Reversible protein phosphorylation is the most common mechanism for cellular regulation in eukaryotic systems. Studies have demonstrated that serine/threonine phosphorylation plays a key role in the regulation of plant growth and development. On the other hand, tyrosine phosphorylation, despite its overwhelming importance in animals, has been largely neglected because a typical tyrosine kinase was not found in plants. Recent studies have characterized several protein tyrosine phosphatases (PTPs) from Arabidopsis and other species (1–3). Furthermore, a diverse group of about 20 genes encoding putative tyrosine phosphatases have been identified from the Arabidopsis genome, implying that tyrosine phosphorylation and dephosphorylation may serve important functions in plant biology. It is timely and exciting to learn in this issue of PNAS that tyrosine phosphatase activity is involved in the regulation of stomatal movement, a highly regulated process pivotal for plant survival (4). Finding a role for tyrosine phosphatases in stomatal regulation provides critical evidence that tyrosine phosphatases not only exist but also play an important role in higher plants. This work, together with those of others, begins to modify the earlier tenets on tyrosine phosphorylation in plant cell signaling and regulation.
Tyrosine phosphatase activity is involved in the regulation of stomatal movement.
Considerable differences are apparent when animals and plants are compared regarding protein phosphorylation in signal transduction. It is well established that both Ser/Thr and tyrosine phosphorylation play pivotal roles in cell signaling in animals. In particular, protein-tyrosine phosphorylation serves as a common mechanism by which growth factors and cytokines regulate cellular proliferation and differentiation in animals (5–7). The level of tyrosine phosphorylation in normal cells is determined by the balanced activity of protein tyrosine kinases (PTKs) and PTPs. Even the slightest tipping of this balance may result in cancer or abnormal cell death. As a result, a typical animal cell expresses a large number of PTKs and PTPs to fine-tune cellular proliferation/differentiation. In contrast, available information indicates that plants produce a large number of Ser/Thr kinases/phosphatases that function in plant signal transduction and regulation, but a typical tyrosine kinase has not yet been characterized from a plant species. Interestingly, bona fide tyrosine-specific protein phosphatases do exist in plants (1, 3). In addition, plants, like animals, produce a large number of protein phosphatases that dephosphorylate phosphoserine/threonine in addition to phosphotyrosine, so-called dual-specificity protein (tyrosine) phosphatases (DsPTPs or DSPs) (2). The DsPTPs have been shown to regulate the mitogen-activated protein kinases (MAPKs) in a variety of signal transduction pathways in both animals and yeast. Because a large number of MAPKs have also been found in plants, PTP/DsPTP regulation of MAPKs could become a common ground where tyrosine phosphorylation/dephosphorylation regulates cellular activities in the different eukaryotic systems. Recent studies have confirmed that this is indeed the case.
MAPKs are activated after mitogen stimulation or environmental stress in mammalian cells (8). At the molecular level, MAPK activation relies on phosphorylation of both tyrosine and threonine residues. A variety of DsPTPs exhibit activity toward activated MAPK isoforms both in vitro and in vivo (9). In each instance, dephosphorylation of a MAPK by a DsPTP leads to loss of kinase activity. It is interesting that multiple DsPTPs can be found in a given cell type as are multiple MAPKs. Studies have shown that each isoform of DsPTPs may dephosphorylate and regulate only one or two MAPKs, a substrate specificity that is closely related to the function of different PTPs, and therefore are referred to as MKPs (for MAP kinase phosphatases) (9). Almost all signaling pathways are turned on transiently and need to be turned off rather quickly after the activation. This is reflected by the activation pattern of MAPKs as well. A signal activates a MAPK rapidly (often reaching peak activity within a few minutes), and the MAPK activity subsequently returns to the basal level to quench the signaling process. It should be emphasized that such transient on-and-off switching is very important for the physiological process regulated by MAPKs. Prolonged or constant activation of a MAPK cascade can have detrimental consequence to the cell as best illustrated by tumorigenesis in mammalian cells that have MAPK constantly on (10). In budding yeast, the Hog1 pathway is required for osmotic stress tolerance, yet a constant MAPK activity of Hog1 actually renders yeast hypersensitive to the stress condition (11). Needless to say, protein phosphatases, especially PTPs and DsPTPs, play a critical role in turning off the activity of MAPKs and are essential for keeping the precise kinetic pattern of MAPK activation and inactivation.
