified for each category.
FTICR-Based Proteomics Can Provide Sensitive and Dynamic Range for Broad Proteome Coverage.
One major challenge for proteomics is the large variation in the relative abundances of proteins (>5 orders of magnitude). A recognized disadvantage of conventional proteomic studies by using two-dimensional polyacrylamide gel electrophoresis separations coupled with protein identification by MS analysis is that, in general, only higher-abundance proteins are detected (23, 24). We have previously shown that ≈20 zmol (≈12,000 molecules) of protein can be detected during an FTICR analysis (25). In this work, capillary LC-FTICR measurements provided an overall dynamic range of 104-105 (16). The further expansion of the dynamic range of measurements by using a new DREAMS technology (26) offers the potential to detect proteins at less than one copy per cell given practical cell populations (108 cells per analysis). For more abundant proteins, broad coverage of peptide fragments was achieved; ribosomal proteins were identified with an average of 9 AMTs. Similarly, DNA polymerase I, considered to be the most abundant polymerase in prokaryotic cells (27), is predicted to be present in about 400 copies per cell in E. coli and was identified with six AMTs providing 20% coverage of the protein.
Another method for estimating protein abundance in a cell population is the use of codon adaptation index (CAI) (28) or predicted highly expressed (PHX) (29) proteins, two separate algorithms devised to estimate protein abundance on the basis of codon usage. Although many of the proteins fall into a range corresponding to proteins expressed at high abundance, about 50% of the proteins we detected have both CAI and PHX scores predicting low abundance. If codon usage were the only indicator of protein abundance, the increase in AMT coverage per ORF with higher CAI and PHX values (Table 3) would suggest a slight and expected bias toward detection of high abundance proteins. However, the significant number of ORFs identified with low PHX and CAI values also suggests that this approach effectively identifies lower abundance proteins.
Table 3.
Detected ORFs for different CAI and PHX values
| CAI | Total | Obs | Cov | PHX | Total | Obs | Cov |
|---|---|---|---|---|---|---|---|
| 0.1–0.2 | 24 | 9 | 10 | 0–0.6 | 542 | 370 | 15 |
| 0.2–0.3 | 94 | 33 | 15 | 0.6–0.7 | 729 | 436 | 17 |
| 0.3–0.4 | 292 | 120 | 16 | 0.7–0.8 | 657 | 387 | 19 |
| 0.4–0.5 | 786 | 404 | 15 | 0.8–0.9 | 451 | 256 | 19 |
| 0.5–0.6 | 1,239 | 801 | 16 | 0.9–1 | 204 | 114 | 21 |
| 0.6–0.7 | 546 | 424 | 24 | 1–1.25 | 233 | 142 | 23 |
| 0.7–0.8 | 120 | 109 | 50 | 1.25–1.5 | 86 | 56 | 25 |
| 0.8–0.9 | 12 | 9 | 58 | 1.5–2 | 96 | 72 | 35 |
| 2.0–3 | 21 | 19 | 43 |
Total number of ORFs with codon adaptation index (27) or predicted highly expressed proteins (28) for the CAI or PHX bin ranges indicated.
Number of observed ORFs with CAI or PHX values indicated.
Percentage of observed proteins represented by AMTs for all the proteins predicted in a single bin.
Protein Expression Profiles Change with Varied Culture Conditions.
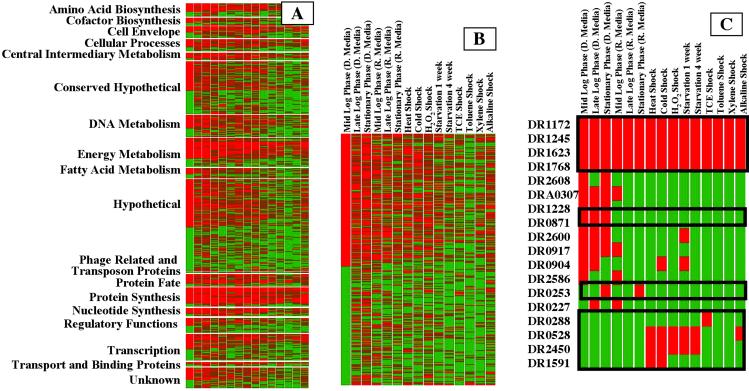
The increased throughput provided by the use of AMTs allows the investigation into the pattern of detected proteins, under a range of environmental conditions (Fig. 2A). These results illustrate that many predicted proteins associated with “housekeeping” functions are detected under all conditions evaluated (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). Although detection of an AMT provides high confidence, the presence of the protein, its absence cannot exclude its presence at a low level. Examination of the expression patterns can suggest potential functions for ORFs having no homology to other known proteins (Fig. 2B). For example, many of these hypothetical proteins are detected under every culture condition, suggesting that they may fill “housekeeping” functions (e.g., DR1172, DR1245, DR1623, and DR1768; Fig. 2C). DR0253 (Fig. 2C) was detected only in poststationary phase (regardless of the culture condition), whereas DR0871 and DR1228 were detected only in minimal medium; DR0528, DR1591, and DR2450 were detected only when cells were exposed to various “stress” conditions. Although limited quantities of such information cannot unequivocally reveal specific protein function, they can help narrow the range of possible functions and identify ORFs for further study.
