Abstract
The nuclease hypersensitivity element III1 upstream of the P1 promoter of c-MYC controls 85–90% of the transcriptional activation of this gene. We have demonstrated that the purine-rich strand of the DNA in this region can form two different intramolecular G-quadruplex structures, only one of which seems to be biologically relevant. This biologically relevant structure is the kinetically favored chair-form G-quadruplex, which is destabilized when mutated with a single G → A transition, resulting in a 3-fold increase in basal transcriptional activity of the c-MYC promoter. The cationic porphyrin TMPyP4, which has been shown to stabilize this G-quadruplex structure, is able to suppress further c-MYC transcriptional activation. These results provide compelling evidence that a specific G-quadruplex structure formed in the c-MYC promoter region functions as a transcriptional repressor element. Furthermore, we establish the principle that c-MYC transcription can be controlled by ligand-mediated G-quadruplex stabilization.
Expression of the c-MYC oncogene is linked to potentiation of cellular proliferation and to inhibition of differentiation, leading to its association with a number of human and animal malignancies, including carcinomas of the breast, colon, and cervix, as well as small-cell lung cancer, osteosarcomas, glioblastomas, and myeloid leukemias (1–4). Simply blocking c-MYC expression with antisense oligonucleotides has been shown to induce differentiation of myelocytes and myeloid leukemia cells, further cementing the inverse relationship between expression of this oncogene and differentiation (5–7). In transfection experiments, c-MYC was demonstrated to immortalize normal fibroblasts, and, in concert with a cooperating oncogene such as v-ABL or v-RAS, it causes malignant transformation (4). Much of this activity stems from the ability of c-MYC to act as both a transcriptional activator and repressor, inducing genes involved in proliferation, such as CAD, CDC25A, ODC, and hTERT, and repressing genes involved in growth arrest, such as GADD45 (4). It also has been found that c-MYC plays a role in the apoptotic response (1, 4, 8). For all of these reasons, c-MYC has emerged as an attractive target for anti-cancer therapeutic agents.
The transcriptional regulation of c-MYC expression is complex and involves multiple promoters and transcriptional start sites. P1 and P2 seem to be the predominant promoters (for reviews, see refs. 2, 3, and 9). The nuclease hypersensitivity element (NHE) III1 of the c-MYC promoter controls 85–90% of c-MYC transcription and has been the subject of considerable research over the past 2 decades (10–20). Originally identified as a major site of DNase I hypersensitivity (12), the NHE III1 is a 27-bp sequence located −142 to −115 bp upstream from the P1 promoter (Fig. 1). Comprising a pyrimidine-rich coding strand and a purine-rich noncoding strand, the NHE III1 is capable of engaging in a slow equilibrium between a typical duplex helix structure and both unwound and non-B-form regions of DNA called paranemic structures (21), which are associated with sensitivity to S1 nuclease digestion (13). Indeed, oligonucleotides representing the coding and noncoding strands can adopt i-motif and G-quadruplex structures, respectively (14, 15). It has been proposed that both types of structures might play a role in c-MYC transcription, but the involvement of either has yet to be substantiated.
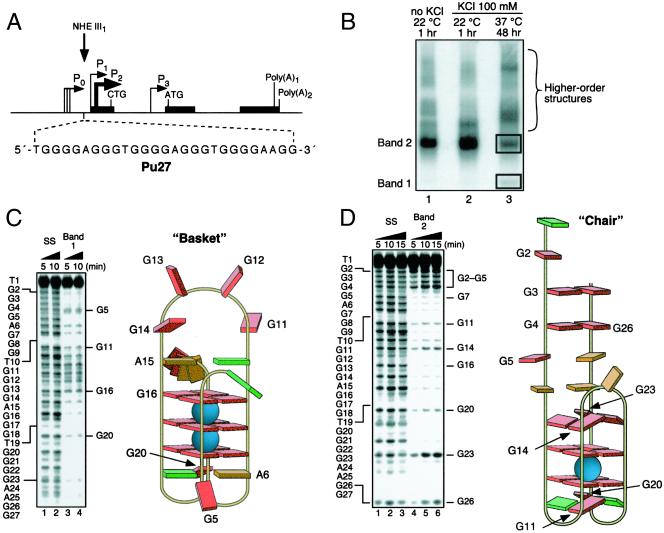
Figure 1.