Perhaps the first blow to the current paradigm that tyrosine phosphorylation is not important in plant cells comes from the finding of MAPK cascades in numerous signal transduction pathways in plants (reviewed in refs. 12–15). Included are those pathways responsible for transmitting biotic and abiotic stress and hormonal signals. Consistent with the diversity of MAPK functions, a large number of genes in the Arabidopsis genome have been identified to encode components in the MAPK cascades including MAPKs (about 20 genes), MAPKKs (10 genes), and MAPKKKs (>25 genes) (14, 15). A unique feature of MAPK activation in animal and yeast cells is dual phosphorylation at a closely spaced pair of threonine and tyrosine. Earlier studies have shown that activation of plant MAPKs also correlates with phosphorylation of tyrosine residue(s). A detailed biochemical analysis confirms that a plant MAPK (AtMPK4) is phosphorylated at a tyrosine residue during activation process (16). Dephosphorylation of the phosphotyrosine by AtPTP1, a tyrosine-specific PTP from Arabidopsis (1), resulted in the loss of MAPK activity. This study demonstrates that tyrosine phosphorylation is essential for the activation of plant MAPKs (16).
Because tyrosine phosphorylation of MAPKs is required for activation of the kinase, dephosphorylation of the tyrosine residue often occurs to bring the enzymes back to the inactive state and turn off the signaling process. As discussed earlier, such an on-and-off pattern is essential for physiological function of these kinases in animal and yeast. It is conceivable that tyrosine-specific and dual-specificity PTPs in plants function to regulate the activation pattern of MAPKs and possibly other target proteins in these organisms (1–3). In vitro biochemical analysis of AtPTP1 and AtDsPTP1 confirmed that they both dephosphorylate and inactivate MAPKs (2, 16). Following these studies, genetic analyses have demonstrated a physiological link between MAPK activation and PTP activity. In one such study (17), screens for UV light-sensitive mutants in Arabidopsis led to identification of a putative DsPTP, named AtMKP1, required for UV resistance. The mutant plants appear to have higher MAPK activity under UV illumination, implicating MKP1 as a negative regulator of MAPK(s) in UV response (17). Consistent with the in vitro studies, AtPTP1 is essential for maintaining the activation and inactivation pattern of a MAPK in Arabidopsis (R. Gupta and S.L., unpublished data). Although it remains to be determined whether AtDsPTP1 and other DsPTPs play a role in MAPK activity control in planta, there is little doubt that MAPK regulation serves as a major target for PTP function in plants as well as animals and fungi.
Besides providing evidence for PTP function in plant signal transduction in general, the study by MacRobbie (4) has identified a critical component in the regulatory circuit of ionic fluxes across the tonoplast during turgor regulation in guard cells. It is well known that stomatal closure is a result of solute (especially K+) efflux from the guard cells that form the stomata. More than 90% of solutes in guard cells are stored in the large central vacuole that serves as a predominant compartment for turgor control (18). To reduce the turgor pressure of guard cells and close the stomata, a large portion of solutes needs to be delivered from the vacuole to the cytosol across the tonoplast and finally exported out of the cell through ion channels in the plasma membrane. Our understanding of plasma membrane ion channels and their regulation by signals such as abscisic acid (ABA) has improved significantly in the past decade (reviewed in refs. 19 and 20). However, rather limited progress has been made in the understanding of ion fluxes across the tonoplast. Using radioactive tracer flux assays, MacRobbie has contributed significantly to our knowledge on K+ flux across tonoplast during stomatal closure caused by ABA and other stimuli (ref. 4 and references therein). In particular, it has been shown that ABA triggers a large transient efflux of K+ from the vacuole to the cytosol, increasing the K+ pool in the cytoplasm. This vacuole-cytosol K+ redistribution closely parallels the ABA-induced activation of outward K+ channels in the plasma membrane. Both processes contribute to the K+ export out of the cell and reduction of guard cell turgor leading to stomatal closure. From ABA to the activation of K+ efflux from vacuole, intermediate signaling components are largely unknown except for calcium changes that serve as a second messenger for ABA and activate K+ efflux channels in the tonoplast (18). It remains unclear how calcium plays such a role.