Fig 2.
(A) The qualitative pattern of ORF expression detected using AMTs for various conditions by TIGR assigned functional category (2) (red, AMTs detected; green, AMTs not observed). Each culture condition (as described in Experimental Protocols) was analyzed at least two times. (B) Expansion of the pattern for predicted hypothetical proteins, illustrating similarities and differences under different culture conditions. (C) Expansion showing several representative hypothetical proteins.
Frame Shifts and Reading Frames Can Be Determined by Using AMTs.
We examined data for protein expression by genes with frame shifts designated in the initial annotation (2) and found that D. radiodurans displays both direct and indirect pathways for the synthesis of asparaginyl-tRNA (tRNAASN) and glutaminyl-tRNA (tRNAGLN) (30) despite the frame shift in the tRNAASN synthetase (DR1270) (2) (based on an AMT analysis of peptides from all possible ORFs predicted from a six-frame translation of the D. radiodurans genome). The Comprehensive Microbial Resource database (www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database = gdr) lists 71 additional genes predicted to encode proteins, having probable frame shifts to unknown reading frames, and where only partial protein sequences were predicted. In addition to peptides for asparaginyl tRNA synthetase, we observed several other peptides derived from genes believed to be disrupted by frame shifts. Some of these peptides are markers for functional proteins, suggesting that frame-shift corrections may be occurring, perhaps at the level of transcription. For example, we detected peptides predicted from DR0100 [putative single-stranded DNA-binding protein (SSB)] (2) that is reported to contain three frame shifts, supporting the existence of at least some functional SSB protein. AMTs were also identified for DR1624 (RNA helicase), DR2477 (3-hydroxyacyl-CoA dehydrogenase), and DRC0020 (putative modification methylase). AMTs were detected in all three reading frames of ORF DR2477, suggesting either that D. radiodurans is able to mediate the required two frame shifts or that there is a mistake in the genome sequence. Detected peptides corresponding to ORFs and frames in which they are coded are published as supporting information on the PNAS web site.
Stress Response, DNA Damage Repair, and Recognition Proteins and RecA.
It is predicted that D. radiodurans encodes a spectrum of 148 stress response proteins (four are predicted to contain authentic frame shifts) (3). We identified 74 proteins predicted from the annotated genome with an average of six AMTs per protein corresponding to 24% overall sequence coverage. Two classes of annotated proteins shown to play roles in the detoxification processes are catalase (DR 1998 and DRA0259) and superoxide dismutase (SOD) (DR1279, DR1546, and DRA0202). We confirmed the presence of both predicted catalases with 48 (DR1998) and 37 (DRA0259) AMTs covering ≈80% and ≈50% of their sequences, respectively. Of the SOD proteins, we identified DR 1279 with 14 AMTs, corresponding to 87% coverage, whereas DR1546 and DRA0202 were identified with two and three AMTs, respectively. Of the predicted 75 proteins with potential DNA repair activities (23), we identified 39 with an average of three AMTs per protein corresponding to 12% coverage of the amino acid sequence. RecA is central to homologous recombinational repair of irradiation-induced double-strand breaks in D. radiodurans chromosomal and plasmid DNA (6, 7, 11, 31). Five different RecA AMTs (covering 34% of the sequence) were identified primarily in cells recovering from exposure to ionizing radiation.
