Abstract
Structural genomics is emerging as a principal approach to define protein structure–function relationships. To apply this approach on a genomic scale, novel methods and technologies must be developed to determine large numbers of structures. We describe the design and implementation of a high-throughput structural genomics pipeline and its application to the proteome of the thermophilic bacterium Thermotoga maritima. By using this pipeline, we successfully cloned and attempted expression of 1,376 of the predicted 1,877 genes (73%) and have identified crystallization conditions for 432 proteins, comprising 23% of the T. maritima proteome. Representative structures from TM0423 glycerol dehydrogenase and TM0449 thymidylate synthase-complementing protein are presented as examples of final outputs from the pipeline.
Genomic information provides a tremendous opportunity to study gene function. Ultimately, the three-dimensional structure of each gene product is required to understand its function fully. One of the core endeavors of structural genomics is to elucidate a complete set of structural families into which all proteins can be classified. Parallels in this challenge and the approaches used are often drawn between recent genomic sequencing efforts and emergent structural genomics programs. Over the last 2 years, multi-institutional collaborations have formed as part of the Protein Structure Initiative (PSI; www.nigms.nih.gov/funding/psi.html) to leverage resources and to facilitate group development of specific expertise, novel technologies, and process methods that are required for successful implementation of such a program.
The Joint Center for Structural Genomics (JCSG) has adopted a core-based model in its approach to structural genomics through a consortium of research centers. The JCSG consists of three functional units. The Bioinformatics Core selects, prioritizes, tracks targets, and provides data management support. The Crystallomics Core clones, expresses, purifies, performs crystallization trials, and prepares samples for x-ray structural analysis. The Structure Determination Core processes crystalline samples through x-ray diffraction analysis and ultimately produces three-dimensional protein structures, which are then validated and deposited with the Protein Data Bank (PDB) (1).
It is clear that reproducible and cost-efficient high-throughput (HT) methods must be used to obtain realistically the goal of a complete collection of protein folds or representative members of all protein families. We have developed and implemented HT technologies for each step of the structure determination process from target selection to PDB submission (2, 3). Here, we describe the design and implementation of the full JCSG structural genomics pipeline by means of a summary of the results obtained from processing the Thermotoga maritima proteome.
An HT pipeline requires integrating technology and process development. To this end, a large set of easily accessible genes is required. T. maritima is an attractive target for a structural genomics research program, as its small genome (1,877 genes) makes it practical for isolating the entire recombinant proteome. Even though many of these genes have known structural homologs, they still present an opportunity to test predictions of protein expression, purification, and crystallization. In addition, they provide a sufficient number of targets to test our HT technologies. By choosing to evaluate all protein targets, including those that pose particular challenges such as membrane proteins, we can identify exceptions to predicted behavior. Through establishing a collection of “difficult” proteins, more generalized approaches will be developed for expression and refolding of these more intractable targets. In addition, T. maritima offers some practical advantages. Bacterial proteins are typically easier to express in Escherichia coli, and their thermophilic nature also may improve the likelihood of crystallization. From a biological view, Thermotoga represents one of the deepest lineages among Eubacteria (4). A structural genomics analysis of T. maritima also should provide insights into early organismal and genome architecture, as well as protein evolution.
Materials and Methods
Cloning and Expression.
Primer pairs encoding the predicted 5′ and 3′ ends of all 1,877 ORFs (4) were used to amplify the corresponding genes from T. maritima strain MSB8 genomic DNA. The PCR product was cloned into plasmids pMH1, pMH2, or pMH4 for expression and introduced into the E. coli methionine auxotrophic strain DL41. These vectors encode a purification tag (MGSDKIHHHHHH) at the amino terminus of the full-length protein. The cloning junctions were confirmed by sequencing. Protein expression was performed in TB media (24 g/liter yeast extract/12 g/liter tryptone) containing 1% glycerol (vol/vol) and 50 mM Mops, pH 7.6. Expression was induced by the addition of 0.15% arabinose for 3 h. For TM0423 and TM0449, selenomethionine-containing media (5) was used.
