It is well established that chloroplasts in green and red algae are derived from a primary endosymbiotic event between a cyanobacterium and a eukaryotic organism ≈1 billion years ago (Fig. 1; refs. 1 and 2). Although these two groups account for many of the world's photosynthetic species, most other major taxonomic groups of photosynthetic organisms (stramenopiles—including diatoms, phaeophytes, chrysophytes—and haptophytes) have plastids derived from a photosynthetic eukaryote implying a secondary endosymbiosis (1, 2). Still other groups, such as the dinoflagellates, have more complicated associations believed to be derived from tertiary endosymbioses involving the engulfment of a secondary endosymbiont. Each endosymbiotic event has characteristic structural changes associated with it, the most notable of which is the addition of two membranes surrounding the plastid (the inner representing the cell membrane of the engulfed organism and the outer representing the phagocytosis vacuole membrane) (2). Dinoflagellates, although believed to be tertiary endosymbionts, have only 3 membranes surrounding their plastids (1, 2), suggesting that the acquisition of too many membranes may be functionally unstable and can cause some to be lost.
Long-held theories suggested that primitive dinoflagellates were heterotrophic and the addition of a peridinin-containing plastid occurred relatively recently.
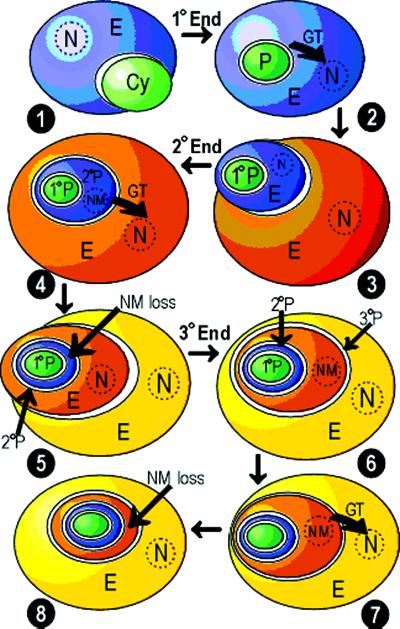
Figure 1.
Theoretical steps of primary, secondary, and tertiary plastid endosymbiosis. (1) A nonphotosynthetic eukaryote engulfs a cyanobacterium. (2) The engulfed cyanobacterium becomes the cell's plastid, and gene transfer occurs between the cyanobacterium cell and the host cell nucleus. The cyanobacterial membrane becomes the inner plastid membrane and the eukaryotic vacuolar membrane becomes the outer plastid membrane (primary endosymbiosis). (3) A nonphotosynthetic eukaryote engulfs a eukaryote with a primary plastid. (4) Four plastid membranes are present from the primary and secondary plastids (see text); the nucleus of the secondary plastid is reduced to a nucleomorph (which is present in the chlorarachniophytes and the cryptophyes); gene transfer occurs between the nucleomorph of the secondary plastid and the nucleus of the host cell (secondary endosymbiosis). (5) A eukaryotic cell with a secondary plastid is engulfed by a nonphotosynthetic eukaryote (tertiary endosymbiosis). The remnant nucleomorph of the secondary plastid is lost. (6) Up to six plastid membranes are theoretically possible (see text). The nucleus of the engulfed cell has been reduced to a nucleomorph. (7) Gene transfer occurs between the nucleomorph of the tertiary plastid and the host cell nucleus. (8) The nucleomorph of the tertiary plastid is lost. Abbreviations: 1° End, primary endosymbiosis; 2° End, secondary endosymbiosis; 3° End, tertiary endosymbiosis; Cy, cyanobacterium; E, eukaryote; GT, gene transfer; N, nucleus; NM, nucleomorph; P, plastid.
