Abstract
Phosphatidylinositol 3-kinase products play a central role in the regulation of several intracellular pathways via adaptor proteins that share the ability to bind to 3′-phosphoinositides with high affinity and specificity. JFC1 is a C2 domain-containing protein involved in cellular trafficking that has been shown to bind 3′-phosphoinositides in vitro. In this work, we demonstrate that the C2A domain of JFC1 is the module responsible for its binding to the plasma membrane via 3′-phosphoinositides in vivo. We show that the C2A domain of JFC1 is the only domain present in this protein that localizes to the plasma membrane in living cells. Moreover, the C2A domain of JFC1 binds 3′-phosphoinositides in vitro with similar specificity as that described for full-length JFC1, suggesting that the domain mediates the specific membrane localization of the full-length protein. Furthermore, the C2A domain of JFC1 colocalized with the pleckstrin homology domain of Akt in vivo, and both the JFC1 C2A domain and the full-length JFC1 dissociated from the membrane in the presence of PI 3-kinase specific inhibitors. We also show that the association of the C2A domain to the membrane is modulated by calcium. From these results we analyze possible mechanisms for the role of JFC1 in cellular trafficking.
Phosphoinositides are specialized phospholipids known to localize to the inner leaflet of the plasma membrane, vesicles, and the nuclear envelope and participate in several signaling pathways (1). One of these phosphoinositides, phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2], plays a central role in the regulation of several intracellular pathways, from actin cytoskeleton modulation and vesicle trafficking (2) to modulation of the transcription factor Tubby protein (3). Furthermore, PtdIns(4,5)P2 is a precursor for the second messenger phosphatidylinositol (3,4,5) trisphosphate [PtdIns(3,4,5)P3] via the phosphoinositide 3-kinases (PI 3-kinases; ref. 4). PI 3-kinases and their products 3′-phosphorylated phosphoinositides (3′-phosphoinositides) are also involved in many crucial cell functions including vesicle trafficking (1, 2), cytoskeleton modulation, and cell survival (5). It is now clear that most of the functions regulated by phosphoinositides are mediated by adaptor proteins that share the ability to bind to PtdIns(4,5)P2 and PI 3-kinase products with high affinity and specificity. These effectors are characterized by the presence of specialized phosphoinositide-binding domains. These domains include the pleckstrin homology (PH) domain (6), the FYVE finger (7), and the recently discovered PHOX homology (PX) domain (8). Furthermore, C2 domains have been shown to bind to phospholipids or phosphoinositides in the presence or absence of calcium (9). Proteins that contain C2 domains could generally be categorized as signal transduction mediators or membrane traffic effectors (10). The most extensively studied proteins included in the last group are the synaptotagmins (Syts). Syts are a group of transmembrane proteins that contain tandem C2 domains and have been found to be involved in neurotransmission. Syt I is considered to be the main Ca2+ sensor in synaptic vesicle exocytosis. Interestingly, Syt I has been described to bind to PtdIns(3,4,5)P3 and PtdIns(4,5)P2 via its C2B domain in a calcium-dependent manner, with the specificity of the binding switching at calcium concentrations similar to those usually found at the synapse in response to a nerve impulse (9). In the same work, this phenomenon has been proposed to be involved in the regulation of exocytosis. Therefore, it is not unlikely that different C2-containing proteins could have similar roles in nonneural tissues.
JFC1 is a tandem C2 domain-containing ATPase (11) that was recently identified in our laboratory by the use of a yeast two-hybrid system with the C-terminal half of the leukocyte NADPH oxidase cytosolic factor p67PHOX as bait (12). The C2A domain of JFC1 is highly homologous to the C2B domain present in Syt I and possesses a close match to the polyphosphate-inositol-binding site K(K/R)KTXXK(K/R) found in several members of this family (12). Interestingly, JFC1 binds to phosphoinositides in vitro, showing preference for PtdIns(3,4,5)P3 over phosphatidylinositol (3,4) bisphosphate [PtdIns(3,4)P2], phosphatidylinositol (3) phosphate [PtdIns(3)P] and PtdIns(4,5)P2 (12). Although the C2A domain of JFC1 is the main candidate thought to mediate the phosphoinositide binding, it is still not clear which domain of JFC1 is involved in this role. Interestingly, in a recent study, JFC1 was found to specifically bind Rab27A in vivo (13), a GTPase known to regulate melanosome transport (14) and secretion in cytotoxic T lymphocytes (15). As mentioned above, PI 3-kinase products have been involved largely in membrane trafficking. Therefore, it is likely that the phosphoinositide-binding capacity of JFC1 is an essential requirement for its accurate function in vivo. In this work, we characterize the phosphoinositide-binding capacity of JFC1 in living cells and identify its phosphoinositide-binding domain.
Experimental Procedures
Cell Cultures and Transfections.