Protein Purification.
Bacteria were lysed by sonication after a freeze-thaw procedure in lysis buffer [50 mM Tris, pH 7.9/50 mM NaCl/1 mM MgCl2/0.25 mM tri(2-carboxyethyl)phosphine hydrochloride (TCEP)/1 mg/ml lysozyme], and cell debris was pelleted by centrifugation at 3,600 × g for 60 min. The soluble fraction was applied to a nickel chelate resin (Invitrogen) previously equilibrated with equilibration buffer [50 mM KPO4, pH 7.8/300 mM NaCl/10% (vol/vol) glycerol/20 mM imidazole/0.25 mM TCEP]. The resin was washed with equilibration buffer containing 40 mM imidazole and protein eluted with elution buffer [20 mM Tris, pH 7.9/10% (vol/vol) glycerol/300 mM imidazole/0.25 mM TCEP]. Buffer exchange was performed to remove imidazole before crystallization, and the protein in crystal buffer (20 mM Tris, pH 7.9/150 mM NaCl/0.25 mM TCEP) was concentrated to approximately 10 mg/ml by centrifugal ultrafiltration. For TM0423 and TM0449, the affinity-purified proteins were equilibrated in buffer Q [20 mM Tris, pH 7.9/25 mM NaCl/5% (vol/vol) glycerol/0.25 mM TCEP] and applied to a Resource Q column (Amersham Pharmacia). Protein was eluted by using a linear gradient to 400 mM NaCl. Appropriate fractions were purified further by size-exclusion chromatography by using Superdex-200 resin (Amersham Pharmacia) with isocratic elution in crystal buffer.
Crystallization.
Proteins were crystallized by using the vapor diffusion method with 50 nl of protein and 50 nl of mother liquor sitting drops on customized microtiter plates (Greiner). Each protein was set up with 480 standard crystallization conditions [Wizard I/II, Cryo I/II (Emerald BioStructures, Bainbridge Island, WA), Crystal Screen, Crystal Screen 2, Crystal Screen Cryo, PEG/Ion Screen, Grid Screen Ammonium Sulfate, Grid Screen PEG 6000, Grid Screen MPD, and Grid Screen PEG/LiCl (Hampton Research, Riverside, CA)] at 20°C. Images of each crystal trial were taken at 0, 7, and 28 days after setup with an Optimag Veeco Oasis 1700 imager. Each image was evaluated by using a crystal detection algorithm (6) and scored for the presence of crystals. Images at days 7 and 28 also were evaluated manually.
Data Collection.
Synchrotron diffraction data sets from flash-cooled crystals were collected at 100 K on the beamlines of the Stanford Synchrotron Radiation Laboratory Structural Molecular Biology/Macromolecular Crystallography Resource. Data collection strategies were optimized depending on sample characteristics and used both multiwavelength anomalous dispersion (MAD) and single-wavelength anomalous dispersion approaches as implemented in the BLU-ICE/DCS data collection environment (http://smb.slac.stanford.edu/blu-ice/). The diffraction images were integrated by using Mosfilm (7) and scaled with SCALA from the CCP4 suite (Collaborative Computational Project No. 4, 1994).
Structure Determination and Refinement.
Structure solution for TM0423 and TM0449 was achieved by using the software packages SNB (8), the CCP4 suite program MLPHARE, and SOLVE (9). Structure refinement was performed by using CNS (10). Detailed descriptions of the data collection and refinement statistics will be presented elsewhere (11).
Validation and Deposition.
Structure models were validated by using a suite of programs including PROCHECK (12), SFCHECK (13), ERRAT (14), PROVE (15), and WHATCHECK (16), which together evaluate model geometry, data quality, and the fit of the model to the data. Refined structures were constrained to have deviations in bond angles and lengths within the expected range of values observed for structures deposited in PDB with comparable resolution. Values for molecular volume also were benchmarked against comparable values from structures of similar quality.