Determination of the structures of the unimolecular G-quadruplexes formed after incubation of the Pu 27 strand (Pu27) in 100 mM KCl for 48 h at 37°C. (A) Promoter structure of the c-MYC gene; shown in Inset is the 27-mer sequence of the purine-rich strand upstream of the P1 promoter (3). (B) Nondenaturing gel analysis (15% polyacrylamide/12.5 mM KCl/NaCl, 4°C, 16 h) of Pu 27 preincubated under the conditions specified in the figure at a strand concentration of ≈25 μM. (C) DMS footprinting of band 1 in B. (Left) DMS treatment of the denatured Pu27 (lanes 1 and 2) and the isolated band (lanes 3 and 4). The Pu27 base sequence is shown to the left. (Right) Proposed structure based upon the footprinting pattern. Guanines showing DMS-induced cleavage are labeled in both Left and Right. Base colors: red, guanine; green, thymine; and orange, adenine. (D) As in C, but for band 2 in B.
Methods
DMS Footprinting.
Each band of interest was excised and soaked in 100 mM KCl solution (300 μl) for 6 h at 4°C. The solutions were filtered (microcentrifuged) and 30,000 cpm (per reaction) of DNA solution was diluted further with 100 mM KCl in 0.1 × TE (10 mM Tris/1 mM EDTA, pH 7.5) to a total volume of 70 μl (per reaction). After the addition of 1 μl of salmon sperm DNA (0.1 μg/μl), the reaction mixture was subjected to 1 μl of dimethyl sulfate solution (DMS:ethanol; 4:1, vol/vol) for the times shown (Fig. 1 C and D). Each reaction was quenched with 18 μl of stop buffer (3 M β-mercaptoethanol:water:NaOAc; 1:6:7, vol/vol). After ethanol precipitation (twice) and piperidine cleavage, the reactions were separated on a preparative gel (16%) and visualized on a PhosphorImager. DMS analysis of unstructured Pu27 was performed essentially in the same way, by using heat-denatured 3′-end-labeled Pu27 in 0.1 × TE buffer.
Site-Directed Mutagenesis.
The Del-4 plasmid was a gift from Bert Vogelstein at Johns Hopkins University. Single guanine mutations were made to the NHE III1 of this plasmid by using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol.
Transfection and Luciferase Assays.
HeLa S3 cells were transfected by using the Effectene lipid-based system (Qiagen, Chatsworth, CA) according to the manufacturer's protocol, using 0.1 μg of pRL-TK (Renilla luciferase reporter plasmid) and 0.9 μg of the Del-4 (wild type) or mutated plasmids. Firefly and Renilla luciferase activities were assayed by using the Dual Luciferase Reporter Assay System (Promega) in a 96-well plate format according to the manufacturer's protocol.
Polymerase Stop Assay.
The procedure is a modification of that used by Weitzmann et al. (ref. 22; see supporting information, which is published on the PNAS web site, www.pnas.org).
Results
Unimolecular G-Quadruplex Structures in NHE III1.
Preparative gel electrophoresis (12.5 mM KCl/NaCl, 16 h, 4°C) of the 3′-end-labeled 27-mer purine-rich strand (Pu27) from the NHE III1 (Fig. 1A) incubated in the presence of 100 mM KCl resulted in the formation of two bands (1 and 2), along with several bands of lower electrophoretic mobility (lane 3, Fig. 1B). Bands 1 and 2 were isolated and subjected to DMS-induced strand cleavage. The pattern of N7 guanine methylation produced by band 1 was consistent with the intramolecular basket quadruplex identified earlier (15), which consists of three stacked G-tetrads, two lateral two-base loops, and a six-base bridging loop (Fig. 1C). For band 2, a distinct and pronounced cleavage pattern suggested to us the formation of an intramolecular chair quadruplex, comprising two stacked G-tetrads, two lateral two-base loops, and an orthogonal three-base bridging loop (Fig. 1D). Because G7 and G16 show strong DMS protection relative to the other two guanines immediately above the top G-tetrad (G14 and G23), we propose a staggered, or split, G-tetrad conformation for these four bases. It is possible that excessive strain imposed by a one-base bridging loop might prevent the adoption of a planar tetrad. DMS footprinting of bands of lower mobility than band 1 (Fig. 1B) were consistent with higher-order quadruplex structures (data not shown). Importantly, an identical DMS footprint to that found for band 2 can be produced from Pu27 exposed to 100 mM KCl for 5 min before DMS treatment (data not shown). Therefore, as anticipated, the chair quadruplex, which results from the simple folding-over of a DNA G-hairpin, is kinetically favored, whereas the basket quadruplex, having a more complex folding pattern associated with higher energy intermediates and a greater number of stacked G-tetrads, is slower to form but is thermodynamically favored (compare lane 3 with lane 1 in Fig. 1B).