The fibroblast embryonic cell line NIH 3T3 (ATCC), was maintained in DMEM supplemented with 10% (vol/vol) FCS (GIBCO/BRL) at 37°C in 5% CO2/air. The human prostate cell line LNCaP (ATCC) was cultured as described (16). Transfections were performed on coverglass slides using FuGene 6 (Roche Molecular Biochemicals) as described by the manufacturer. The transfected DNA comprised 1 μg of the appropriate enhanced green fluorescent protein (EGFP-C2, CLONTECH) fusion constructs described under Cloning. Where specified, the DNA mixture also included 0.5 μg of an expression vector for the activated PI 3-kinase p110 under the control of the CMV promoter (Upstate) or 0.5 μg of the appropriate red fluorescent protein (pDsRed-N1, CLONTECH) fusion constructs also described under Cloning. After transfection (48 h), cells were washed twice and transferred to Hank's balanced salt solution (GIBCO/BRL) at 30°C. Living cells were analyzed by use of a krypton/argon laser-equipped Bio-Rad MRC1024 laser scanning confocal microscope at The Scripps Research Institute. Where appropriate, actin filaments were stained by using phalloidin-rhodamine (Molecular Probes) and fixed as described by the manufacturer. Where indicated, cells were treated with the PI 3-kinase inhibitor LY294002 (Calbiochem; 16 μM, changed every hour) or carrier (DMSO) 2 to 4 h before the analysis of the sample.
Cloning.
The various steps in the cloning of the constructs were performed by standard techniques and all constructs were verified by sequencing (17). The various jfc1 DNA fragments cloned into the EcoRI/SalI sites of the pEGFP-C2 polylinker were generated by PCR by using jfc1 cDNA as template. The oligonucleotides including EcoRI or SalI sites (underlined) used were GGGGAATTCATGCCCCAGAGGGGCCACCCATCGCAA (oligonucleotide 1, coordinates: nucleotides +1 to +27) and GGGGTCGACCCCGTCCTGGGGGCCAGGTTGGTTCT (oligonucleotide 2, coordinates: nucleotides +1686 to +1662) for full-length JFC1 (EGFP-JFC1); oligonucleotide 1 and GGGGTCGACCCGCCCCAGTCCCACGTGTCCAGGGG (oligonucleotide 3, coordinates: nucleotides +1131 to +1108) for amino terminal JFC1 including the C2A domain (EGFP-NH2-C2A); GGGGAATTCAGCCTGTCAGGCGACGCGGAGGCGGTG (oligonucleotide 4, coordinates: nucleotides +766 to +792 and oligonucleotide 3, for the C2A domain (EGFP-C2A); oligonucleotide 4 and oligonucleotide 2, for carboxyl terminal including both C2 domains (EGFP-C2A-C2B) and GGGGAATTCTCCCTCAAGTACGTCCCCGCCGGCTCC (coordinates: nucleotides +1213 to +1239) and oligonucleotide 2, for carboxyl terminal including the C2B domain (EGFP-C2B).
The p40PHOX-PX domain (amino acids 1–147), the Akt1-PH domain (amino acids 1–131) and the phospholipase C-δ (PLCδ) PH domain (amino acids 1–174), cloned into the EcoRI/SalI sites of pDsRed1-N1, were generated by PCR by using p40PHOX cDNA as a template (gift from Lucia Lopes, Universidad de Sao Paulo, Sao Paulo, Brazil), Akt1 cDNA (Upstate), or Human Universal QUICK Clone cDNA (CLONTECH), respectively. For p40PHOX-PX, the oligonucleotides (EcoRI or SalI sites underlined) used were GAATTCCCATGGCTGTGGCCCAGCAGCT (coordinates: nucleotides +1 to +20) and GTCGACCCATGGCCTGGGGCACCTGCTC (coordinates: nucleotides +441 to +421). For DsRed-Akt-PH, the oligonucleotides used were GAATTCCCATGAGCGACGTGGCTATTGTGAAG (coordinates: nucleotides +1 to +24) and GTCGACCCAGCCCCTGAGTTGTCACTGGGTGA (coordinates: nucleotides +393 to +370). For DsRed-PLCδ-PH, the oligonucleotides used were GGGGAATTCATGGACTCGGGCCGGGACTTCCTG (coordinates: nucleotides +1 to +24) and CCCGTCGACTGGATGTTGAGCTCCTTCAG (coordinates: nucleotides +524 to +505).
Nitrocellulose Phospholipids-Binding Assays.
The binding of glutathione S-transferase (GST)-C2A or GST-Rac (negative control) to several phospholipids and phosphoinositides was evaluated by a dot-blot assay, as described (18) with minor modifications. The GST fusion proteins were affinity-purified by the use of glutathione-agarose (Amersham Pharmacia), as described (11, 19). Phospholipids were resuspended at 2 mg/ml according to the manufacturer's specifications and were spotted (4 μg) onto nitrocellulose sheets. GST-C2A or the negative control in Tris-buffered saline containing 0.5% BSA was then used to probe the phosphoinositide-containing nitrocellulose for 30 min at room temperature. The bound protein was visualized by immuno-blot analysis by using an antibody raised against GST (Upstate Biotechnology, Lake Placid, NY). Phosphatidylinositol, phosphatidylcholine, phosphatidylserine, and phosphatidylinositol 4-phosphate [PtdIns(4)P] were obtained from Sigma; PtdIns(4,5)P2 was from Calbiochem or Matreya (Pleasant Gap, PA); and PtdIns(3)P, PtdIns(3,4)P2, and PtdIns(3,4,5)P3 were from Matreya.
Results
JFC1 Associates with the Plasma Membrane via Its C2A Domain in Living Cells.
