Abstract
Phytochromes are photochromic photoreceptors with a bilin chromophore that are found in plants and bacteria. The soil bacterium Agrobacterium tumefaciens contains two genes that code for phytochrome-homologous proteins, termed Agrobacterium phytochrome 1 and 2 (Agp1 and Agp2). To analyze its biochemical and spectral properties, Agp1 was purified from the clone of an E. coli overexpressor. The protein was assembled with the chromophores phycocyanobilin and biliverdin, which is the putative natural chromophore, to photoactive holoprotein species. Like other bacterial phytochromes, Agp1 acts as light-regulated His kinase. The biliverdin adduct of Agp1 represents a previously uncharacterized type of phytochrome photoreceptor, because photoreversion from the far-red absorbing form to the red-absorbing form is very inefficient, a feature that is combined with a rapid dark reversion. Biliverdin bound covalently to the protein; blocking experiments and site-directed mutagenesis identified a Cys at position 20 as the binding site. This particular position is outside the region where plant and some cyanobacterial phytochromes attach their chromophore and thus represents a previously uncharacterized binding site. Sequence comparisons imply that the region around Cys-20 is a ring D binding motif in phytochromes.
Keywords: bilin‖biliprotein‖photochromic‖histidine kinase
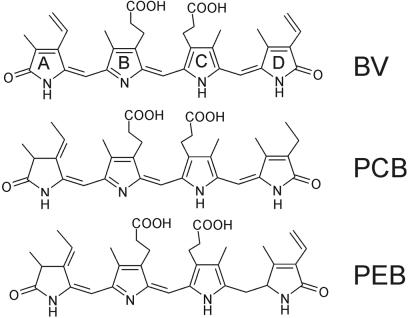
Many developmental processes in plants such as seed germination, de-etiolation, or flowering are controlled by phytochrome photoreceptors (1). The discovery of phytochromes in bacteria (2, 3) showed that these chromoproteins are of prokaryotic origin, which gave great insight into the evolution of phytochromes. Prokaryotic phytochromes offer advantages for biochemical and biophysical studies (4–6) and help to define the role of protein domains and single amino acids (7). Phytochromes carry a bilin chromophore, either phytochromobilin (8), phycocyanobilin (PCB; ref. 9, 10), or biliverdin (BV), as was recently shown for bacteriophytochrome photoreceptor (BphP) of the bacterium Deinococcus radiodurans (ref. 11; chemical structures of chromophores are given in Fig. 1). In plant and most cyanobacterial phytochromes, the chromophore is bound via its ring A ethylidene side chain to a particular Cys residue. However, proteobacteria, Deinococcus, and some cyanobacteria have leucin, valin, isoleucin, or methionine at that position. For Deinococcus BphP, it was postulated that the chromophore is covalently attached to the neighboring, highly conserved, His (3). Phytochromes are synthesized in a red-absorbing form (Pr); a second thermostable far-red absorbing form (Pfr) that absorbs in the longer wavelength region is part of the photocycle. As a result, phytochromes appear as photoreversibly photochromic pigments. For this reason phytochromes were the first plant photoreceptors to be detected and characterized (12). However, Pfr is not always infinitely stable. Some plant phytochromes revert from Pfr to Pr in darkness on a time scale of hours (13, 14). Dark-reversion has significant effects on the activity of plant phytochromes. The physiological activity of Arabidopsis phytochrome B is reduced if dark-reversion is accelerated by a particular mutation (15), and it is increased if dark-reversion is hindered by overexpressed interacting proteins (16). A comparable dark-reversion has so far not been found in bacterial phytochromes. Cph1 of Synechocystis (17) and CphA of Calothrix (18) have a stable Pfr form; reports on other bacterial orthologs are missing so far.
Figure 1.
Chemical structure of biliverdin, phycocyanobilin, and phycoerythrobilin.
Most bacterial phytochromes carry a histidine-kinase module, the first component of “two-component” systems. His-kinase activity is light-modulated; cyanobacterial phytochromes are more active in the Pr form (19–21), whereas phytochrome BphP from the proteobacterium Pseudomonas aeruginosa is more active in the Pfr form (11). In general, His kinases transphosphorylate particular response regulators (22); this mechanism also has been shown for bacterial phytochromes (11, 19, 21).
The genome of the soil bacterium Agrobacterium tumefaciens has recently been published (23, 24). It contains two genes that code for phytochrome-homologous proteins (11). Agrobacterium is well known among plant scientists because it can transform tumor-inducing genes into plants and can be used as a shuttle system for plant transformation (23). The question of what role a photoreceptor might play in an organism of such agricultural and genetic importance prompted us to begin analyzing the biochemical properties of recombinant Agrobacterium phytochrome. Biliverdin and PCB yielded products with the spectral characteristics of Pr that photoconverted to Pfr. Quite interestingly, both adducts showed Pfr-to-Pr dark-reversion, as is the case for some plant phytochromes. Studies on chromophore binding revealed a new Cys-binding site in the N terminus of the protein. This finding might have implications for the study of chromophore interaction and photoconversion of all phytochromes.