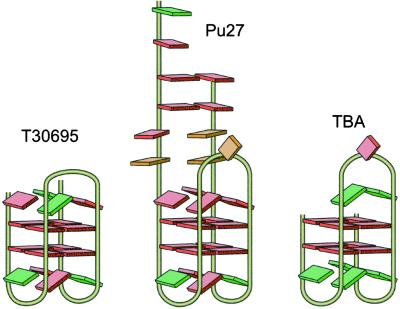
Further evidence for the facile formation of this chair conformation of the G-quadruplex structure is the selection of a thrombin-binding aptamer (TBA; ref. 23) and an HIV-integrase-binding oligonucleotide (ref. 24; typified by T30695; Scheme S1), which have very similar structures, from libraries of DNA oligomers. These two chair quadruplexes have been extensively characterized by x-ray diffraction and NMR studies (25–28). Both aptamers consist of two stacked G-tetrads with two lateral two-base loops, but whereas the bridging loop of TBA contains three bases, for T30695 there are only two. The chair structure proposed for Pu27 incorporates a TGTG lateral loop motif, identical to the loop region of T30695, which may form an ordered tetrad arrangement (28).
Scheme 1.
Structures of the aptamers T30695 (24) and TBA (23) in comparison to that proposed for the chair G-quadruplex from Pu27 (Center).
The G-Quadruplex Is the Repressor Element in the NHE III1.
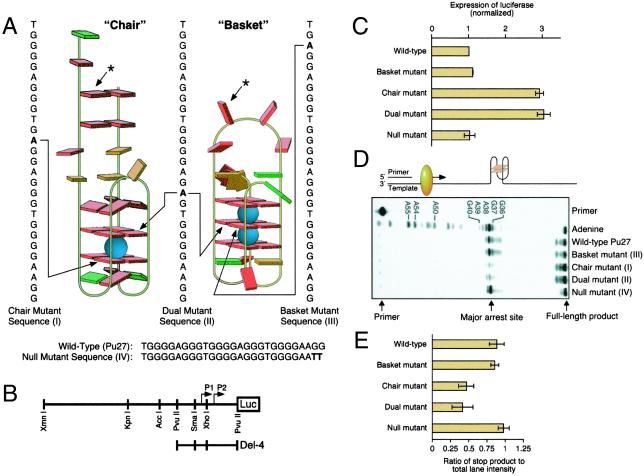
To evaluate the potential biological significance of the two intramolecular G-quadruplex structures, single- or double-base mutations of Pu27 were designed (Fig. 2A), and the various constructs were evaluated for both promoter activity in a luciferase reporter assay (Fig. 2 B and C) and for the ability to form G-quadruplex structures using a Taq polymerase stop assay (Fig. 2 D and E). In the luciferase reporter assay, single-base mutations, which destabilized a single tetrad uniquely associated with either the basket or chair G-quadruplex (Fig. 2A), had quite different effects (Fig. 2C). In comparison to the wild-type sequence, a single-base mutation of a tetrad unique to the chair form resulted in a 3-fold increase in transcriptional activity, whereas a corresponding mutation to the basket form had a negligible effect. A single-base mutation, which eliminated a G-tetrad in both chair and basket forms, also caused a 3-fold increase in transcriptional activation, whereas a null double-base mutation, which has no effect on either G-quadruplex structure, had the same activity as the wild type. These results have two important implications: (i) only the chair G-quadruplex structure is biologically relevant, and (ii) disruption of the chair G-quadruplex structure results in a significant increase in transcriptional activation (3-fold), implicating formation of this G-quadruplex structure as a repressor element to transcriptional activation of c-MYC.
Figure 2.
Effect of mutations of Pu27 on transcriptional activity and stability of the G-quadruplexes. Site-directed mutagenesis was performed as described in Methods. (A) Cartoon showing the mutations that destabilize the chair (I), basket (III), and chair and basket (II) alongside the wild-type (Pu27) and null-mutant (IV) sequences. The positions of the mutants in the chair and basket G-quadruplex regions are shown by the arrows. The arrows from the asterisks point to the corresponding positions in the non-G-quadruplex regions of the alternative basket or chair structure (the mutant bases in I–IV are shown in bold) in which mutations occur. (B) Diagram of the c-MYC-luciferase reporter vector (modified from ref. 46). The Del-4 plasmid, a gift from Bert Vogelstein, places ≈850 bp of c-myc sequence, including the two major promoters (P1 and P2), in control of a luciferase expression cassette. (C) Expression of luciferase normalized to the wild-type and various mutant sequences shown in A. Transfection and luciferase assays were performed as described in Methods. Each data point is the average of three experiments. (D) Polymerase stop assay (29) for determination of the effect of mutations on the stability of the G-quadruplex structure at 45°C in the presence of 10 mM KCl. Each data point is the average of three experiments. The cartoon above the gel illustrates the principle of the assay. The G-quadruplex structure is shown in proximity to the pause site for Taq polymerase. (E) Quantitation of the results from the polymerase stop assay shown in D.