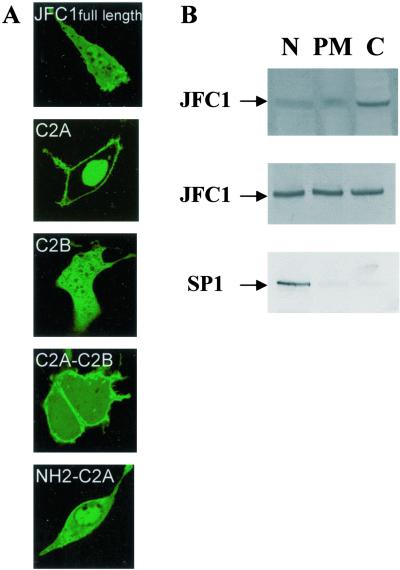
As we reported, JFC1 is peripherally associated with the plasma membrane (12). To determine which domain of JFC1 is responsible for this association, we generated various chimeras consisting of a fusion between the EGFP protein and various JFC1 domains. When expressed in NIH 3T3 cells, the C2A was the only domain of JFC1 to localize exclusively to the plasma membrane in vivo (Fig. 1A). The presence of non-C2A JFC1 domains (either amino- or carboxyl-terminal extensions) in the fusion proteins increased the presence of the proteins in the cytosol (Fig. 1A). Similar results have been observed when the constructs were expressed in 3T3-L1 and LNCaP cells (not shown). When expressed in NIH 3T3 cells, the EGFP-C2A also could be detected in the nucleus, but the physiological relevance of this later observation is still unknown. This data strongly suggests that the C2A domain is responsible for the membrane-binding capacity of JFC1. The C2B domain was the other obvious candidate to be responsible for the membrane association of JFC1. However, the fusion containing the C2B but not the C2A domain of JFC1 localized mainly in the cytosol (Fig. 1A), indicating that the C2B domain is not involved in the membrane localization, at least under unstimulated conditions. The EGFP-JFC1 full-length fusion was codistributed in the membrane, the cytosol and the nucleus in the NIH 3T3 cell line (Fig. 1A) and LNCaP cells (see Fig. 5). Correspondingly, endogenous or overexpressed JFC1 was equally distributed in the soluble fraction as well as in the membrane and nuclear fraction of LNCaP or JFC1-transfected HeLa S3 cells, respectively, when detected by immunoblotting (Fig. 1B). Interestingly, JFC1 has been shown to be present in the particulate fraction but not in the cytosol in human neutrophils (12). This difference is probably attributable to a basal level of activation of those cells as a result of the isolation process or to the relatively low level of expression of JFC1 in neutrophils.
Figure 1.
The peripheral binding of JFC1 to the plasma membrane is mediated by the C2A domain. (A) JFC1 domains were expressed as EGFP-fusion proteins in NIH 3T3 cells. Cells were analyzed by confocal laser scanning microscopy. Similar data were obtained in at least five different experiments. (B) Western blot analysis of the distribution of JFC1 after cell fractionation. Cell fractionation was performed exactly as described (3). (Top) Endogenous JFC1 in LNCaP cells. (Middle) Overexpressed JFC1 in HeLa S3. (Bottom) Control experiment using an antibody (Upstate) raised against the Sp1 transcription factor with nuclear localization (ref. 3; HeLa S3 cells). Similar results were obtained for control experiments by using LNCaP cells. N, nuclear fraction; PM, plasma membrane; and C, cytosol.
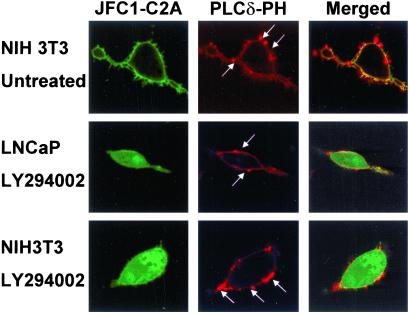
Figure 5.
The C2A domain of JFC1 but not the PtdIns(4,5)P2-binding PH domain of PLCδ dissociates from the membrane after treatment with LY294002. NIH 3T3 cells (Top and Bottom) or PTEN-deficient LNCaP cells (Middle) were cotransfected with the EGFP-C2A (JFC1-C2A) and the DsRed-PLCδ-PH (PLCδ-PH). The cells were either untreated (Top) or treated with the PI 3-kinase inhibitor LY294002, as described in Fig. 4. Samples were analyzed by confocal laser scanning microscopy. The arrows indicate the characteristic membrane blebs (25) that are enriched in PLCδ-PH but not in JFC1-C2A. (Left, Center, and Right, respectively) Distribution of EGFP-C2A (JFC1-C2A), DsRed-PLCδ-PH (PLCδ-PH), and the merged image, where yellow indicates colocalization. The result is representative of at least five different experiments.
The C2A Domain of JFC1 Binds to 3′-Phosphoinositides in Vitro.