Results and Discussion
Proteome Analysis.
The T. maritima genome has 1,877 predicted ORFs (4). Over 1,400 T. maritima proteins could be considered structural genomics targets because their sequence identity to any known structure is less than 30%. A majority of these protein folds can be predicted by using more sensitive tools, such as fold recognition or threading algorithms such as PSI-BLAST (17) and FFAS (18). By these criteria, a maximum of 448 T. maritima ORFs could potentially have uncharacterized folds. Of these, 292 have significant homology to functionally characterized proteins. A quarter of the total predicted ORFs (445) contain predicted transmembrane helices. These proteins typically are difficult to express and crystallize. Indeed, none of these predicted membrane proteins crystallized in our initial screen. Additional refinements or alternate strategies will be required to address this important class of proteins. The complete proteome analysis of the T. maritima genome is available through the target-tracking pages at www.jcsg.org.
The T. maritima proteome serves as an effective target list for technology and methods development. However, a prioritization scheme was developed to focus continued efforts on proteins with proposed unique structural significance. The target prioritization scheme was a multistep selection procedure based on analysis of the T. maritima proteome (details are available at the Data Acquisition Prioritization System at the JCSG web site, www.jcsg.org). The target selection procedure was designed to complete coverage of the entire fold-space of the organism with the minimal number of structures. The total number of structures to be determined depends on the number of predicted novel folds and on how well one can model potentially similar folds from distantly related sequences. The target selection process initially seeks to identify sequences that have functional annotations but are of unknown structure by clustering the genome into related sequences with PSI-BLAST. The families are filtered to remove sequences with more than 30% identity to PDB depositions. These targets are further filtered for projected crystallization success as determined by size, absence of transmembrane regions, and the absence of low complexity and coiled-coil regions. Once coverage is attained at a coarse level, targets of interest can be investigated at finer levels of granularity.
Genes to Crystals Pipeline.
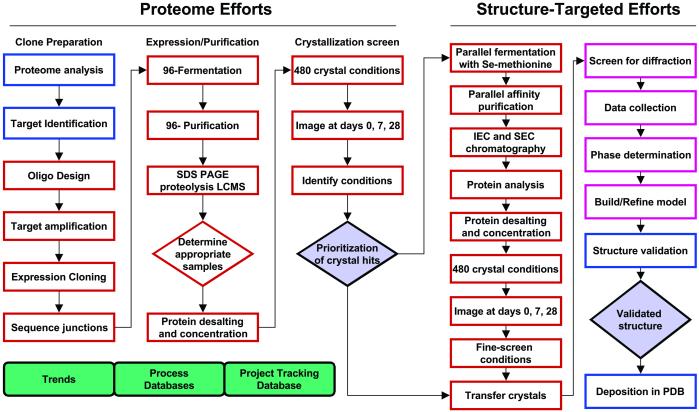
A summary of the HT pipeline process and strategy is presented in Fig. 1. Converting genomic information into expression-ready clones for the genes of interest is the first step. Full-length ORFs were chosen for expression based on TIGR predictions (4). The individual steps, outlined in Fig. 1, were typically performed in parallel with 96 samples comprising 96 individual targets. Scheduling was performed in two phases. The first phase involved molecular biology tasks for accumulating expression clones. Those that failed two cloning attempts were bypassed, to be addressed later with more focused efforts. The second phase involved protein expression, purification, and crystallization tasks. Large-scale samples were processed at a rate of 96–192 per week. Fermentation through the crystallization trial setups were performed in 7 days or less to minimize protein aggregation and degradation. The proteome collection of 1,376 expression clones was processed in 4 months by using this regimen. This aggressive timeframe was made possible by custom automation of expression, purification, and crystallization tasks (see Fig. 2). Table 1 provides throughput success rates in processing individual T. maritima genes through the HT crystal production pipeline. Although attrition was observed at each processing step, the overall pipeline output resulted in the identification of crystallization conditions for 23% of the proteins comprising the T. maritima proteome.