Dinoflagellates are fascinating organisms that have intrigued researchers for many years. They are most well known for toxic blooms associated with red tides and symbiotic relationships with corals (zooxanthellae) (2). They contain an astounding array of unique features that has been the impetus for continued evolutionary studies. One is their close phylogenetic link with apicomplexans, organisms that are best known for causing some of our most deadly infectious diseases (3, 4). Another is the diverse array of light harvesting pigments within the group. Peridinin is a xanthophyll found exclusively in dinoflagellates and, together with chl a (ubiquitous among photosynthetic organisms), makes up the light harvesting complex found in most species. Dinoflagellates with other combinations of plastid pigments are also known, including chl b (also in green algae), fucoxanthin, chl c1 and c2 (also in stramenopiles and haptophytes) and chl c1 and phycobilins (also in cryptophytes), and are believed to be the products of further endosymbioses with species from those groups (5–8). In this issue of PNAS, Yoon et al. (9) provide startling new evidence that implicates dinoflagellate plastids containing fucoxanthin and chl c1 and c2 (derived from a haptophyte ancestor) as being ancestral to those with peridinin. This new paradigm in the relationship of these species forces us to rethink many aspects of dinoflagellate evolution.
In many respects, the findings by Yoon et al. (9) allow a more parsimonious view of dinoflagellate evolution. Previously long-held theories suggested that primitive dinoflagellates were heterotrophic and the addition of a peridinin-containing plastid occurred relatively recently (10), which was borne out by the fact that approximately 50% of dinoflagellate species are heterotrophic (1) and that many photosynthetic dinoflagellates have retained heterotrophic behavior (11). However, recent studies have demonstrated that photosynthetic dinoflagellates are clearly ancestral, and that heterotrophy has been independently derived numerous times within the group (8). Fucoxanthin and chl c1 and c2 are the predominant form of light harvesting pigments among stramenopiles and haptophytes. With the finding that haptophytes are the sister group to dinoflagellates in phylogenetic analyses and that dinoflagellates with fucoxanthin and chl c1 and c2 are apparently ancestral to the peridinin-containing species, there is no need to hypothesize an independent origin of these pigments nor a later endosymbiosis of a haptophyte in dinoflagellates.
Selection of the proper gene for examination of deep phylogenetic relationships was critical for carrying out this study. The gene encoding the carbon assimilation protein Rubisco, rbcL, has frequently been used to examine deep phylogenetic relationships (12–14). However, recent studies have demonstrated that there are several problems associated with using rbcL for this purpose. First, there appears to have been rampant horizontal gene transfer among various algal groups (14, 15). Reliance on such a gene to determine potential endosymbiotic relations would be fraught with potential misinterpretations. Second, peridinin-containing dinoflagellates have a nuclear encoded form II Rubisco (L2 structure rather than the L8S8 structure typical of most eukarotic plastids) that is similar in sequence homology to anaerobic bacteria (16–18). Third, multiple copies of Rubisco have been found in some organisms (14). Although identical or nearly identical copies of photosynthesis related genes are often found in cyanobacteria (19–21), both form I and form II Rubisco have been found in some species.
The genes chosen by Yoon et al. (9) were psaA (encoding the photosystem I P700 apoprotein A1) and psbA (encoding the photosystem II thylacoid protein D1). These genes are intimately tied to the photosystem reaction centers and have a low rate of base substitution (nonsynonomous replacement rate KA = 0.022 and 0.009 for psaA and psbA, respectively; ref. 22) compared with other genes such as rbcL (KA = 0.041, considered slowly evolving; ref. 22), so that significant phylogenetic signal should be expected even at these deep evolutionary positions. The resulting phylogenies were rigorously examined using various positions of the protein codons, amino acid sequences, and statistical testing, all pointing to a haptophyte being the sister group to both peridinin and fucoxanthin, chl c1 and c2 dinoflagellates. Supporting analyses were also performed with 16S rRNA data, although data sets were not as complete.