Materials and Methods
Computer Science.
Database searches were performed on National Center for Biotechnology Information (NCBI) BLASTP (http://ncbi.nlm.nih.gov); the Agrobacterium phytochrome homologues (Agp1 and Agp2) were found with Cph1 as template (17). Both Agrobacterium sequences are already mentioned in an earlier publication (11). Searches for heme oxygenase and bilin reductase genes in the Agrobacterium genome were performed by using known protein sequences as templates (25, 26). Phytochrome protein domains were identified by the SMART computer tool at the European Molecular Biology Laboratory (EMBL; http://smart.embl-heidelberg.de); the “PHY” domain was identified by the PFAM tool of the Sanger Centre (www.sanger.ac.uk/Software/Pfam/). Protein alignments were performed with CLUSTALX V.1.8 (27) with the default parameters, except in the case of “gap opening” and “gap extension”, which were set to 50 and 0.5, respectively. For secondary structure prediction, the protein sequences were analyzed with the program PHD (28, 29) at the EMBL Predict-Protein server (www.embl-heidelberg.de/predictprotein/).
Cloning of Agp1 and Mutants.
Agp1 was amplified by PCR from A. tumefaciens, strain C58, by using TaKaRa Ex Taq polymerase (Takara Shozu, Otsu, Japan). The primers were GGAATTCATTAAAGAGGAGAAATTAACTATGCAAAGAGAGCGGCTGGAG and GGGAGATCTGGCAATTTTTTCCTCTTCAACTTTC. The PCR product was digested with EcoRI and BglII and cloned into EcoRI-BglII-digested expression vector pQE12 (Qiagen, Hilden, Germany). The resulting ORF begins with the original start codon and ends with six additional His codons. The insert was sequenced and proved identical with the sequence from the database. The C20A, C279S, and C295A mutants were cloned with the QuikChange site-directed mutagenesis kit (Stratagene), according to the instructions of the manufacturer. The mutations also were confirmed by sequencing.
Chromophores, Protein Expression, Extraction, and Purification.
Biliverdin was purchased from Frontier Scientific (Carnforth, U.K.). Phycocyanobilin and PEB were extracted from Spirulina geitlerie and Porphyridium cruentum, respectively, and purified by high-pressure liquid chromatography, as described (20). Bilin stock solutions were prepared in methanol, and concentrations of stock solutions were estimated spectroscopically (20). Agp1 was expressed as a polyhistidine-tagged apoprotein in E. coli as described for the phytochrome Cph1 from Synechocystis PCC 6803 (20). After affinity chromatography, Agp1 apoprotein (Apo-Agp1) was purified further by size-exclusion chromatography (SEC) on a 3 × 100 cm Sephacryl S-300 (Amersham Pharmacia, Freiburg, Germany) column in 50 mM Tris⋅Cl/300 mM NaCl/5 mM EDTA, pH 7.8. To yield holoprotein saturated with chromophore, purified Apo-Agp1 was mixed with 2-fold molar excess of bilin. The holoprotein then was separated from the free bilin by Sephacryl SEC. Analytical SEC, chromophore assembly, SDS/PAGE, Zn2+-fluorescence, and iodoacetamide blocking were performed as described for Cph1 (20). Blocking with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) was done with a 2 mM final concentration for 20 min. Before chromophore addition, the protein was separated from free DTNB by using NAP 10 (Amersham Pharmacia) desalting columns.
Chromophore–Protein Interaction.
For testing chromophore–protein interaction in the native state, 10 μM Agp1 in TE buffer (50 mM Tris/5 mM EDTA, pH 7.8) and 4 μM bilin from a 1 mM methanol stock solution were mixed and incubated for 5 min in darkness at room temperature. An aliquot of the protein then was separated from free bilin by using NAP-5 desalting-columns (Amersham Pharmacia). The columns were equilibrated in TE buffer; thereafter, 0.5 ml of the protein–chromophore mix was applied and allowed to enter the gel. The solution that eluted after application of 1 ml buffer was collected. This fraction contained protein and was depleted of low-molecular compounds such as free bilin. Spectra were recorded from 250 to 900 nm with a Uvikon 931 photometer (Kontron/Biotek, Milano) and compared with those of the control (the sample before the separation). The protein peak at 280 nm was used to normalize both samples; the chromophore peak in the range above 550 nm was used for estimating the ratio of chromophore that eluted together with the protein. To test for covalent chromophore–protein interaction, the chromophore–protein mix was incubated with 1% SDS (final concentration) before the separation to dissociate noncovalently bound chromophore. The desalting columns were run in the presence of 1% SDS; the procedure was otherwise as described above. In control runs without the protein, 7 ± 2% of the bilin appeared in the front fraction. This background value and slight variations between different runs made exact quantifications difficult if the fraction of bound chromophore was very low. However, comparisons between the different bilins were possible, and the results were confirmed by repeated experiments. For data presentation, the background value was subtracted in those cases where the apparent value was low. The measurements performed without SDS were not critical in this respect, because free chromophore bound tightly to the matrix, and it was thus quantitatively separated from the protein.