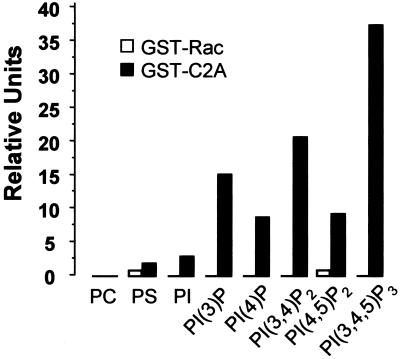
In a previous work, we demonstrated that JFC1 binds to 3′-phosphoinositides in vitro (12). Because the C2A domain of JFC1 seemed to be responsible for its membrane-binding capacity in vivo, we evaluated whether this domain binds phosphoinositides in vitro, similarly to that observed for the full-length JFC1. Fig. 2 shows that GST-C2A bound to 3′ phosphoinositides in a dot-blot assay. The C2A domain of JFC1 bound mainly to PtdIns(3,4,5)P3 and, to a lesser extent, to PtdIns(3,4)P2 and PtdIns(3)P, indicating that the fully phosphorylated inositol head group enhances the binding capacity of this domain. JFC1-C2A binding to PtdIns(4,5)P2 was only 24.9% of that observed for PtdIns(3,4,5)P3 and 44.8% of that for PtdIns(3,4)P2, suggesting that phosphorylation in position 3′ plays a central role in the specificity of the binding.
Figure 2.
The C2A domain of JFC1 binds preferentially to 3′ phosphoinositides in vitro. The binding of GST-C2A or GST-Rac (negative control) to several phospholipids spotted on nitrocellulose was evaluated by a dot-blot assay. The bound protein was detected by using an antibody raised against GST. PC, phosphatidylcholine; PS, phosphatidylserine; PI, phosphatidylinositol; PI(3)P, PtdIns(3)P; PI(4)P, PtdIns(4)P; PI(3,4)P2, PtdIns(3,4)P2; PI(4,5)P2, PtdIns(4,5)P2; and PI(3,4,5)P3, PtdIns(3,4,5)P3. Images were collected by a Fluor-S MultiImager (Bio-Rad) and quantified by QUANTITY ONE software (Bio-Rad). The result is representative of three different experiments.
JFC1-C2A and Full-Length JFC1 Bind to PI 3-Kinase Products in Living Cells.
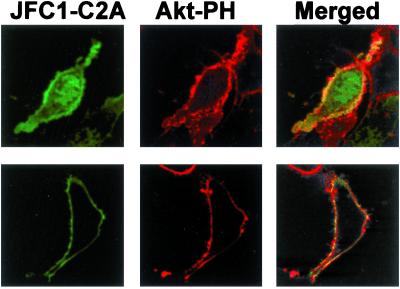
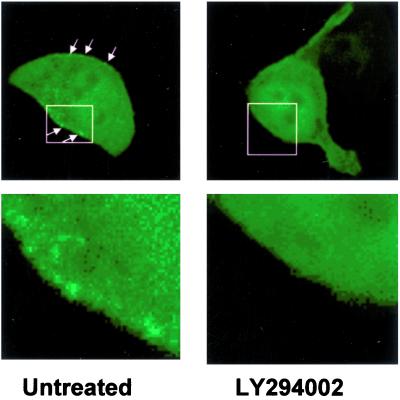
The PH domain of Akt is known to bind specifically to the PI-3 kinase products PtdIns(3,4)P2 and PtdIns(3,4,5)P3 (6, 20). To evaluate whether the C2A domain of JFC1 shares the plasma membrane docking point with the Akt PH domain, we cotransfected expression vectors for the JFC1 C2A domain and the Akt PH domains as fusion proteins with the EGFP and the DsRed, respectively. When NIH 3T3 cells were cotransfected with the Akt PH domain, the C2A domain of JFC1 and an active form of the PI 3-kinase, both phosphoinositide-binding domains colocalized at the plasma membrane level (Fig. 3 Upper). We then used a similar approach to evaluate the distribution of JFC1-C2A in LNCaP cells. This prostate carcinoma cell line has elevated PI 3-kinase activity and 3′-phosphoinositides levels because of a deficiency of the lipid phosphatase and tensin homolog (PTEN) (21, 22). Noticeably, when the Akt PH domain and the JFC1-C2A are expressed in LNCaP cells, they again colocalized at the plasma membrane (Fig. 3 Lower). Furthermore, when the EGFP-JFC1 full-length fusion was expressed in LNCaP cells, a speckled distribution was observed at the plasma membrane (Fig. 4). This pattern vanished when the LNCaP cells were treated with the specific PI 3-kinase inhibitor LY294002, suggesting that JFC1 localizes to areas of the plasma membrane that are enriched in 3′-phosphoinositides.
Figure 3.
The C2A domain of JFC1 colocalizes with the PH domain of Akt in PTEN-deficient LNCaP cells and in NIH 3T3 cells cotransfected with PI 3-kinase. (Upper) NIH 3T3 cells were transfected with EGFP-C2A, DsRed-Akt-PH, and an active PI 3-kinase expression vector. (Lower) PTEN-deficient LNCaP cells were transfected with the same constructs as in Upper, except that the PI 3-kinase vector was omitted. The cells were analyzed by confocal microscopy. (Left, Center, and Right, respectively) Distribution of EGFP-C2A (JFC1-C2A), DsRed-Akt-PH (Akt-PH), and the merged image, where yellow indicates colocalization. Similar data were obtained in at least five other cells.
Figure 4.