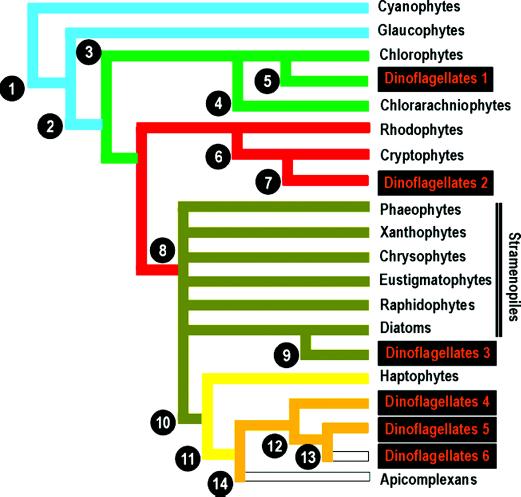
What are the relationships among stramenopiles, haptophytes, and dinoflagellates? It seems clear that stramenopile plastids form a monophyletic group and were derived from a red alga-like progenitor by means of a secondary endosymbiosis (9) (Fig. 2). Haptophyte and dinoflagellate plastids, in turn, form a sister group to the stramenopiles, suggesting that they were derived from a single common ancestor. It cannot be ruled out that this ancestor was more red alga-like (23), but given the features shared between stramenopiles and haptophytes (four membrane layers surrounding the plastid and similar pigment composition among other characters) (2), it is likely these share a close relationship. It also seems clear that dinoflagellate (and possibly apicomplexan) plastids are derived from haptophyte ancestors (Fig. 2). Yoon et al. (9) hypothesize a separate origin for the plastids within apicomplexans, yet a recent study suggests a common origin for the plastids of both groups (4). If so, it is likely that both are derived from a haptophyte progenitor.
Figure 2.
Hypothetical evolutionary relationships among algal groups and derivations of the various dinoflagellate lineages based on possible plastid endosymbiosis events. (1) Initial symbiosis of a cyanophyte/glaucophyte progenitor (see text). (2) Loss of residual peptidoglycan cell wall. (3) Loss of phycobilins and gain of chl b. (4) Secondary endosymbiosis of green alga with retention of algal nucleus as nucleomorph between second and third layers of plastid membrane. (5) Secondary endosymbiosis of green alga by dinoflagellate (Dinoflagellates 1). (6) Secondary endosymbiosis of red alga by cryptophyte with retention of algal nucleus (nucleomorph) between second and third layers of plastid membrane. (7) Tertiary endosymbiosis of cryptophyte by dinoflagellate (Dinoflagellates 2). (8) Secondary endosymbiosis of red alga; loss of phycobilins with gain of chl c1 and c2 as light harvesting pigments; gain of xanthophylls (predominantly fucoxanthin) as accessory pigments. (9) Tertiary endosymbiosis of diatom by dinoflagellate (Dinoflagellates 3). (10) Possibly a tertiary endosymbiosis of stramenopile by haptophyte (see text). (11) Possibly a tertiary or quaternary endosymbiosis of haptophyte by dinoflagellate (Dinoflagellates 4) (see text and figure 3 of ref. 9 for further details). (12) Loss of fucoxanthin and chl c1 and c2 after development of peridinin as light harvesting pigment (Dinoflagellates 5). (13) Loss of plastids; roughly half of peridinin-containing dinoflagellates (Dinoflagellates 6). (14) Loss of photosynthesis in apicomplexans.
This then begs the question, how many endosymbiotic events are required to account for all of the diversity present among photosynthetic algae? This is a difficult question that we may never fully understand. However, evidence from nuclear small subunit RNA sequences may help shed light on the answer (24, 25). Taken at face value, red algae, stramenopiles, haptophytes, and dinoflagellate/apicomplexans form four distinct and unrelated clades. Given the plastid phylogeny (Fig. 2), this would suggest that plastids of red algae are primary endosymbionts and those of stramenopiles are secondary endosymbionts (both consistent with previous theories) (1, 2). However, if haptophyte plastids are derived from a primitive stramenopile plastid, as believed, its occurrence would necessarily represent a tertiary endosymbiotic event. Similarly then, it follows that the dinoflagellate (and apicomplexan) plastid could conceivably represent a quaternary endosymbiotic event! Such a sequence of events seems contrary to morphological and physiological patterns in these groups, especially among the haptophytes.