Photoconversion and Extinction Coefficients.
Spectra were recorded with a Uvikon 931 photometer at 18°C. For recording photoconversion kinetics and dark-reversion, the intensity of the measuring light was reduced so that apparently no photoconversion was induced within 20 min. For actinic irradiation, light from a tungsten-halogen projector was passed through DAL-interference filters (Schott, Mainz, Germany) of 637 ± 12 nm, 681 ± 12 nm, or 755 ± 10 nm and a side window of the photometer onto the measuring cuvette. The Pfr content was observed at 720 nm for PCB-Cph1 and at 755 nm for Agp1 holoprotein (Holo-Agp1); the absorbance after saturating red irradiation was set to unity. The relative Pfr content ϕ at photoequilibrium was estimated according to Butler et al. (30); the quantum efficiency Φ of photoconversion was calculated from the initial rate of photoconversion according to Mancinelli (14). An extinction coefficient ɛ at λmax,R of 85 mM−1 cm−1 was used for PCB-Cph1-Pr (20); 90 mM−1 cm−1 was used for both PCB-Agp1-Pr and biliverdin adduct of Agp1 (BV-Agp1)-Pr. The latter values were obtained from limited assembly as for Cph1 (20). To yield the initial rate of the Pfr-to-Pr photoconversion, the initial rate of dark-reversion was subtracted from the initial conversion rate under actinic light.
Phosphorylation.
Autophosphorylation was performed as described for Cph1 (20). Before the addition of [γ-32P]ATP, PCB-Cph1 and PCB-Agp1 were either irradiated with saturating far-red or red light from light-emitting diodes of 730 and 660 nm, respectively, and subsequently incubated in darkness at 20°C. Radioisotope imaging was performed with a fluorescent image analyzer FLA 2000 (Fuji). The signal intensity was quantified by integrated analyses software. In each autoradiogram, the intensity of Agp1-Pr was set to 100.
Results and Discussion
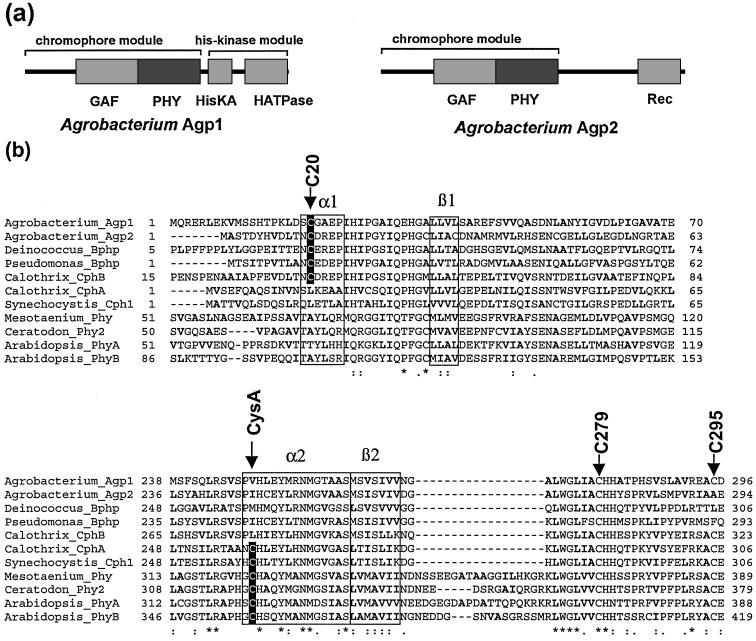
The genome of A. tumefaciens (23, 24) contains two genes that code for phytochrome-homologous proteins with the NCBI GI nos. 15889282 and 15889444, termed Agp1 (Agrobacterium phytochrome) and Agp2, respectively. SMART and PFAM protein analysis tools showed that Agp1 has a domain arrangement typical for bacterial phytochromes such as Cph1 from the cyanobacterium Synechocystis PCC 6803 (17) or BphP from Deinococcus radiodurans; it contains the chromophore-binding GAF-domain, a so-called PHY-domain, and a His-kinase module (Fig. 2a). Agp2 is unusual in so far as it also contains the GAF- and PHY-domains but no His-kinase module. Instead, Agp2 carries a response regulator at its C terminus (Fig. 2a). In plant and some cyanobacterial phytochromes, the C-3 side chain of ring A of the chromophore is covalently bound to a particular Cys side chain. These phytochromes are termed CysA- (Cys ring A) phytochromes here. At the homologous position (see arrow “CysA” in the alignment of Fig. 2b), Agp1 and Agp2 carry a valin and isoleucin instead. The Cys also is lacking some cyanobacterial phytochromes (21) and all known proteobacterial orthologs (11, 31).