The membrane distribution of the full-length JFC1 in PTEN-deficient cells is abolished by LY294002. PTEN-deficient LNCaP cells were transfected with the full-length EGFP-JFC1 construct. Where indicated, cells were treated with the PI 3-kinase inhibitor LY294002 (16 μM). Cells were analyzed by confocal microscopy. The arrows show a speckled distribution of full-length JFC1 in the plasma membrane of untreated cells. (Lower) Result of a magnified image. Similar data were obtained in five different experiments.
Does the JFC1 C2A Domain Bind to PtdIns(4,5)P2 in Living Cells?
The cellular levels of PtdIns(4,5)P2 are approximately 50 times larger than that of PtdIns(3)P and 1,000 times larger than that of PtdIns(3,4,5)P3 in unstimulated cells (23). Thus, although JFC1-C2A principally binds to 3′-phosphoinositides in vitro, the low-level binding of PtdIns(4,5)P2 in vitro could be significant, and thus, the possibility that PtdIns(4,5)P2 could act as a docking point for JFC1-C2A in vivo should not be ruled out before examination. To check this hypothesis, we used the PH domain of PLCδ, which is known to specifically bind to PtdIns(4,5)P2 (24). When DsRed-PLCδ-PH was coexpressed with EGFP-C2A in untreated NIH 3T3 cells, localization of both fusion proteins at the plasma membrane level was observed (Fig. 5 Top). However, it has been established that the overexpression of PtdIns(4,5)P2 binding proteins decreases the local PtdIns(4,5)P2 concentration (25). Therefore, the number of binding interactions between PtdIns(4,5)P2 and cytoskeletal proteins diminishes and so does the adhesion energy, leading to the formation of membrane blebs (25). In this work, the overexpression of PLCδ-PH but not JFC1-C2A induced the formation of these structures (Fig. 5). Moreover, PLCδ-PH but not JFC1-C2A localized to the membrane blebs. These data establish a difference in the distribution of these domains, although it could be as a result of differences in their affinity for PtdIns(4,5)P2 rather than by the recognition of a different docking point by JFC1-C2A. To resolve this issue, we evaluated the effect of PI 3-kinase inhibitors on the distribution of both domains. As discussed above, JFC1-C2A localizes at the plasma membrane in both NIH 3T3 and LNCaP cells (Figs. 1 and 3, respectively). When NIH 3T3 or LNCaP cells were treated with the PI 3-kinase inhibitor LY294002 (Fig. 5 Middle and Bottom) after being cotransfected with JFC1-C2A and PLCδ-PH, only the former domain was dissociated from the plasma membrane. These results confirm that the JFC1 C2A domain mainly binds to a PI 3-kinase inhibitor-sensitive basal pool of lipids, presumably PtdIns(3,4,5)P3 or PtdIns(3,4)P2, even in unstimulated cells. Nevertheless, even after the continuous treatment with LY294002, a small fraction of JFC1-C2A remains in the plasma membrane (Fig. 5), so it is possible that this domain, when highly expressed, may bind to some extent to a PI 3-kinase inhibitor-insensitive pool of phosphoinositides.
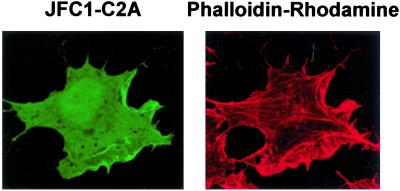
It is well documented that overexpression of PtdIns(4,5)P2 binding proteins can disrupt actin polymerization (25), and it has been demonstrated that the overexpression of the PtdIns(4,5)P2-sequestering protein PLCδ-PH dramatically reduces the adhesion energy and disrupts polymerized actin in NIH 3T3 cells (25). In an attempt to use a different approach to demonstrate that PtdIns(4,5)P2 is not the major membrane-binding site for JFC1-C2A, we determined whether the overexpression of JFC1-C2A induces the disruption of the actin cytoskeleton in NIH 3T3 cells. As shown in Fig. 6, overexpression of JFC1-C2A did not significantly affect actin polymerization, suggesting again that PtdIns(4,5)P2 is not the main docking point of JFC1-C2A in the plasma membrane.
Figure 6.
The overexpression of the C2A domain of JFC1 does not disrupt actin polymerization. NIH 3T3 cells were transfected with the EGFP-C2A construct, and polymerized actin was visualized by using rhodamine-labeled phalloidin. (Left) Fluorescence image of the C2A domain of JFC1. (Right) Fluorescence image of phalloidin-rhodamine. The less apparent membrane staining of JFC1-C2A observed in this figure was caused by the fixation process. This appearance has been described for other membrane-binding proteins (25) and does not affect the validation of the result.
The Membrane Localization of JFC1-C2A Is Modulated by Calcium.