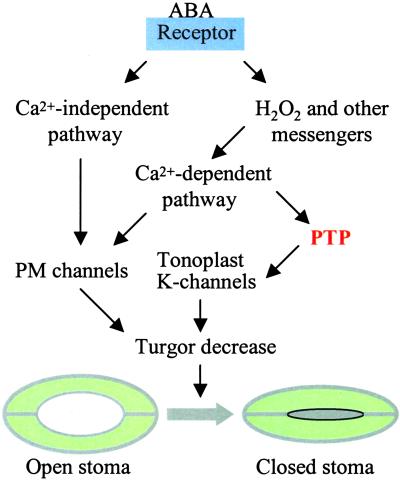
Using several specific PTP inhibitors, MacRobbie (4) demonstrates that PTP activity is essential for stomatal closure induced by four different factors including ABA, external calcium, darkness, and H2O2. Clearly, each of these factors may cause changes in intracellular calcium homeostasis that, in turn, alter the K+ channel activity in both plasma membrane and tonoplast (18). It is well known that ABA signaling in guard cells involves a calcium-dependent pathway (19, 20). H2O2 has been shown to serve as a downstream messenger for ABA and activates an inward calcium channel in the plasma membrane of guard cells (19). Darkness elicits calcium fluxes across chloroplast membranes (21) that may also affect calcium concentration in the cytosol. It is therefore possible that the PTP component lies downstream from calcium and upstream of K+ channel activity in the plasma membrane and tonoplast. The study by MacRobbie (4) has identified efflux from vacuole, but not fluxes across the plasma membrane, as the target process for the PTP inhibitors, suggesting that a protein specifically related to tonoplast K+ channels is regulated by tyrosine phosphorylation and dephosphorylation (Fig. 1). As discussed earlier, the major targets for PTPs in plants are MAPKs that participate in numerous signaling processes. Several previous studies have also shown that MAPK activities are involved in stomatal regulation (22, 23). However, MAPK does not appear to regulate the ABA-induced K+ efflux from vacuole, as a MAPK inhibitor does not interfere with the process (4).
Figure 1.
A simplified model of ABA-induced stomatal closure (see more detail in refs. 19 and 20). ABA receptors are possibly located in the plasma membrane and/or the cytoplasm. ABA signal is transmitted through calcium-dependent and calcium-independent pathways, leading to the regulation of ion channel activities in the plasma membrane (PM) and tonoplast. A tyrosine phosphatase (PTP) appears to be located downstream of calcium and regulates tonoplast K-channels responsible for K efflux from the vacuole. Vacuole K efflux and PM channels contribute to turgor decrease, leading to stomatal closure.
What could be the target for the PTP activity in ABA-induced K+ flux in guard cells? An interesting parallel has been drawn between the leaf movement in Mimosa and stomatal regulation in Commelina (4). A recent study (24) shows that tyrosine phosphorylation status of actin correlates with the leaf movement in Mimosa. Tyrosine phosphatase inhibitors including phenylarsine oxide used in MacRobbie's study (4) block the tyrosine dephosphorylation of actin and leaf closure, suggesting that tyrosine dephosphorylation of actin is essential for petiole bending (24). Leaf movement, a classical movement phenomenon in plants, is driven mainly by K+ fluxes in and out of the motor cells located in the petiole of small leaves. It is conceivable that tyrosine phosphorylation of actin is somehow related to the regulation of K+ fluxes in motor cells—a PTP activity seems to be upstream of a K+ transport process resembling the pathway in guard cells. How is actin related to K+ channel activity? In guard cells, several studies have correlated ABA effect on stomatal aperture with actin organization. For example, ABA treatment disrupts actin filaments, leading to stomatal closure (25). In this process, changes in actin organization may result from altered activity of small G proteins such as Rho-like AtRac1 in Arabidopsis (26). One study shows that actin organization correlates with K+ channel activity in the plasma membrane (27). However, it remains unknown how actin filament may affect the activity of tonoplast K+ channels. Besides actin, an actin-interacting protein profilin has been shown to be phosphorylated at tyrosine, although the functional significance of this event is not clear (28). It is known that MAPKs are autophosphorylated and phosphorylated by MAPKK at the tyrosine residue. One intriguing question concerns the molecular nature of the kinase(s) that phosphorylates actin and profilin at the tyrosine residues. Are they tyrosine-specific kinases or dual-specificity kinases? Identification of these kinases will open a new area of research in plant signal transduction.