Figure 2.
(a) Domain structure of Agrobacterium Agp1 and Agp2 according to SMART and PFAM. HisKA, histidine-kinase-substrate domain; HATPase, histidin-kinase-ATPase domain; Rec, response-regulator-domain. (b) Protein alignment (CLUSTALX) around the N terminus of Agp1 (Upper) and the core of the GAF-domain (Lower) with Agp2, further bacterial phytochromes from Pseudomonas, Deinococcus (3), Calothrix (18), and Synechocystis (2) and plant phytochromes from Mesotaenium (35), Ceratodon (36), and Arabidopsis (37). The upper five proteins are denominated as non-CysA, and the lower six are denominated as CysA-phytochromes in the text. Positions of the Agp1 cysteines and the chromophore-binding cysteine of CysA phytochromes are indicated by arrows. The frames labeled with α1, α2, β1, and β2 indicate regions where α-helix or β-sheet were predicted by the program PHD (see text). Hydrophobic amino acids with aliphatic side chains (A, I, L, V) are printed in bold letters. The characters in the bottom line denote highly conserved amino acids, according to CLUSTALX.
In Vitro Assembly and Chromophore Binding of Agp1.
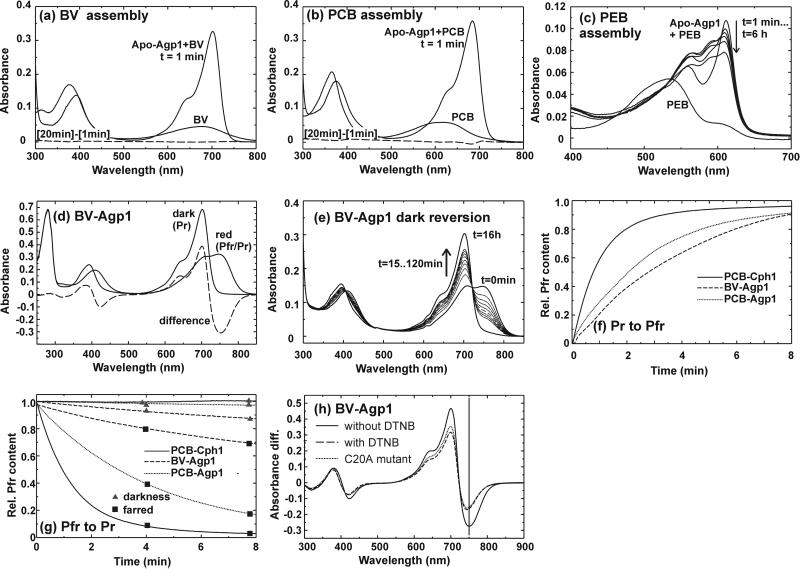
The Agp1 gene was amplified by PCR, cloned into the pQE12 vector, and expressed as a polyhistidine-tagged protein in E. coli; the yields were ≈150 mg of soluble protein per liter of culture. Agp1 was readily purified by Ni-NTA affinity chromatography. A preparative SEC step that was included in the purification procedure removed aggregated protein quantitatively. On analytical SEC, purified Apo-Agp1 and Holo-Agp1 eluted with an apparent molecular size of the dimer (data not shown). We tested the assembly of Agp1 with BV, PCB, and phycoerythrobilin (PEB) by taking spectra before and after mixing chromophore with purified Apo-Agp1. With BV, we observed a rapid spectral change within 1 min after mixing (Fig. 3a); no further changes were noted thereafter (see [20 min]−[1 min] difference spectrum of Fig. 3a). With PCB, comparable rapid spectral changes were observed (Fig. 3b). These were followed by subtle alterations, as indicated by the difference spectrum in Fig. 3b. With PEB, a spectrally unusual species appeared immediately after mixing, with two peaks at 560 and 611 nm. Absorbance around 600 nm decreased during subsequent hours, whereas an increase and band broadening were observed in the spectral region around 550 nm (Fig. 3c). The assembly pattern of all three bilins contrasts strongly with the results we obtained for Cph1 (20). Nevertheless, spectra of the BV- and PCB-Agp1 adducts indicate the formation of photoactive holophytochrome species. Compared with PCB adducts of CysA-phytochromes such as Synechocystis Cph1, PCB-Agp1 (Fig. 3b) has a red-shifted absorbance maximum of 685 nm (see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org) such as the non-CysA phytochromes Deinococcus BphP (3) and Calothrix CphB (18). Compared with PCB-Agp1, BV-Agp1 is further red-shifted to 701 nm (Fig. 3a; Table 3), in agreement with the further extended system of conjugated double bonds (Fig. 1). BLASTP searches identified a putative heme-oxygenase (25) gene in the Agrobacterium genome (GI accession no. 15157689), whereas no homologues of bilin-reductases, such as PcyA (26) were found. Heme oxygenase catalyses the formation of BV, and bilin reductases convert BV into PCB, PEB, or phytochromobilin. Therefore, we assume that BV is the natural chromophore of Agp1. This assumption is supported by spectral measurements on extracts from Agrobacterium (C. Schellenberg, N.M., and T.L., unpublished data).