The same wild-type and mutant sequences of the Pu27 were used in a Taq polymerase stop assay (29), which can be used to evaluate the ability of a DNA sequence to form stable G-quadruplex structures. In this assay (Fig. 2D), primer extension using Taq polymerase leads to premature stops at G-quadruplex structures. The results show that significant arrest occurs with the wild-type insert at 45°C. The primary arrest site occurs at G37, which corresponds to G7 in Pu27. The single-base mutant in the basket G-quadruplex and the null double-base mutant are equivalent to the wild type, whereas the single-base mutant in the chair and the mutant that affects the stability of both chair and basket structures lead to loss of the G-quadruplex-mediated polymerase arrest (Fig. 2E). This result is exactly complementary to that found in the promoter assay, in which only those mutations that result in destabilization of the chair quadruplex result in enhancement of c-MYC transcriptional activation.
Stabilization of the G-Quadruplex Leads to Repression of c-MYC.
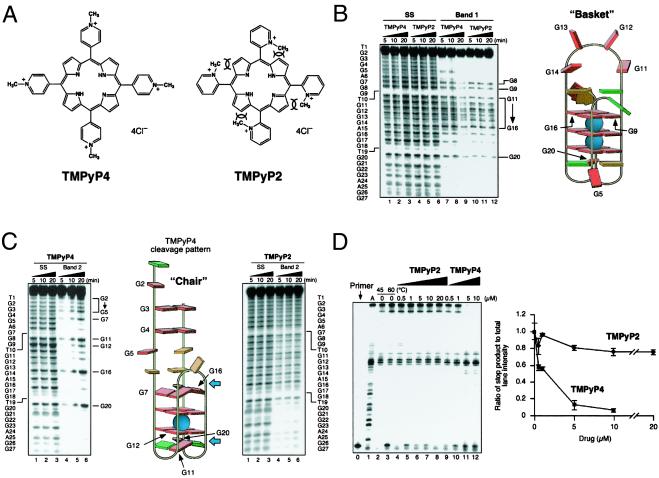
In previous studies, we have demonstrated that whereas the cationic porphyrin TMPyP4 (Fig. 3A) is able to bind to G-quadruplex structures, TMPyP2 (Fig. 3A) is much less able to do so because of restricted rotation around the meso bond, which is required for either insertion into or external stacking to the G-tetrad structure (30). Consequently, the biochemical effects, such as inhibition of telomerase (31) and inhibition of helicase-mediated unwinding of G-quadruplex structures (32), are only associated with TMPyP4. Corresponding biological effects, such as production of anaphase bridges (33) and in vivo antitumor activity (34), are also only associated with TMPyP4. TMPyP4, but not TMPyP2, has been demonstrated to lower c-MYC transcription and protein expression, as well as downstream genes, such as hTERT, ODC, and CDC25A (34). To determine whether these effects could be directly related to TMPyP4 binding to the biologically relevant G-quadruplex structure in the NHE III1, the interaction of TMPyP4 and TMPyP2 with both the chair and basket G-quadruplex structures was investigated (Fig. 3 B and C). Both TMPyP4 and TMPyP2 catalyze the oxidation of DNA upon exposure to light, which results in DNA strand breakage in proximity to the binding sites. Consequently, the specific cleavage patterns of G-quadruplex DNA by these compounds have been used to infer their binding modes and sites (32). The results of TMPyP4 and TMPyP2 photoinduced cleavage of the basket G-quadruplex are shown in Fig. 3B. An identical cleavage pattern was produced by both cationic porphyrins for the basket quadruplex. DNA damage was centered on the constituent guanines of the six-base bridging loop, inferring that both porphyrins are able to bind to this region, but in a nonspecific way that is neither intercalating nor end-stacking. Conversely, only TMPyP4 gave rise to a specific cleavage pattern when the chair G-quadruplex structure was subjected to photocleavage, which is most likely associated with partial end-stacking of TMPyP4 to the external G-tetrads, that is, at the positions shown by the blue arrows (Fig. 3C; ref. 32). If TMPyP4 lowers c-MYC transcription by interacting with a quadruplex structure, the implication from the photocleavage data (Fig. 3 B and C) is that it is the chair and not the basket G-quadruplex that is the molecular target. To determine whether the selectivity of TMPyP4 vs. TMPyP2 in lowering c-MYC is caused by specific stabilization of the chair conformation by TMPyP4, we performed the Taq polymerase stop assay at an elevated temperature (60°C) to destabilize partially the arresting quadruplex structure, but now in the presence of increasing concentrations of TMPyP2 and TMPyP4 (Fig. 