A closer examination of the nuclear phylogeny trees (24, 25) indicates weak branch support for the lineages that separate haptophytes and stramenopiles. It is therefore possible that haptophytes are an ancient derivation from stramenopiles after fucoxanthin and chl c1 and c2 evolved in the lineage. This relationship is supported by Yoon et al. (9) in that the haptophyte lineage is sister to that of stramenopiles rather than from within it as might be expected if resulting from a tertiary endosymbiosis.
Separation of haptophytes and dinoflagellates is similarly weakly supported in nuclear phylogenetic trees (24, 25). Might these disparate organisms also share a common ancestor? This possibility seems less likely. Structurally, the plastids are different in the two groups (as pointed out previously) with dinoflagellates having only three membranes surrounding the plastid and the outer most membrane not being continuous with that of the nuclear envelope (1). Plastid chromosomes are also apparently different; photosystem genes in dinoflagellates are found separate on minicircular DNA chromosomes rather than a single large chromosome typical of most plastid DNA (26, 27). This occurrence may also help to explain why Yoon et al. (9) found dinoflagellate plastid DNA to evolve at a rate much faster than other plastid DNAs. A third strike against a common ancestry is that most dinoflagellates have retained a heterotrophic life history; most species still gain nourishment through phagocytosis of other organisms (11), perhaps indicating an incomplete transfer of nuclear-encoded plastid genes from their endosymbiotic eukaryotic plastid (see below). Although the evidence is less strong, the fossil record also suggests a different origin. Dinoflagellate-like organisms are present as early as in Silurian sediments (≈400 million years ago), whereas modern dinoflagellate fossils are only found in sediments from the late Triassic (200 million years ago), well after haptophytes had become established in the Carboniferous to Triassic periods (290 to 220 million years ago) (2).
Another indication that dinoflagellate plastids are derived from a separate endosymbiosis is their propensity toward forming endosymbiotic relationships. Several examples exist of dinoflagellates acquiring extra symbionts that eventually displace the original plastid symbiont (note multiple positions of dinoflagellates in the plastid tree in Fig. 2). The recruitment of a new plastid does not appear to be linked to strictly heterotrophic organisms; heterotrophic species are scattered throughout the dinoflagellate phylogeny (8) and some taxa that have undergone plastid replacement have maintained their former plastid as a three membrane-bound eyespot within the cell (1, 2). Examples of the secondary endosymbiosis of green algae by a dinoflagellate are found in Lepidodinium viride and Gymnodinium chlorophorum (8). The tertiary endosymbiosis of diatoms has been well studied in Kryptoperidinium foliaceum (6, 7) and Durinskia baltica (8, 28), and of cryptophytes in Gymnodinium acidotum (5). A final conjectured example is the transient symbiont of an anaerobic bacterium hypothesized to have been engulfed by an early dinoflagellate, apparently while the haptophyte plastid was still in place (9), and contributed its form II rbcL gene to the nuclear genome (16–18).
Acquisition of new plastids for photosynthesis is only one of several necessary changes for an organism to become completely autotrophic. Much of the cyanobacterial genome responsible for photosynthesis and other metabolic functions was transported from the plastid to the nuclear genome in the early endosymbiont. Should this organism have been taken up as a secondary endosymbiont, it would similarly have been necessary to move these genes from the nuclear genome of the primary symbiont to the nucleus of the secondary symbiont. Our review of the literature has determined that over 300 nuclear-encoded plastid genes are present in flowering plant systems, nearly three times the number of genes encoded in plant chloroplast DNA (29, 30). Although not functionally photosynthetic, it is likely that apicomplexans have maintained their plastids for these other functions (31).
It is a tribute to dinoflagellates that they have readily taken up additional symbionts in so many different lineages. Such a proclivity must also have a readily activated system for moving genes from the symbiont to the nuclear genome of the host. Characteristics such as these make dinoflagellates kings of symbioses.
Footnotes
See companion article on page 11724.