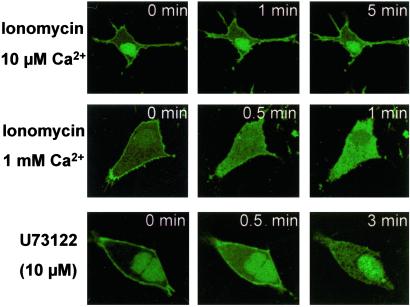
It is well established that the capability of several proteins to bind to phosphoinositides is affected by calcium (9). In this work, the effect of calcium on the ability of the JFC1 C2A domain to localize to the plasma membrane has been evaluated in NIH 3T3 cells. The C2A domain of JFC1 was dissociated from the plasma membrane after calcium influx induced by ionomycin (Fig. 7). Interestingly, the effect was differentially manifested proportionally to the extra-cellular calcium concentration used in the assay (Fig. 7), showing a low effect at 10 μM Ca2+ and total dissociation at 1 mM Ca2+. In a recent work (26), the effect of a calcium influx trigger by ionomycin on the binding of GAP1IP4BP to PtdIns(4,5)P2 was evaluated. In that study, it was suggested that a Ca2+-mediated PLC activation was responsible for the release of the protein from its docking point, which is supported by the fact that the PLC inhibitor U73122 prevented the effect. Nevertheless, a similar approach failed to show the release of the PtdIns(4,5)P2-binding protein Tubby from the plasma membrane (3). Importantly, in this work, the PLC inhibitor U73122 did not prevent the effect elicited by Ca2+/ionomycin (not shown). Therefore, in this case, the calcium-mediated membrane dissociation of JFC1-C2A does not seem to be mediated by the activation of a PLC as demonstrated for GAP1IP4BP (26). Moreover, the inhibitor U73122 was found to mimic the effect triggered by Ca2+/ionomycin (Fig. 7), which correlates with the described capacity of U73122 to induce the release of Ca2+ from intracellular deposits (27). Altogether, these data suggest that the C2A domain of JFC1 may be dissociated from the membrane by calcium-mediated electrostatic interference rather that by a Ca2+-mediated PLC activation. This suggestion is also supported by the fact that, although 10 μM calcium has been demonstrated to elicit maximum PLC activation (28), only slight C2A membrane dissociation was observed in this work at this calcium concentration. Moreover, neither the phosphoinositide-PLC inhibitor ET-18-OCH3, the calmodulin-dependent kinase II inhibitor AIP-myristoylated, or the calpain inhibitors III and V (Calbiochem) were able to prevent the effect elicited by Ca2+ on the dissociation of JFC1-C2A from the plasma membrane (not shown).
Figure 7.
The C2A domain of JFC1 is released from the membrane in response to an increase in intracellular calcium. NIH 3T3 cells were transfected with the EGFP-C2A construct and analyzed 48 h after transfection. The cells were stimulated with ionomycin at various Ca2+ concentrations (Top and Middle) at room temperature. (Bottom) The effect of U73122 in the absence of ionomycin or extracellular calcium. Images were collected before (0 min) and after the addition of the indicated stimuli. Similar data were obtained in six different experiments.
Discussion
In this work, we present strong evidence that the C2A domain of JFC1 is responsible for the binding of JFC1 to the plasma membrane via 3′-phosphoinositides. This observation is supported by three different findings. First, the C2A domain of JFC1 is the only domain present in this protein that localizes to the plasma membrane and minimally to the cytosol. Moreover, the solubility of JFC1 fusion increased with the inclusion of other domains (either amino- or carboxyl-terminal extensions). Secondly, in this work, the C2A domain of JFC1 binds 3′-phosphoinositides in vitro with specificity similar to that described for JFC1 (12), indicating that this domain is sufficient for the specific membrane localization of the full-length protein. Finally, the JFC1 C2A domain colocalized with the PtdIns(3,4,5)P3-binding domain, Akt-PH, and both, the JFC1-C2A and the full-length JFC1, dissociated from the membrane in the presence of PI 3-kinase-specific inhibitors.
In vitro binding analysis performed in this work indicates that, similar to that described for the full-length JFC1 (12), the JFC1 C2A domain binds preferentially to 3′-phosphoinositides over PtdIns(4,5)P2. However, the concentration of PI 3-kinase products is known to be relatively low, especially in unstimulated cells (23). Therefore, it is considered that to bind to PtdIns(3,4,5)P3 exclusively, such a domain must exhibit an affinity for PtdIns(3,4,5)P3 at least one order of magnitude larger than for PtdIns(4,5)P2 (24). Not surprisingly, although more than 100 PH domain-containing proteins have been described, only two domains have been found to specifically bind to PtdIns(4,5)P2 or PtdIns(3,4,5)P3 in an exclusive manner (6). These are the PH domain of PLCδ and the PH domain of the general receptor for phosphoinositides-1 (GRP1), respectively. In terms of phosphoinositide recognition, promiscuity is not restricted to PH domains. The C2B domain of Syt I has been shown to bind to PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in vitro (9). In that case, the specific binding to more than one pool of phosphoinositides was considered to have direct physiological implications. It has been proposed that Syt I is a bimodal calcium sensor, switching its affinity for PtdIns(4,5)P2 and PtdIns(3,4,5)P3 accordingly with the intracellular calcium concentration and, thus, regulating neurotransmitter release (9). The binding of the Syt I C2B domain to PtdIns(3,4,5)P3 represents the priming state of exocytosis and, in response to an increase in the intracellular calcium, the Syt I-C2B would switch to the PtdIns(4,5)P2 pool, characterizing the fusion-competent conformation (9). Because of the similarities observed between the JFC1 C2A domain and the Syt I C2B domain (12), it could be speculated that JFC1 plays a similar role. However, from the data presented here, it becomes apparent that JFC1 binds to PtdIns(3,4,5)P3 but not to PtdIns(4,5)P2 in vivo. This reasoning is supported by several observations. In the presence of the specific PI 3-kinase inhibitor LY294002 (Fig. 5), the C2A domain of JFC1 is dissociated from the plasma membrane under circumstances that the PtdIns(4,5)P2-binding domain PLCδ-PH remains bound to the membrane. Interestingly, we observed similar results in unstimulated NIH 3T3 cells, suggesting that JFC1-C2A might recognize basal levels of 3′-phosphoinositides. Noticeably, there is increasing evidence that basal levels of PI 3-kinase activity are crucial in some regulatory pathways. For instance, the activation of Ras by mitogenic concentrations of EGF has been found to depend on basal, rather than stimulated, PI 3-kinase activity (29). In addition, the lack of PtdIns(4,5)P2 binding by JFC1-C2A is also supported by its inability to disrupt actin polymerization significantly or to induce the formation of membrane blebs when overexpressed in NIH 3T3 cells, as demonstrated for other PtdIns(4,5)P2-binding proteins (25). Finally, as discussed above, the Syt I C2B domain binds PtdIns(4,5)P2 preferentially over PtdIns(3,4,5)P3 only at high calcium concentrations (9). Conversely, we show in this work that JFC1-C2A dissociates from the membrane in the presence of calcium (Fig. 7), indicating that in contrast to that observed for Syt I-C2B, JFC1-C2A does not have high affinity for PtdIns(4,5)P2 even in the presence of Ca2+. This discrepancy could be attributed to the fact that although Syt I-C2B and JFC1-C2A are highly homologous, they differ in residues that have been shown to be involved in the recognition of phosphoinositol groups (12).