Figure 3.
(a–c) Spectral changes during Agp1 assembly with BV, PCB, and PEB. The spectrum of the free bilin is shown in comparison with spectra after mixing with Agp1. In the case of BV and PCB, negligible or subtle spectral changes occurred after t = 1 min. These changes are shown by the dashed lines, which represent difference spectra between t = 20 min and t = 1 min. (d) Spectra of BV-Agp1 after dark incubation, red irradiation, and the difference spectrum between both (dashed line). Before the spectra were recorded, assembled BV-Agp1 was purified by size-exclusion chromatography. (e) Dark-reversion of BV-Agp1 after saturating red irradiation. Spectra were taken in 15-min intervals until t = 120 min; a final spectrum was taken at t = 16 h. (f) Photoconversion kinetics from Pr-to-Pfr by red irradiation (637 nm, 10 μmol m−2 s−1). Pfr was recorded at 720 nm for Cph1 and at 755 for BV-Agp1 and PCB-Agp1; the value of 1 is equivalent to Pfr at photoequilibrium. (g) Dark-reversion (▴) and photoinduced reversion from Pfr-to-Pr (■). In the latter case, samples were irradiated with 730 nm/15 μmol m−2 s−1 Cph1 or 755 nm/12 μmol m−2 s−1 BV-Agp1 and PCB-Agp1. (h) Pr − Pfr difference spectrum of BV-Agp1 after DTNB blocking (dashed line) and blocking of the C20A mutant in comparison with the nonpretreated control (solid line).
Both the BV and PCB adduct converted into the far-red absorbing form Pfr upon irradiation with red light. No photoconversion was observed in the case of PEB-Agp1, which correlates with PEB adducts of other phytochromes (32). The Pfr form of PCB-Agp1 also was red-shifted compared with PCB adducts of CysA-phytochromes, and BV-Agp1-Pfr was again further red shifted (Table 3).
Photoconversion Rates and Dark-Reversion.
During trials to revert Pfr back to Pr by light, we noted that this process is very inefficient for BV-Agp1, as it required very long irradiation times. We also found that the Pfr form of either adduct is not stable in darkness and undergoes dark-reversion to Pr (see Fig. 3e for BV-Agp1). Examples of photoconversion and dark-reversion traces are shown for both adducts and Cph1 in Fig. 3f and g. Based on such measurements, we calculated the relative Pfr content ϕ at photoequilibrium under red light, the initial rate of dark-reversion, and the photoconversion quantum efficiencies for both directions. These values and the extinction coefficients ɛ are summarized in Table 1. A rather high ϕ value of 0.96 was calculated for BV-Agp1 under saturating red light. With this value, an almost pure Pfr spectrum should be obtained, which contrasts, however, with the spectra we obtained under such conditions (e.g., Fig. 3d). Assuming that the shape of the Agp1 Pfr-spectrum resembles other phytochromes such as Cph1 (17), we estimated that ϕ is ≈0.88. This number was obtained empirically by calculating the Pfr spectrum for different ϕ; spectra that were calculated with higher or lower ϕ values looked untypical. The discrepancy shows that in addition to the observed dark-reversion, a rapid dark-reversion has to be considered. A similar, but less exaggerated, discrepancy was found for the PCB adduct (Table 1).
Table 1.
Extinction coefficients (ɛ), relative Pfr content (ϕ) at photoequilibrium with 647 nm actinic light, photoconversion quantum efficiencies (Φ), and initial rates of dark-reversion of PCB-Cph1, BV-Agp1, and PCB-Agp1
| Chromophore | ɛ at λmax,R | ϕ | Φ(Pr-Pfr) | Φ(Pfr-Pr) | Dark-reversion |
|---|---|---|---|---|---|
| PCB-Cph1 | 85 mM−1 cm−1 | 0.68 | 0.15 | 0.12 | 0 |
| BV-Agp1 | 90 mM−1 cm−1 | 0.96/0.88* | 0.078 | 0.004 | 3.4 10−4 s−1 |
| PCB-Agp1 | 90 mM−1 cm−1 | 0.84/0.77* | 0.13 | 0.048 | 5.7 10−5 s−1 |
Calculated and empirically obtained values, respectively (see text).