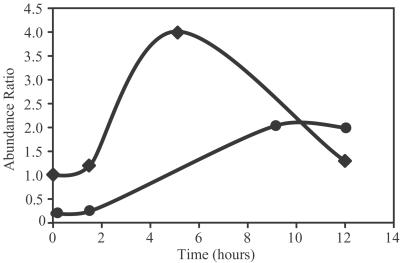
Stable isotope labeling allows the quantitation of changes in protein expression levels. The protein expression pattern of D. radiodurans recovering from a dose of 17.5 kGy of ionizing radiation (growing on 14N media) was compared with the protein expression pattern from control cells (growing on 15N media) (Fig. 3). Initial studies indicate that the expression of RecA as well as DNA-directed RNA polymerase I is significantly induced during recovery from 17.5 kGy irradiation, consistent with previous observations (32).
Fig 3.
Change in relative abundance of (•) RecA and (♦) DNA-directed RNA polymerase in cells after exposure to 17.5 kGy of ionizing radiation obtained be analysis of a mixture of cells grown on unlabeled media and the 15N-labeled reference proteome (control, nonirradiated cells). Cells were prepared at 0, 3, 7, 9, and 12 hr after exposure.
Discussion
Comprehensive proteome analyses can be achieved with greater confidence by using AMTs for protein identifications because of the combined use of sequence-related information and accurate mass measurement. Although a catalog of proteins from a particular organism is useful for gene verification, the analysis of protein expression patterns under various culture conditions can be used to infer function, deduce metabolic pathways, identify regulatory networks, etc.
When D. radiodurans is exposed to ionizing radiation, a dose-dependent delay of cellular replication suggests the existence of a DNA damage checkpoint (33). During this delay, several phases of cellular detoxification are hypothesized (34), but details remain unclear. One detoxification process in D. radiodurans involves removal of the activated oxygen species by catalase and SOD. Although the biochemical function of these two enzymes is known (35), their role in radiation resistance is unclear. Basal levels of catalase in exponentially growing cultures of D. radiodurans have been shown to be over 100-fold greater than for E. coli (10, 35). Genetic inactivation of members of either protein group render D. radiodurans cells more sensitive to ionizing radiation at doses that exceed 16,000 Gy (36), whereas mutations in DR1279 result in a greater sensitivity to ionizing radiation compared with mutations in DR1998. We confirmed the presence of both catalases under most culture conditions, suggesting constitutive expression. Of the SOD proteins, we identified DR1279 under many culture conditions but detected DR1546 and DRA0202 proteins in only a few culture conditions and in relatively low abundance. Thus, DR1279 apparently dominates SOD function and also appears to be constitutively expressed.
Additional detoxification processes in D. radiodurans export substantial portions of its DNA as large or small oligonucleotides from the cell after irradiation (37), presumably to prevent reincorporation into the genome. Homologs of UvrA (including UvrA1) are believed to play a role in these processes because they have been linked to ABC transporter proteins (2), and UvrA serves as the site of attachment for nucleotide excision repair proteins to the cell membrane in E. coli (2, 38). The dramatic DNA damage resistance of D. radiodurans has been attributed to exceedingly efficient DNA repair mechanisms, although such mechanisms remain poorly understood (3, 34). We observed most of the predicted proteins associated with nucleotide excision repair (including the UvrA1, UvrB, UvrC, Mfd, and PolA) in both stressed and unstressed cultures, which may indicate that these processes are continuously removing damaged nucleotides from the cell. UV DNA damage endonuclease and DNA helicase II (UvrD) were not detected, suggesting that these proteins may be expressed only at low basal levels. The detoxification process originally ascribed to the activity of the MutT Nudix protein family has been refuted by Xu et al., who found low levels of MutT activity for proteins putatively identified with MutT domains (39), consistent with the present observation of the MutT Nudix protein family predominantly in unstressed cells.
Although D. radiodurans can express a suite of stress response proteins, expression patterns have initially revealed few correlations between expression and stress response. One exception is for the hypothetical protein DR0070, which is observed only after alkaline treatment (Fig. 2). Closer examination of this ORF reveals a limited similarity to alkaline protease of Bacillus subtilis (39), surrounding the catalytic serine residue, and illustrates the use of such data to verify or suggest protein function.
One protein that has been closely linked with the DNA repair ability of D. radiodurans is RecA. RecA is detected predominantly in cells recovering from ionizing radiation and also at low levels in nonstressed cells, suggesting a low level of constitutive expression, contrary to previously published work. Previous studies using conventional PAGE with a RecA immunoprobe were not sufficiently sensitive to detect these low levels of RecA in D. radiodurans grown under nonstress conditions (32). Besides RecA, few proteins typically associated with presynaptic or postsynaptic recombination events were detected.