3D). Whereas TMPyP2 only modestly stabilized the G-quadruplex arrest site, even at 20 μM, the addition of TMPyP4 led to almost 50% arrest at 0.5 μM and virtually total arrest at 5 μM. Consequently, we have shown that TMPyP4, but not TMPyP2, exhibits a marked binding and stabilizing effect on the chair quadruplex. This finding is in complete agreement with the differential effects of the two compounds on c-MYC expression in vitro and sensitivity in vivo to tumors that overexpress c-MYC (MX-1 and PC-3; ref. 34). Although G-quadruplex-forming units have been located in a number of oncogene promoters, including c-ABL, c-FOS, and c-MYB (15), it is only in the promoter region of the human insulin gene that indirect experimental evidence has been obtained (35). In this promoter region, the insulin-linked polymorphic region has been implicated as a factor in insulin-dependent diabetes mellitus. The insulin-linked polymorphic region is composed of a variable number of tandemly repeated variants of the sequence 5′-ACAGGGGTGTGGGG-3′. Variations in the sequence and number of repeats alters transcriptional activity, which correlates with the ability of these sequences to form intra- and intermolecular G-quadruplexes (35).
Figure 3.
Photocleavage and stabilization of the basket and chair G-quadruplexes formed from Pu27 as a result of interaction with TMPyP4 and TMPyP2. (A) Structures of TMPyP4 and TMPyP2. The overlapping arcs shown in the TMPyP2 structure demonstrate the steric clash between the 2-methylpyridyl groups and the 3-pyrrol hydrogens. (B) Photo-induced cleavage of the denatured (lanes 1–6) and isolated (lanes 7–12) basket G-quadruplex structures with TMPyP4 (lanes 1–3 and 7–9) and TMPyP2 (lanes 4–6 and 10–12). Photocleavage was carried out for the times shown at the top of the gel. Photocleavage of bands 1 and 2 (see Fig. 1B), each with TMPyP4 and TMPyP2, was conducted essentially as outlined in ref. 31, except that salmon sperm DNA was used instead of calf thymus DNA in the stop buffer. Guanines cleaved by TMPyP4 and TMPyP2 are shown to the right of the gel and are identified with arrows on the cartoon on the right. Base colors: red = guanine; green = thymine; orange = adenine. (C) As for B except that TMPyP4 and TMPyP2 were evaluated on separate gels, and the chair G-quadruplex was used in place of the basket. Blue arrows show the implied positions for TMPyP4 end-stacking to the two intact G-tetrads. (D Left) The polymerase stop assay was used to compare the stabilization of the G-quadruplex structure by TMPyP2 (lanes 4–8) and TMPyP4 (lanes 9–12) by using increasing concentrations of TMPyP2 (0.5–20 μM) and TMPyP4 (0.5–10 μM) at 60°C (10 mM KCl). “A” refers to an adenine sequencing reaction. (Right) The ratio of the full-length product to the major arrest product was plotted against concentrations. Each data point is the average of three experiments.
Down-Regulation of c-MYC by TMPyP4 Requires the NHE III1.
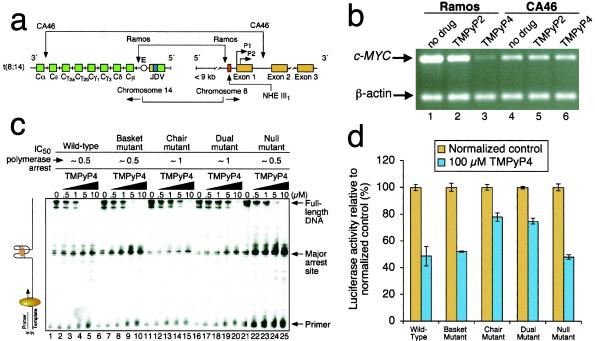
To assess directly the importance of the NHE III1 in mediating the c-MYC transcriptional inhibition by TMPyP4, two Burkitt's lymphoma cell lines with different translocation break points within the c-MYC and Ig loci were selected. Whereas the Ramos cell line retains the NHE III1 during the translocation, the CA46 cell line has lost this element, together with the P1 and P2 promoters (Fig. 4a) (4, 36). As anticipated, when the NHE III1 element was deleted, as in the CA46 cell line, TMPyP4 had no effect on c-MYC transcriptional activation, whereas in the Ramos cell line, which retains this element, TMPyP4, but not TMPyP2, lowered c-MYC expression (Fig. 4b). This result is consistent with TMPyP4 mediating its transcriptional inhibitory effect on c-MYC by interaction with the NHE III1 found upstream of the P1 promoter.