Dark-reversion has long been known for plant phytochromes (13, 14, 16) but is missing in cyanobacterial PCB-Cph1 and PCB-CphA (18). It is likely that the ancestors of phytochromes were photoreceptors with only one thermostable form, and that the lifetime of the designated Pfr intermediate was later increased. Thus, Agp1 might represent one step in the evolution of photochromicity. The low quantum yield of the Pfr-to-Pr photoreaction of BV-Agp1 points in the same direction. However, it is also possible that during Agrobacterium evolution, Agp1 was optimized as a photoreceptor with these specific properties to fulfil specific requirements. A comparative analysis of several bacterial phytochromes will show whether or not the properties of Agp1 are unique.
Chromophore Binding.
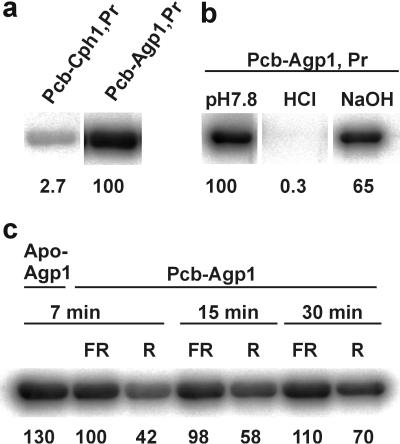
In general, the phytochrome chromophore is covalently bound. However, covalent binding seems not to be required for full spectral activity, as shown for CphB, for example (18). Covalent associations of biliproteins are usually analyzed by SDS/PAGE and Zn2+-induced fluorescence (33). However, it is difficult to quantify the amount of covalently bound chromophore in this assay. Therefore, we used an alternative method that utilizes prepacked desalting columns (see Materials and Methods). This method allows testing for chromophore binding both in the native and the SDS-denatured state. In the native state, BV seemed tightly bound to the Agp1 protein, and the binding of PCB was only slightly weaker. Phycoerythrobilin was rather weakly bound; it was largely removed from the protein during the separation (Table 2). After SDS-denaturation, a different pattern was obtained: whereas biliverdin was still quantitatively bound (Table 2), PCB was largely removed from the protein, and only 5% seemed to be covalently attached. The fraction of bound PEB was 8% and, thus, greater than that of PCB (Table 2). The chemical structure of the chromophores (Fig. 1) leads us to propose that covalent attachment occurs via a ring D side group, because PCB differs from the other two bilins here. This difference can explain the inefficient covalent binding of PCB. If chromophores attached via ring A, where BV differs from PCB/PEB, the covalent binding of PCB should be greater than that of PEB because of the much stronger noncovalent binding of the former.
Table 2.
Binding of BV, PCB, and PEB to Agp1 in the native state (“without SDS”) and after SDS-denaturation
| Chromophore | Without SDS | With SDS | After DTNB with SDS | Mutant C20A with SDS |
|---|---|---|---|---|
| BV | 99 ± 1% | 98 ± 1% | 0 ± 2%* | 1 ± 3%* |
| PCB | 95 ± 3% | 5 ± 2%* | n.d. | n.d. |
| PEB | 36 ± 2% | 8 ± 2%* | n.d. | n.d. |
After mixing bilin and apo-Agp1, free bilins were separated from the protein using pre-packed desalting columns, and the fraction that remained attached to the protein was determined by UV-VIS spectroscopy. Mean values of three separate chromophore/protein mixes from one apoprotein sample ± S.E.; n.d., not determined. When the value of an SDS-treated sample was low (*), the background value of the free chromophore without protein was subtracted (see Materials and Methods).
With the SDS/PAGE–Zn2+-fluorescence approach, we tested whether chromophore attachment can be inhibited by thiol-reactive agents, as has been shown for Cph1 (20). Biliverdin attachment was blocked by iodoacetamide and DTNB (data not shown), compounds that react with Cys side chains (34). The DTNB block was confirmed by the desalting column method (Table 2). The BV adduct that was formed after the block was spectrally similar to the control and was fully photoreversible, but it had a reduced extinction coefficient, blue-shifted Pfr maximum, and a reduced ratio of Pfr:Pr absorption (Fig. 3h and Table 3 for DTNB). Spectra of PCB adducts were not affected by these chemical treatments (Table 3). The blocking experiments show that BV is covalently bound to a Cys side chain. Agp1 has three cysteines at positions 20, 279, and 295 (see Fig. 2b). We mutated these cysteines to either Ser or Ala. The spectra of BV adducts of the C279S and C295A mutants were indistinguishable from wild-type BV-Agp1, and both retained their ability for covalent BV attachment (data not shown). However, spectra of the BV adduct of the C20A mutant were very similar to DTNB-blocked BV-Agp1 (Fig. 3h, Table 3). The mutant lost its ability for covalent BV binding, as was shown by the desalting column method (Table 2). These results show that in wild-type Agp1, BV is covalently bound to C20.