Initially, we isolated and cloned JFC1 in our laboratory as a protein that associates with the NADPH oxidase cytosolic factor p67PHOX. In the original work, we suggested that because of the presence of two C2 domains in tandem, its capacity to bind to phosphoinositides in vitro and the expression of its mRNA in tissues with secretory functions, JFC1 could be involved in cellular trafficking (12). Noticeably, a specific interaction between JFC1 and Rab27A has been demonstrated recently by Seabra and coworkers (13). Moreover, similar results have been described for a mouse homologous for JFC1 by a different group (30). Because it is well established that the Rab family of proteins function in membrane trafficking by determining the specificity of vesicle transport in eukaryotic cells (31), the findings described above confirm our previous suggestions. In particular, Rab27A has been implicated in the mechanism of melanosome movement on cytoskeletal tracks mediated by its interaction with the unconventional motor protein myosin Va and the effector melanophilin (32). Interestingly, mutations in Rab27A or myosin Va lead to the autosomal recessive disease known as Griscelli syndrome (33), which is characterized by partial albinism originated by deficient melanosome transport (14) and immune deficiency caused by abnormal regulated-secretion in cytotoxic T lymphocytes (CTL; ref. 15). In the same way, mutations in melanophilin cause the melanosome transport defects observed in leaden mice (34). Similarly to JFC1, melanophilin presents homology to the Rab3a-binding domain of Rabphilin-3 (34) and binds specifically to Rab27A via this domain. Interestingly, based on the observation that Rab27A mutations but not melanophilin mutations lead to CTL granule exocytosis deficiency, it has been suggested that Rab27A associates with different effectors in nonmelanocytic cells (35). As proposed (13), JFC1 is an obvious candidate to play this role. Importantly, PI 3-kinase products have been largely involved in regulated secretory mechanisms by taking part in the segregation of Rab proteins and their effectors into different cell compartments (36). In this work, we demonstrate that JFC1-C2A binds preferentially to PtdIns(3,4)P2 and PtdIns(3,4,5)P3, two phosphoinositides that localize to the plasma membrane (1). On the other hand, JFC1-C2A did not colocalize with the PtdIns (3)P binding domain of p40PHOX-PX that localizes in early endosomes (not shown). Because JFC1 seems to be a specific effector of Rab27A, we propose that JFC1 could act as a key molecule to position the Rab-containing vesicle into a specific docking point on the plasma membrane by binding to Rab27A by its amino-terminal domain and to PtdIns(3,4,5)P3 via its C2A domain. This proposal correlates with the suggestion that the defect in secretion in CTL's lytic granules in the Rab27A-deficient ashen mice is caused by the granule's inability to move toward the docking point at the plasma membrane (15) and with the demonstration that granule exocytosis in CTLs is mediated by PI 3-kinase activation (37). Moreover, the mobilization of intracellular calcium takes place downstream from PI 3-kinase activation during exocytosis in CTLs (37). Therefore, from the data presented here, it is possible that JFC1-C2A plays a dual role during exocytosis. First, docking the vesicle to a PtdIns(3,4,5)P3-enriched membrane microenvironment and then dissociating the exocytosis complex from the plasma membrane after the calcium increase and vesicle fusion.
Acknowledgments
We thank Dr. M. Wood for his technical assistance. This work was supported in part by U.S. Public Health Service Grants AI44434, AI24227, and AI28479.