Figure 1.
T. maritima structural genomics pipeline.
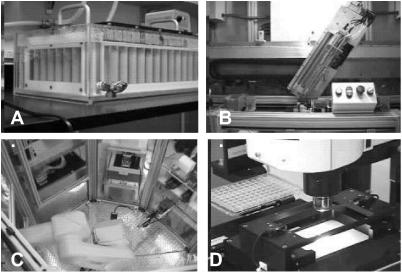
Figure 2.
Custom instrumentation used to process T. maritima proteome. (A) A 96-tube fermentor for high-cell density growth. (B) Purification robot that processes cell pellets from fermentation through affinity purification. (C) Nano-drop crystallization robotics used to set up crystal trials. (D) Plate imaging robotics used to analyze crystal trials.
Table 1.
Summary of results to date for HT protein expression, purification, and crystallization efforts with T. maritima proteome
| T. maritimaHT pipeline | No. | % |
|---|---|---|
| Targets | 1877 | 100 |
| Amplifers | 1791 | 95 |
| Expression clones | 1376 | 73 |
| Proteins attempted expression | 1376 | 73 |
| Proteins prepared for crystallography | 542 | 29 |
| Proteins crystallized | 432 | 23 |
Generic methods for parallel protein expression and purification require purification tags. Despite their potential interference with the crystallization process, the capacity for rapid protein purification and the uniformity of recombinant protein processing outweighs the potential problems. Our standard expression system introduces a small 12 amino acid tag at the amino terminus of targets. The initial six amino acids, selected to provide enhanced and more homogeneous expression of recombinant proteins, are followed by six histidines for purification via a nickel-affinity resin. Although such short tags can interfere with crystallization and proper protein folding, we have not observed this problem to be generic.
Crystallization experiments traditionally require large amounts of protein. Tens of milligrams of highly purified protein may be necessary to complete initial crystallization trials. Nano-droplet crystallization robotics substantially mitigate this requirement. Nonetheless, even these amounts are beyond the expression level attainable with the small-scale expression systems typically used with HT instrumentation. Therefore, instrumentation was developed that allows 96 parallel fermentations to be carried out to high cell densities (OD600 = 20–40) (19). On average, >6.5 mg of protein was isolated after affinity purification from a single 65-ml fermentation for those cells expressing soluble T. maritima proteins. Such expression levels, combined with the crystallization robotics, often permit over 1,000 crystal trials per protein sample. For the T. maritima proteome, 480 crystallization trials were performed on each protein sample.
Protein purity correlates to the success of crystallization. Histidine-tag purification provides the generic route needed for our HT applications. To facilitate parallel purification on the required scale, purification robotics were developed to process cultures directly from a fermentation run through affinity purification. This automation (19) integrates the centrifugation, sonication, and aspirate/dispense functions necessary to lyse cells and separate the soluble from the insoluble material before chromatography. In addition to the soluble fractions, the robotic system purifies the insoluble portions of each sample through detergent extractions for use in refolding studies. Secondary purification is often required to obtain high-quality crystals for x-ray diffraction. This step is achieved with multiple liquid chromatography systems by using autosamplers and standardized chromatographic protocols. Typically, anion exchange is followed by size-exclusion chromatography. In total, these systems provide the necessary throughput for large-scale expression and purification required for structural genomics.