Figure 4.
Effect of cationic porphyrins on c-MYC mRNA synthesis in two different Burkitt's lymphoma cell lines and on the stability of mutant G-quadruplex structures and luciferase expression mediated by wild-type and mutant promoters. (a) Diagram of the rearrangements involved in the Ramos and CA46 Burkitt's lymphoma cell lines (modified from ref. 2). Vertical arrows indicate the breakage and rejoining points between chromosomes 14 and 8 for each translocation. (b) RT-PCR for c-MYC and β-actin in Ramos (lanes 1–3) and CA46 (lanes 4–6) cell lines after no treatment (lanes 1 and 4) and treatment with 100 μM TMPyP2 (lanes 2 and 5) and TMPyP4 (lanes 3 and 6) for 48 h (for experimental details, see ref. 33). The experiment was repeated, with comparable results. (c) Polymerase stop assay (29) for determination of the effect of TMPyP4 on the stabilization of the G-quadruplex in the wild-type (lanes 1–5) and various mutant (see Fig. 2A) sequences (lanes 6–25). TMPyP4 was added at the concentrations shown, and the experiment was carried out under the same conditions as in Fig. 2D. Above the gel are the approximate IC50 values for polymerase arrest by TMPyP4 determined in this experiment. (d) Luciferase expression assays to determine the effect of TMPyP4 on promoter activity of the wild-type and various mutants (see Fig. 2A). HeLa S3 cells were transiently transfected as described earlier. Twenty-four hours after transfection, cells were exposed to 100 μM TMPyP2 or TMPyP4 or an equivalent volume of water as a control added to the growth medium. Treatments lasted for 24 h, after which the cells were lysed, and the lysates were tested for luciferase activity, as in Fig. 2B. Experiments were performed in duplicate. Error bars represent 1 SD above and below the mean % luciferase activity. The numbers are given relative to treatment with TMPyP2, which, like TMPyP4, shows about a 20% nonspecific inhibition of luciferase activity in both these plasmids, as well as in an unrelated vector (pGL3 control).
Although the deletion experiments using the CA46 cell line were suggestive of the involvement of the NHE III1 in mediating the effect of TMPyP4 on c-MYC gene expression, an additional experiment using the mutant promoter–luciferase constructs in concert with TMPyP4 would provide greater insight into the identity of the target receptor. Before determining the effects of TMPyP4 on luciferase expression under the control of the mutant NHE III1 elements, the ability of TMPyP4 to stabilize the mutant quadruplex structures was evaluated by using the polymerase stop assay (Fig. 4c). As anticipated, TMPyP4 stabilized the arrest site of the null and basket mutants to an equivalent degree as the wild type. Although the chair and dual mutants produced only a minimal polymerase arrest in the absence of TMPyP4, significant arrest was induced at high drug concentrations, presumably because the destabilizing effect of the single-base mutations can be annulled partially by the reinforcing stabilization by TMPyP4. The IC50 values for polymerase arrest are shown above the gel in Fig. 4c. In the luciferase expression experiment (Fig. 4d), the addition of TMPyP4 produced similar results for both the null and basket mutants as with the wild-type promoter. In the case of both the chair and dual mutants, TMPyP4 produced a modest reduction in luciferase expression, in accord with the stabilization effects on the mutant quadruplexes observed in the polymerase stop assay. Moreover, if the effect of TMPyP4 had been mediated through a receptor other than that formed by the NHE III1, we would have expected equal effects on luciferase expression, regardless of the ability to form the biologically relevant chair quadruplex. This result, alongside the differential effects on CA46 and Ramos cell lines, provide compelling support that the c-MYC-lowering effect of TMPyP4 is mediated through a specific G-quadruplex in the NHE III1 of the c-MYC promoter.