This finding was rather surprising, because this amino acid lies far outside the GAF domain, which extends from amino acid 142 to 320 in Agp1. The GAF domain is thought to form the chromophore-binding pocket, because it contains the chromophore-binding Cys of the CysA phytochromes. Our findings show that the region around C20 of Agp1 is also part of the chromophore pocket. A comparison with the sequences of other phytochromes indicated the importance of this region. Within the 150 N-terminal amino acids of Agp1, the sequences from position 18 to 50 of Agp1 had the highest degree of homology (see alignment in Fig. 1a, which shows amino acids 1 to 70 of Agp1). Furthermore, C20 was found to be conserved in all non-CysA phytochromes with known spectral activity, whereas none of the CysA phytochromes had a Cys at the homologous position (Fig. 2b). This finding suggests that other non-CysA phytochromes also might use the C20-homologous Cys as a BV-attachment site. For Deinococcus BphP, it was shown by mass spectrometry that the chromophore is attached to a His residue (3), contrary to our assumption regarding Cys coupling. However, the BphP results were obtained for the PCB chromophore, because at the time of these analyses, it was not clear that the natural chromophore is BV.
We submitted all sequences shown in Fig. 2b to the Predict-Protein server at EMBL for secondary structure prediction. Striking similarities between the regions around Agp1-C20 and the CysA cysteine were found by the PHD algorithm (28, 29). The respective cysteines or the orthologous amino acids were found to be located within a predicted α-helix (α1 and α2 in Fig. 2b). In addition, 16 to 18 amino acids further toward the C terminus, a β-sheet structure (β1 and β2 in Fig. 2b) was predicted. Both β regions are also characterized by their high content of hydrophobic amino acids. Based on these similarities and the two possible sites of covalent attachment, we propose that the A ring of the chromophore interacts with amino acids of the α2/β2 subdomain, and that the D ring interacts with amino acids of the α1/β1 subdomain. This model can now be tested by site-directed mutagenesis, chemical and biophysical approaches.
His-Kinase.
Light-regulated His autophosphorylation has been demonstrated for several bacterial phytochromes. Sequence homology implies that Agp1 also functions as a His kinase. In our assays, Agp1 phosphorylated rather efficiently. Compared with Cph1, it incorporated about 30 times more phosphate in the Pr form (Fig. 4). His-phosphate is stable in alkaline but labile in acid solutions (22). Accordingly, phosphate was washed away with HCl but not with NaOH (Fig. 4). Different incubation times also were tested; this demonstrated that phosphorylation of PCB-Agp1-Pr was already saturated after 7 min or earlier. With Agp1, phosphorylation was stronger in the Pr form (Fig. 4); thus, it matches with the cyanobacterial orthologs (19–21) and not with Pseudomonas BphP (11). Quite interestingly, Apo-Agp1 incorporated more phosphate than Holo-Agp1. Chromophore incorporation seems to inhibit phosphorylation, and this inhibition is strengthened upon photoconversion to Pfr. His kinases always act as homodimers: the ATPase of one subunit phosphorylates the substrate site of the other. The inhibition upon chromophore incorporation and photoconversion may easily be explained by the enlarged distance between both subunits.
Figure 4.
Autogradiogramms after [γ-32P]ATP labeling, SDS/PAGE, and blotting. Relative intensities (%) are indicated under the bands. (a) Comparison between PCB-Cph1 and PCB-Agp1 (each 5 μg), 30-min [γ-32P]ATP incubation as Pr. (b) PCB-Agp1-Pr (5 μg) blot-strips treated for 2 h with 1 M HCl, 2 M NaOH, or pH 7.8 buffer as control. (c) Comparison between Apo-Agp1 and PCB-Agp1 (5 μg each). The latter was preirradiated with 730 nm (Pr) or 645 nm light-emitting diodes (Pfr/Pr) and incubated for different times, as indicated.
Supplementary Material
Acknowledgments
We thank Sabine Buchert and Sabine Artelt for technical help, Karola Rück-Braun for helpful discussions, Dieter Weichart and Regine Hengge Aronis for allowing us to use the PhosphorImager, and Timo Thoms for correcting the English. This work was supported by the Deutsche Forschungsgemeinschaft (Sfb 498).