References
- 1.Van Den Hoek C, Mann D G, Jahns H M. Algae: An Introduction of Phycology. Cambridge, U.K.: Cambridge Univ. Press; 1995. [Google Scholar]
- 2.Graham L E, Wilcox L W. Algae. Englewood Cliffs, NJ: Prentice–Hall; 2000. [Google Scholar]
- 3.McFadden G I, Reith M E, Munholland J, Lang-Unnasch N. Nature (London) 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 4.Fast N M, Kinninger J C, Roos D S, Keeling P J. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox L W, Wedemeyer G J. J Phycol. 1984;20:236–242. [Google Scholar]
- 6.Chesnick J M, Morden C W, Schmeig A M. J Phycol. 1996;32:850–857. [Google Scholar]
- 7.Chesnick J M, Kooistra W H C F, Wellbrock U, Medlin L K. J Eukaryotic Microbiol. 1997;44:314–320. doi: 10.1111/j.1550-7408.1997.tb05672.x. [DOI] [PubMed] [Google Scholar]
- 8.Saldarriaga J F, Taylor F J R, Keeling P J, Cavalier-Smith T. J Mol Evol. 2001;53:204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- 9.Yoon H S, Hackett J D, Bhattacharya D. Proc Natl Acad Sci USA. 2002;99:11724–11729. doi: 10.1073/pnas.172234799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor F J R. BioSystems. 1980;13:65–108. doi: 10.1016/0303-2647(80)90006-4. [DOI] [PubMed] [Google Scholar]
- 11.Schnepf E, Elbraechter M. Eur J Protistol. 1999;28:3–24. doi: 10.1016/S0932-4739(11)80315-9. [DOI] [PubMed] [Google Scholar]
- 12.Morden C W, Golden S S. J Mol Evol. 1991;32:379–395. doi: 10.1007/BF02101278. [DOI] [PubMed] [Google Scholar]
- 13.Morden C W, Delwiche C F, Kuhsel M, Palmer J D. BioSystems. 1992;28:75–90. doi: 10.1016/0303-2647(92)90010-v. [DOI] [PubMed] [Google Scholar]
- 14.Delwiche C F, Palmer J D. Mol Biol Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 15.Palmer J D. BioEssays. 1995;17:1005–1008. doi: 10.1002/bies.950171202. [DOI] [PubMed] [Google Scholar]
- 16.Morse D, Salois P, Markovic P, Hastings J W. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 17.Palmer J D. Plant Cell. 1996;8:343–345. doi: 10.1105/tpc.8.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowan R, Whitney S M, Fowler A, Yellowlees D. Plant Cell. 1996;8:539–553. doi: 10.1105/tpc.8.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden S S, Brusslan J, Haselkorn R. EMBO J. 1986;5:2789–2798. doi: 10.1002/j.1460-2075.1986.tb04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golden S S, Stearns G W. Gene. 1988;67:85–96. doi: 10.1016/0378-1119(88)90011-x. [DOI] [PubMed] [Google Scholar]
- 21.Morden C W, Golden S S. Nature (London) 1989;337:382–385. doi: 10.1038/337382a0. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe K H. In: Cell Culture and Somatic Cell Genetics of Plants. Bogorad L, Vasil I K, editors. 7B. London: Academic; 1991. pp. 467–482. [Google Scholar]
- 23.Durnford D G, Deane J A, Tan S, McFadden G I, Green B R. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 24.Van de Peer Y, de Wachter R. J Mol Evol. 1997;45:619–630. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- 25.Van de Peer Y, Baldauf S L, Doolittle W F, Meyer A. J Mol Evol. 2000;51:565–576. doi: 10.1007/s002390010120. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Green B R, Cavalier-Smith T. Nature (London) 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 27.Barbrook A C, Howe C J. Mol Gen Genet. 2000;263:152–158. doi: 10.1007/s004380050042. [DOI] [PubMed] [Google Scholar]
- 28.Chesnick J M, Cox E R. BioSystems. 1987;21:69–78. doi: 10.1016/0303-2647(87)90007-4. [DOI] [PubMed] [Google Scholar]
- 29.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayshida N, Metsubayasha T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun C-R, Meng B-Y, et al. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 31.Palmer J D. Science. 1997;275:790–791. doi: 10.1126/science.275.5301.790. [DOI] [PubMed] [Google Scholar]