Discussion
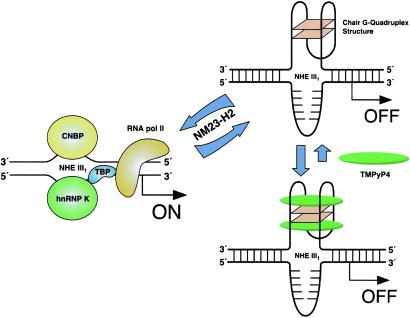
On the basis of new molecular insights gained through the experiments described in this paper, and building on previous studies of the factors affecting transcriptional activation of c-MYC (12–20, 37), a model can be proposed for how the NHE III1 controls c-MYC expression (Scheme S2). First, the G-quadruplex structure is a negative regulator of c-MYC because a single base mutation, which destabilizes the chair form, increases c-MYC expression 3-fold. Furthermore, compounds that stabilize this G-quadruplex structure have the anticipated opposite effect: decrease in c-MYC expression (34). The paranemic forms of the NHE III1—that is, the chair G-quadruplex formed by Pu27 and possibly an i-motif structure formed in the complementary strand—need to be converted to unstructured purine and pyrimidine single-stranded forms before c-MYC can be transcriptionally activated (20). The interconversion between the paranemic forms and duplex DNA is proposed to require NM23-H2 as an accessory factor (20). This hexameric c-MYC transcription factor binds to both the duplex and both paranemic forms of NHE III1 with similar stoichiometry (20), although there is some disagreement with this conclusion (38). In addition to its sequence-specific DNA-binding property, NM23-H2 catalyzes two other seemingly unrelated functions (39). The first is sequence-specific double-strand cleavage of the DNA backbone, which involves a Schiff-base intermediate between the ɛ-amino group of Lys-12 of NM23-H2 and C1′ of the ribose in DNA (39). The second is an NDP kinase activity that generates dNTPs. Significantly, both functions use the same catalytic pocket, suggesting that NM23-H2 may play a reversible role, that is, strand cleavage and repair synthesis (40). Thus, NM23-H2 would play an accessory, or transacting, role rather than be directly involved in the on/off gene switch (41). CNBP and hnRNP K, on the other hand, would play a more immediate role in transcriptional activation by binding to the Pu- and Py-rich strands of the duplex DNA directly (17–19). Indeed, hnRNP K has been shown to interact with RNA polymerase II via the TATA binding protein to activate c-MYC expression (42). In a more general sense, if paranemic forms of DNA, such as G-quadruplexes and i-motifs, which are associated with polypurine/polypyrimidine tracts, are important elements of on/off gene switches in regulatory regions of DNA (43), their inherently quite different molecular recognition properties to those associated with duplex DNA make them attractive molecular targets for the design of small molecules to selectively interfere with oncogene expression (44, 45). The formation of similar G-quadruplexes in other promoters of growth regulatory genes (unpublished results), such as PDGF-A, c-myb, and Ki-ras, suggest that this phenomenon will be more general in genes associated with growth and proliferation. The sequestration of the active form of the promoter as a G-quadruplex rather than in a nucleosome may have advantages for a rapid response required for genes involved in proliferation and may have been an ancient mechanism for controlling gene expression.
Scheme 2.
Model for the activation and repression of gene transcription involving the accessory role of NM23-H2 in interconversion of the unstructured purine and pyrimidine single-stranded DNA forms to the paranemic secondary DNA structures. Interaction of the G-quadruplex structure with TMPyP4 stabilizes the gene-off form by inhibition of conversion to the single-stranded gene-on forms (see text for further details).
Supplementary Material
Acknowledgments
We thank Dr. Haiyong Han and Mr. Mu-Yong Kim for their helpful advice and Dr. Daniel Von Hoff for his encouragement. We also thank Dr. David M. Bishop for preparing, proofreading, and editing the manuscript and for preparing the final version of the figures and schemes. This research was supported by grants from the National Institutes of Health and the Arizona Disease Control Research Commission. L.H.H. is founder and Scientific Director of Cyternex, Inc.