Although this is the first study, to our knowledge, to demonstrate such comprehensive (>61%) proteome coverage, an additional significance of our approach lies in the breadth of proteomic studies now enabled. The present results provide a large set of AMT “protein biomarkers” for quantitative expression studies that can be conducted with greater sensitivity and higher throughput due to elimination of the need for MS/MS measurements. Such studies use cells grown on stable-isotope-labeled media whose proteins (and their peptide AMTs) serve as internal standards to establish relative expression levels (40, 41). Several studies have now demonstrated that precision of better than 10–20% is achievable (41–43), a significant improvement over conventional proteomics approaches or gene expression arrays. The observation that both RNA polymerase I and RecA were induced after exposure to ionizing radiation (Fig. 3), although not surprising, illustrates that the use of AMTs for quantitation also provides the rapid throughput that will be necessary to extract biological insights from global proteomic studies.
Supplementary Material
Acknowledgments
This research was supported by the U.S. Department of Energy, Office of Biological and Environmental Research. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy through contract DE-AC06-76RLO 1830. A portion of this work was also supported by Grants DE-FG02-97ER62492 (from the Natural and Accelerated Bioremediation Research Program) and DE-F602-01ER63220 (from the Microbial Cell Program), Office of Biological and Environmental Research, U.S. Department of Energy (to M.J.D.).
Abbreviations
LC, liquid chromatography
FTICR, Fourier transform ion cyclotron resonance
PMT, potential mass tag
AMT, accurate mass tag
MS/MS, tandem MS
TCA, tricarboxylic acid cycle
MMA, mass measurement accuracy
log, logarithm
SOD, superoxide dismutase
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 10943.
References
- 1.Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1325.11181995 [Google Scholar]
- 2.White O., Eisen, J. A., Heidelberg, J. F., Hickey, E. K., Peterson, J. D., Dodson, R. J., Haft, D. H., Gwinn, M. L., Nelson, W. C., Richardson, D. L., et al. (1999) Science 286, 1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V. & Daly, M. J. (2001) Microbiol. Mol. Biol. Rev. 65, 44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly M. J. (2000) Biotechnology 11, 280-285. [DOI] [PubMed] [Google Scholar]
- 5.Smith K. C. & Martignoni, K. D. (1976) Photochem. Photobiol. 24, 515-523. [DOI] [PubMed] [Google Scholar]
- 6.Daly M. J., Ouyang, L., Fuchs, P. & Minton, K. W. (1994) J. Bacteriol. 176, 3508-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minton K. W. (1994) Mol. Microbiol. 1, 9-15. [DOI] [PubMed] [Google Scholar]
- 8.Lange C. C., Wackett, L. P., Minton, K. W. & Daly, M. J. (1998) Nat. Biotechnol. 16, 929-933. [DOI] [PubMed] [Google Scholar]
- 9.Fredrickson J., Kostandarithes, H., Li, S., Plymale, A. & Daly, M. (2000) Appl. Environ. Microbiol. 66, 2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattimore V. & Battista, J. R. (1996) J. Bacteriol. 178, 633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayama S. & Matsuyama, A. (1971) Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 19, 13-19. [DOI] [PubMed] [Google Scholar]
- 12.Minton K. W. & Daly, M. J. (1995) Bioessays 17, 457-464. [DOI] [PubMed] [Google Scholar]
- 13.Venkateswaran A., McFarlan, S. C., Ghosal, D., Minton, K. W., Vasilenko, A., Makarova, K., Wackett, L. P. & Daly, M. J. (2000) Appl. Environ. Microbiol. 66, 2620-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y., Tolić, N., Zhao, R., Paša-Tolić, L., Li, L., Berger, S. F., Harkewicz, R., Anderson, G. A., Belov, M. E. & Smith, R. D. (2001) Anal. Chem. 73, 3011-3021. [DOI] [PubMed] [Google Scholar]