Abbreviations
- PCB
phycocyanobilin
- BV
biliverdin
- BphP
bacteriophytochrome photoreceptor
- Pr
red-absorbing form of phytochrome
- Pfr
far-red absorbing form of phytochrome
- Agp1
-2, Agrobacterium phytochrome 1, -2
- EMBL
European Molecular Biology Laboratory
- Apo-Agp1
Agp1 apoprotein
- SEC
size-exclusion chromatography
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- PEB
phycoerythrobilin
- BV-Agp1
BV adduct of Agp1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. 2nd Ed. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- 2.Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Börner T. Nature (London) 1997;386:663–663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 3.Davis S J, Vener A V, Vierstra R D. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 4.van Thor J J, Borucki B, Crielaard W, Otto H, Lamparter T, Hughes J, Hellingwerf K J, Heyn M P. Biochemistry. 2001;40:11460–11471. doi: 10.1021/bi002651d. [DOI] [PubMed] [Google Scholar]
- 5.Remberg A, Lindner I, Lamparter T, Hughes J, Kneip K, Hildebrandt P, Braslavsky S E, Gärtner W, Schaffner K. Biochemistry. 1997;36:13389–13395. doi: 10.1021/bi971563z. [DOI] [PubMed] [Google Scholar]
- 6.Heyne K, Herbst J, Stehlik D, Esteban B, Lamparter T, Hughes J, Diller R. Biophys J. 2002;82:1004–1016. doi: 10.1016/S0006-3495(02)75460-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S H, Lagarias J C. Biochemistry. 2000;39:13487–13495. doi: 10.1021/bi001123z. [DOI] [PubMed] [Google Scholar]
- 8.Rüdiger W, Thümmler F. In: Photomorphogenesis in Plants. 2nd Ed. Kendrick R E, Kronenberg G H M, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 51–69. [Google Scholar]
- 9.Wu S H, McDowell M T, Lagarias J C. J Biol Chem. 1997;272:25700–25705. doi: 10.1074/jbc.272.41.25700. [DOI] [PubMed] [Google Scholar]
- 10.Hübschmann T, Börner T, Hartmann E, Lamparter T. Eur J Biochem. 2001;268:2055–2063. doi: 10.1046/j.1432-1327.2001.02083.x. [DOI] [PubMed] [Google Scholar]
- 11.Bhoo S H, Davis S J, Walker J, Karniol B, Vierstra R D. Nature (London) 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 12.Butler W L, Norris K H, Siegelman H W, Hendricks S B. Proc Natl Acad Sci USA. 1959;45:1703–1708. doi: 10.1073/pnas.45.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler W L, Lane H C, Siegelman H W. Plant Physiol. 1963;38:514–519. doi: 10.1104/pp.38.5.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancinelli A. In: Photomorphogenesis in Plants. 2nd Ed. Kendrick R E, Kronenberg G H M, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 211–269. [Google Scholar]
- 15.Elich T D, Chory J. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schäfer E, Harter K. Science. 2001;294:1108–1111. doi: 10.1126/science.1065022. [DOI] [PubMed] [Google Scholar]
- 17.Lamparter T, Mittmann F, Gärtner W, Börner T, Hartmann E, Hughes J. Proc Natl Acad Sci USA. 1997;94:11792–11797. doi: 10.1073/pnas.94.22.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorissen H J, Quest B, Remberg A, Coursin T, Braslavsky S E, Schaffner K, Tandeau de Marsac N, Gärtner W. Eur J Biochem. 2002;269:2662–2671. doi: 10.1046/j.1432-1033.2002.02928.x. [DOI] [PubMed] [Google Scholar]
- 19.Yeh K C, Wu S H, Murphy J T, Lagarias J C. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 20.Lamparter T, Esteban B, Hughes J. Eur J Biochem. 2001;268:4720–4730. doi: 10.1046/j.1432-1327.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 21.Hübschmann T, Jorissen H J, Börner T, Gärtner W, Tandeau de Marsac N. Eur J Biochem. 2001;268:3383–3389. doi: 10.1046/j.1432-1327.2001.02229.x. [DOI] [PubMed] [Google Scholar]
- 22.Stock A M, Robinson V L, Goudreau P N. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 23.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman B S, Cao Y, Askenazi M, Halling C, et al. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 24.Wood D W, Setubal J C, Kaul R, Monks D E, Kitajima J P, Okura V K, Zhou Y, Chen L, Wood G E, Almeida N F, Jr, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 25.Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman H M. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankenberg N, Mukougawa K, Kohchi T, Lagarias J C. Plant Cell. 2001;13:965–978. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rost B, Sander C, Schneider R. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 29.Rost B, Sander C. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler W L, Hendricks S B, Siegelman H W. Photochem Photobiol. 1964;3:521–528. [Google Scholar]
- 31.Jiang Z Y, Swem L R, Rushing B G, Devanathan S, Tollin G, Bauer C E. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Murphy J T, Lagarias J C. Biochemistry. 1995;34:7923–7930. doi: 10.1021/bi00024a017. [DOI] [PubMed] [Google Scholar]
- 33.Berkelman T R, Lagarias J C. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 34.Lottspeich F, Zorbas H. Bioanalytik. Berlin: Springer; 1998. [Google Scholar]
- 35.Lagarias D M, Wu S H, Lagarias J C. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 36.Hughes J, Lamparter T, Mittmann F. Plant Physiol. 1996;112:446–446. [Google Scholar]
- 37.Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.