Abbreviations
- PtdIns(3,4)P2
phosphatidylinositol 3,4-bisphosphate
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4,5)P3
phosphatidylinositol 3,4,5-trisphosphate
- PtdIns(3)P
phosphatidylinositol (3) phosphate
- PH
pleckstrin homology
- PX
phox homology
- Syt
synaptotagmin
- EGFP
enhanced green fluorescent protein
- GST
glutathione S-transferase
- PTEN
phosphatase and tensin homolog
- CTL
cytotoxic T lymphocytes
- PI 3-kinase
phosphoinositide 3-kinase
References
- 1.Stephens L, McGregor A, Hawkins P. In: Biology of Phosphoinositides. Cockcroft S, editor. Oxford: Oxford Univ. Press; 2000. pp. 32–108. [Google Scholar]
- 2.Martin T F. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- 3.Santagata S, Boggon T J, Baird C L, Gomez C A, Zhao J, Shan W S, Myszka D G, Shapiro L. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Leevers S J, Panayotou G, Waterfield M D. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 5.Roymans D, Slegers H. Eur J Biochem. 2001;268:487–498. doi: 10.1046/j.1432-1327.2001.01936.x. [DOI] [PubMed] [Google Scholar]
- 6.Kavran J M, Klein D E, Lee A, Falasca M, Isakoff S J, Skolnik E Y, Lemmon M A. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 7.Gaullier J M, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. Nature (London) 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 8.Cheever M L, Sato T K, de Beer T, Kutateladze T G, Emr S D, Overduin M. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 9.Schiavo G, Gu Q M, Prestwich G D, Sollner T H, Rothman J E. Proc Natl Acad Sci USA. 1996;93:13327–13332. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizo J, Sudhof T C. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 11.Catz S D, Johnson J L, Babior B M. Proc Natl Acad Sci USA. 2001;98:11230–11235. doi: 10.1073/pnas.191369598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAdara Berkowitz J K, Catz S D, Johnson J L, Ruedi J M, Thon V, Babior B M. J Biol Chem. 2001;276:18855–18862. doi: 10.1074/jbc.M011167200. [DOI] [PubMed] [Google Scholar]
- 13.Strom M, Hume A N, Tarafder A K, Barkagianni E, Seabra M C. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 14.Bahadoran P, Aberdam E, Mantoux F, Busca R, Bille K, Yalman N, Saint-Basile G, Casaroli-Marano R, Ortonne J P, Ballotti R. J Cell Biol. 2001;152:843–850. doi: 10.1083/jcb.152.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stinchcombe J C, Barral D C, Mules E H, Booth S, Hume A N, Machesky L M, Seabra M C, Griffiths G M. J Cell Biol. 2001;152:825–834. doi: 10.1083/jcb.152.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catz S D, Johnson J L. Oncogene. 2001;20:7342–7351. doi: 10.1038/sj.onc.1204926. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Klein D E, Lee A, Frank D W, Marks M S, Lemmon M A. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 19.Lopes L R, Hoyal C R, Knaus U G, Babior B M. J Biol Chem. 1999;274:15533–15537. doi: 10.1074/jbc.274.22.15533. [DOI] [PubMed] [Google Scholar]
- 20.Frech M, Andjelkovic M, Ingley E, Reddy K K, Falck J R, Hemmings B A. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Senechal K, Neshat M S, Whang Y E, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Cheville J C, Pan Y, Roche P C, Schmidt L J, Tindall D J. J Biol Chem. 2001;276:38830–38836. doi: 10.1074/jbc.M103632200. [DOI] [PubMed] [Google Scholar]
- 23.Stephens L R, Jackson T R, Hawkins P T. Biochim Biophys Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- 24.Czech M P. Cell. 2000;100:603–606. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- 25.Raucher D, Stauffer T, Chen W, Shen K, Guo S, York J D, Sheetz M P, Meyer T. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 26.Cozier G E, Lockyer P J, Reynolds J S, Kupzig S, Bottomley J R, Millard T H, Banting G, Cullen P J. J Biol Chem. 2000;275:28261–28268. doi: 10.1074/jbc.M000469200. [DOI] [PubMed] [Google Scholar]
- 27.Jan C R, Ho C M, Wu S N, Tseng C J. Life Sci. 1998;63:895–908. doi: 10.1016/s0024-3205(98)00346-4. [DOI] [PubMed] [Google Scholar]
- 28.Allen V, Swigart P, Cheung R, Cockcroft S, Katan M. Biochem J. 1997;327:545–552. doi: 10.1042/bj3270545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wennstrom S, Downward J. Mol Cell Biol. 1999;19:4279–4288. doi: 10.1128/mcb.19.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda T S, Fukuda M, Ariga H, Mikoshiba K. J Biol Chem. 2002;277:9212–9218. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 31.Zerial M, McBride H. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 32.Hammer J A, III, Wu X S. Curr Opin Cell Biol. 2002;14:69–75. doi: 10.1016/s0955-0674(01)00296-4. [DOI] [PubMed] [Google Scholar]
- 33.Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffraat N, Bianchi D, Fischer A, Le Deist F, et al. Nat Genet. 2000;25:173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 34.Matesic L E, Yip R, Reuss A E, Swing D A, O'Sullivan T N, Fletcher C F, Copeland N G, Jenkins N A. Proc Natl Acad Sci USA. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hume A N, Collinson L M, Hopkins C R, Strom M, Barral D C, Bossi G, Griffiths G M, Seabra M C. Traffic. 2002;3:193–202. doi: 10.1034/j.1600-0854.2002.030305.x. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer S R. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 37.Fuller C L, Ravichandran K S, Braciale V L. J Immunol. 1999;162:6337–6340. [PubMed] [Google Scholar]