- 15.Eng J. K., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- 16.Smith R. D., Anderson, G. A., Lipton, M. S., Paša-Tolić, L., Shen, Y., Conrads, T. P., Veenstra, T. D. & Udseth, H. R. (2002) Proteomics 2, 513-523. [DOI] [PubMed] [Google Scholar]
- 17.Harkewicz R., Belov, M., Anderson, D., Paša-Tolić, L., Masselon, C., Prior, D., Udseth, H. & Smith, R. (2001) J. Am. Soc. Mass Spectrom. 13, 144-154. [DOI] [PubMed] [Google Scholar]
- 18.Bruce J. E., Anderson, G. A., Brands, M. D., Paša-Tolić, L. & Smith, R. D. (2000) J. Am. Soc. Mass Spectrom. 11, 416-421. [DOI] [PubMed] [Google Scholar]
- 19.Marshall A. G., Hendrickson, C. L. & Jackson, G. S. (1998) Mass Spectrom. Rev. 17, 1-35. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y., Zhao, R., Belov, M. E., Conrads, T. P., Anderson, G. A., Tang, K., Paša-Tolić, L., Veenstra, T. D., Lipton, M. S., Udseth, H. R. & Smith, R. D. (2001) Anal. Chem. 73, 1766-1775. [DOI] [PubMed] [Google Scholar]
- 21.Li L., Masselon, C., Anderson, G. A., Paša-Tolić, L., Lee, S.-W., Shen, Y., Zhao, R., Lipton, M. S., Conrads, T. P., Tolić, N. & Smith, R. D. (2001) Anal. Chem. 73, 3312-3322. [DOI] [PubMed] [Google Scholar]
- 22.Conrads T. P., Anderson, G. A., Veenstra, T. D., Paša-Tolić, L. & Smith, R. D. (2000) Anal. Chem. 72, 3349-3354. [DOI] [PubMed] [Google Scholar]
- 23.Wilm M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T. & Mann, M. (1996) Nature (London) 379, 466-469. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- 25.Belov M., Gorshkov, M., Udseth, H. & Smith, R. (2001) J. Am. Soc. Mass Spectrom. 12, 1312-1319. [DOI] [PubMed] [Google Scholar]
- 26.Belov M. E., Anderson, G. A., Angell, N. H., Shen, Y., Tolić, N., Udseth, H. R. & Smith, R. D. (2001) Anal. Chem. 73, 5052-5060. [DOI] [PubMed] [Google Scholar]
- 27.Patel P. H., Suzuki, M., Adman, E., Shinkai, A. & Loeb, A. (2001) J. Mol. Biol. 308, 823-827. [DOI] [PubMed] [Google Scholar]
- 28.Sharp P. M. & Li, W.-H. (1987) Nucleic Acids Res. 15, 1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlin S. & Mrazek, J. (2000) J. Bacteriol. 182, 5238-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curnow A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Soll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moseley B. E. & Evans, D. M. (1983) J. Gen. Microbiol. 129, 2437-2445. [DOI] [PubMed] [Google Scholar]
- 32.Carroll J. D., Daly, M. J. & Minton, K. W. (1995) J. Bacteriol. 178, 130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattimore V., Udupa, K. S., Berne, G. A. & Battista, J. R. (1995) J. Bacteriol. 177, 5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battista J. R. (1997) Annu. Rev. Microbiol. 51, 203-224. [DOI] [PubMed] [Google Scholar]
- 35.Chou F. I. & Tan, S. T. (1990) J. Bacteriol. 172, 2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markillie L. M., V. S., Hradecky, P. & Wong, K. K. (1999) J. Bacteriol. 181, 666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vukovic-Nagy B., Fox, B. W. & Fox, M. (1974) Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 25, 329-337. [DOI] [PubMed] [Google Scholar]
- 38.Bauche C. & Laval, J. (1999) J. Bacteriol. 181, 262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W., Shen, J., Dunn, C. A., Desai, S. & Bessman, M. J. (2001) Mol. Microbiol. 39, 286-290. [DOI] [PubMed] [Google Scholar]
- 40.Gygi S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H. & Aebersold, R. (1999) Nat. Biotechnol. 17, 994-999. [DOI] [PubMed] [Google Scholar]
- 41.Paša-Tolić L., Jensen, P. K., Anderson, G. A., Lipton, M. S., Peden, K. K., Martinovic, S., Tolić, N., Bruce, J. E. & Smith, R. D. (1999) J. Am. Chem. Soc. 121, 7949-7950. [Google Scholar]
- 42.Gygi S. P., Rist, B. & Aebersold, R. (2000) Biotechnology 11, 396-401. [DOI] [PubMed] [Google Scholar]
- 43.Jensen P. K., Paša-Tolić, L., Anderson, G. A., Horner, J. A., Lipton, M. S., Bruce, J. E. & Smith, R. D. (1999) Anal. Chem. 71, 2076-2084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.