In addition to actin cytoskeleton, a more direct target for PTP function in guard cells could be the tonoplast K+ channel itself. Recent studies in animal cells have revealed the regulation of K+ channel activity by tyrosine phosphorylation of the channel protein. For example, ROMK1 activity is down-regulated by increased tyrosine phosphorylation status (29). The PTP inhibitor phenylarsine oxide (PAO) specifically reduces the activity of ROMK1, implicating a PTP activity in the up-regulation of ROMK1 (29). The highly phosphorylated form of ROMK1 is susceptible to dynamin-dependent endocytosis that internalizes the channel protein and reduces the number of plasma membrane channels. It remains to be determined whether PAO reduces K+ efflux from vacuole in guard cells through reducing channel activity or channel number in the tonoplast (4). Nevertheless, finding a PAO-sensitive process in stomatal regulation provides critical evidence that a PTP(s) plays a role in a major signaling network of plants. Further identification of the target PTP and its substrates will be important steps toward understanding the signaling network for ABA-induced stomatal closure, a fundamental process for plant adaptation to the environment.
Footnotes
See companion article on page 11963.
References
- 1.Xu Q, Fu H, Gupta R, Luan S. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta R, Huang Y, Kieber J J, Luan S. Plant J. 1998;16:581–590. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- 3.Fordham-Skelton A P, Skipsey M, Evans I M, Edwards R, Gatehouse J A. Plant Mol Biol. 1999;39:593–605. doi: 10.1023/a:1006170902271. [DOI] [PubMed] [Google Scholar]
- 4.MacRobbie E A C. Proc Natl Acad Sci USA. 2002;99:11963–11968. doi: 10.1073/pnas.172360399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1993;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 6.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 7.Darnell J E. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 8.Chang L, Karin M. Nature (London) 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 9.Keyse S M. Semin Cell Dev Biol. 1998;9:143–152. doi: 10.1006/scdb.1997.0219. [DOI] [PubMed] [Google Scholar]
- 10.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Wounde G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 11.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizoguchi T, Ichimura K, Shinozaki K. Trends Biotechnol. 1997;15:15–19. doi: 10.1016/S0167-7799(96)10074-3. [DOI] [PubMed] [Google Scholar]
- 13.Ligterink W, Hirt H. Int Rev Cytol. 2001;201:209–275. doi: 10.1016/s0074-7696(01)01004-x. [DOI] [PubMed] [Google Scholar]
- 14.Tena G, Asai T, Chiu W, Sheen J. Curr Opin Plant Biol. 2001;4:392–400. doi: 10.1016/s1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Klessig D F. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Li H, Gupta R, Luan S, Kieber J J. Plant Physiol. 2000;122:1301–1310. doi: 10.1104/pp.122.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulm R, Revenkova E, di Sansebastiona G-P, Bechtold N, Paszkowski J. Genes Dev. 2001;15:699–709. doi: 10.1101/gad.192601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward J M, Pei Z M, Schroeder J I. Plant Cell. 1995;7:833–844. doi: 10.1105/tpc.7.7.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder J I, Kwak J M, Allen G J. Nature (London) 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 20.Luan S. Plant Cell Environ. 2002;25:229–237. doi: 10.1046/j.1365-3040.2002.00758.x. [DOI] [PubMed] [Google Scholar]
- 21.Sai J, Johnson C H. Plant Cell. 2002;14:1279–1291. doi: 10.1105/tpc.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luan S, Ting T, Gupta R. New Phytol. 2001;151:155–164. doi: 10.1046/j.1469-8137.2001.00168.x. [DOI] [PubMed] [Google Scholar]
- 23.Burnett E C, Desikan R, Moser R C, Neill S J. J Exp Bot. 2000;51:197–205. doi: 10.1093/jexbot/51.343.197. [DOI] [PubMed] [Google Scholar]
- 24.Kameyama K, Kishi Y, Yoshimura M, Kanzawa N, Sameshima M. Nature (London) 2000;407:37. doi: 10.1038/35024149. [DOI] [PubMed] [Google Scholar]
- 25.Eun S-O, Lee Y. Plant Physiol. 1997;115:1491–1498. doi: 10.1104/pp.115.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemichez E, Wu Y, Sanchez J-P, Mettouchi A, Mathur J, Chua N-H. Genes Dev. 2001;15:1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K, Luan S. Plant Cell. 1998;10:1957–1970. doi: 10.1105/tpc.10.11.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillen G, Valdes-Lopez V, Noguez R, Olivares J, Rodriguez-Zapata L C, Perez H, Vidali L, Villanueva M A, Sanchez F. Plant J. 1999;19:497–508. doi: 10.1046/j.1365-313x.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- 29.Sterling H, Lin D, Gu R, Dong K, Hebert S C, Wang W. J Biol Chem. 2002;277:4317–4323. doi: 10.1074/jbc.M109739200. [DOI] [PMC free article] [PubMed] [Google Scholar]