Abbreviation
- NHE
nuclease hypersensitivity element
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pelengaris S, Rudolph B, Littlewood T. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 2.Spencer C A, Groudine M. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 3.Marcu K B, Bossone S A, Patel A J. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 4.Facchini L M, Penn L Z. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 5.Adachi S, Obaya A J, Han Z, Ramos-Desimone N, Wyche J H, Sedivy J M. Mol Cell Biol. 2001;21:4929–4937. doi: 10.1128/MCB.21.15.4929-4937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canelles M, Delgado M D, Hyland K M, Lerga A, Richard C, Dang C V, Leon J. Oncogene. 1997;14:1315–1327. doi: 10.1038/sj.onc.1200948. [DOI] [PubMed] [Google Scholar]
- 7.Holt J T, Redner R L, Nienhuis A W. Mol Cell Biol. 1988;8:963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prochownik E V, Kukowska J, Rodger C. Mol Cell Biol. 1988;8:3683–3695. doi: 10.1128/mcb.8.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levens D, Duncan R C, Tomonaga T, Michelotti G A, Collins I, Davis-Smyth T, Zheng T, Michelotti E F. Curr Top Microbiol Immunol. 1997;224:33–46. doi: 10.1007/978-3-642-60801-8_3. [DOI] [PubMed] [Google Scholar]
- 10.Sakatsume O, Tsutsui H, Wang Y, Gao H, Tang X, Yamauchi T, Murata T, Itakura K, Yokoyama K K. J Biol Chem. 1996;271:31322–31333. doi: 10.1074/jbc.271.49.31322. [DOI] [PubMed] [Google Scholar]
- 11.Cooney M, Czernuszewicz G, Postel E H, Flint S J, Hogan M E. Science. 1988;214:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 12.Siebenlist U, Henninghausen L, Battey J, Leder P. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 13.Boles T C, Hogan M E. Biochemistry. 1987;26:367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- 14.Simonsson T, Pribylova M, Vorlickova M. Biochem Biophys Res Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 15.Simonsson T, Pecinka P, Kubista M. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji L, Arcinas M, Boxer L M. J Biol Chem. 1995;270:13392–13398. doi: 10.1074/jbc.270.22.13392. [DOI] [PubMed] [Google Scholar]
- 17.Tomonaga T, Levens D. Proc Natl Acad Sci USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bossone S A, Asselin C, Patel A J, Marcu K B. Proc Natl Acad Sci USA. 1992;89:7452–7456. doi: 10.1073/pnas.89.16.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelotti E F, Tomonaga T, Krutzsch H, Levens D. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 20.Postel E H, Berberich S J, Rooney J W, Kaetzel D M. J Bioenerg Biomembr. 2000;32:277–284. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 21.Watson J D, Crick F H. Cold Spring Harbor Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Weitzmann M N, Woodford K J, Usdin K. J Biol Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 23.Bock L C, Griffin L C, Latham J A, Vermaas E H, Toole J J. Nature (London) 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 24.Rando R F, Ojwang J, Elbaggari A, Reyes G R, Tinder R, McGrath M S, Hogan M E. J Biol Chem. 1995;270:1754–1760. doi: 10.1074/jbc.270.4.1754. [DOI] [PubMed] [Google Scholar]
- 25.Schultze P, Macaya R F, Feigon J. J Mol Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 26.Kelly J A, Feigon J, Yeates T O. J Mol Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 27.Jing N J, Gao X L, Rando R F, Hogan M E. J Biomol Struct Dyn. 1997;15:573–585. doi: 10.1080/07391102.1997.10508967. [DOI] [PubMed] [Google Scholar]
- 28.Jing N J, Hogan M E. J Biol Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 29.Han H, Salazar M, Hurley L H. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han F X, Wheelhouse R T, Hurley L H. J Am Chem Soc. 1999;121:3561–3570. [Google Scholar]
- 31.Sun D, Thompson B, Cathers B E, Salazar M, Kerwin S M, Trent J O, Jenkins T C, Neidle S, Hurley L H. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 32.Han H, Rangan A, Langley D R, Hurley L H. J Am Chem Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 33.Izbicka E, Nishioka D, Marcell V, Raymond E, Davidson K K, Lawrence R A, Wheelhouse R T, Hurley L H, Wu R S, Von Hoff D D. Anticancer Drug Des. 1999;14:355–366. [PubMed] [Google Scholar]
- 34.Grand C L, Han H, Muñoz R M, Weitman S, Von Hoff D D, Hurley L H, Bearss D J. Mol Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 35.Lew A, Rutter W J, Kennedy G C. Proc Natl Acad Sci USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonsson T, Henriksson M. Biochem Biophys Res Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
- 37.Postel E H, Berberich S J, Flint S J, Ferrone C A. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 38.Hildebrandt M, Lacombe M-L, Mesnildrey S, Véron M. Nucleic Acids Res. 1995;23:3858–3864. doi: 10.1093/nar/23.19.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postel E H, Abramczyk B A, Gursky S K, Xu Y. Biochemistry. 2002;41:6330–6337. doi: 10.1021/bi025606+. [DOI] [PubMed] [Google Scholar]
- 40.Postel E H, Abramczyk B M, Levit M N, Kyin S. Proc Natl Acad Sci USA. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma D, Xing Z, Liu B, Pedigo N G, Zimmer S G, Bai Z, Postel E H, Kaetzel D M. J Biol Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 42.Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. FEBS Lett. 1997;403:113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 43.Wells R D. J Biol Chem. 1988;263:1095–1098. [PubMed] [Google Scholar]
- 44.Hurley L H. J Med Chem. 1989;32:2027–2033. doi: 10.1021/jm00129a001. [DOI] [PubMed] [Google Scholar]
- 45.Hurley L H. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 46.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.