The key to a successful structure determination pipeline is the ability to generate crystals from target proteins. An empirical screen of a sparse matrix of conditions provides the greatest likelihood of obtaining protein crystals (20). This approach is most successful when widely varying conditions (coarse screen) are followed by refinement around successful conditions (fine screen) to produce larger and better-diffracting crystals. The large numbers of such tests typically require significant amounts of protein and labor. Crystallization robotics with miniaturized volume assemblies were used to setup crystallization trials in specialized 96-well microtiter trays containing 50 nl of protein solution and 50 nl of crystallization solution in a sitting-drop format. For the T. maritima proteome, 480 conditions were screened per protein at 20°C, representing 260,160 individual crystallization experiments for the 542 T. maritima proteins examined to date. Each 100-nl droplet was imaged for the appearance of crystals by custom imaging robotics. Individual images were obtained at approximately days 0, 7, and 28 after setup and analyzed by using a combination of automated image analysis and manual evaluation. The resulting 780,480 images were analyzed for the presence of crystals. Crystals of harvestable size (>20–50 μm) were passaged through cryoprotectant solution and mounted onto cryo-loops 28 days after setup and stored in liquid nitrogen until screened for diffraction. The main purpose of the coarse screening effort was to identify proteins that expressed well and gave preliminary crystallization conditions. A fortunate unexpected outcome was the production of many diffraction quality crystals from the initial screen. Nanoliter crystallization trials are often considered to be too small to provide diffraction quality crystals. Recent studies (21), however, have identified numerous advantages to microcrystallization, including more rapid equilibrium and conservation of materials. To date, our effort has produced over 1,000 protein crystals for screening, mainly from 100-nl droplets, many of which give sufficient diffraction for structure determination. Tables 1 and 2 contain summaries of the purification efforts and coarse screening results for the T. maritima proteome.
Table 2.
Summary of crystal screening results to date for T. maritima proteome
| T. maritima current screening status | No. |
|---|---|
| Crystals screened | 1053 |
| Protein targets represented | 122 |
| Diffraction resolution of 3.0 Å or better | 220 |
| Datasets collected | 54 |
| Targets phased and traced | 24 |
Pipeline Analysis.
Our approach uses technology and parallel processing to maximize throughput and structural output. A two-tiered strategy was adopted by using an initial proteome screen followed by focused crystallization attempts. In the first tier, the expression, purification, and initial crystallization screen of the entire proteome was undertaken. From this effort, we identified proteins that express well, are soluble, and crystallize readily under standard conditions. At a minimum, these results provide a measure of what subset of the proteome is experimentally tractable. Comparative analysis of this subset can be used for target prioritization. As an opportunistic outcome, many of the proteins from the initial screening produced crystals of sufficient quality for data collection and structure determination. Beyond practical considerations, analysis of this first tier of experiments provides a large volume of data that can be mined for further insights about the protein properties that correlate with enhanced expression levels, solubility, and crystallizability.
The ability to routinely express soluble proteins in recombinant form is one of the major hurdles for structure determination. For T. maritima, 40% (542/1,376) of the proteins tested were soluble and expressed in sufficient quantity to pursue crystallization trials (Table 1). A cursory analysis of the results from the T. maritima (TM) proteome screen (Fig. 3) indicates some bias toward smaller molecular weight proteins (average TM protein = 35,778 Da; average soluble TM protein = 31,964 Da). However, proteins up to 100,000 Da were expressed and crystallized. Some bias toward acidic proteins also was observed (average TM protein pI = 7.16; average soluble TM protein pI = 6.54). This bias may be because of the internal pH of E. coli (pH 7.4–7.8; ref. 22) which might select against solubility of proteins with pIs near this pH range. Another important factor influencing our results is the insolubility of proteins as a result of missing protein partners. Most proteins in the cell interact with at least one other protein, typically through hydrophobic interfaces. Without the appropriate partner, many such proteins would be insoluble. Although it might be possible to predict some interactions, many proteins of unknown function need to be tested in two-hybrid studies (23) to identify potential partners for coexpression.
Figure 3.
Predicted molecular mass and isoelectric point. (A) All predicted T. maritima proteins. (B) Proteins that expressed in soluble form suitable for crystallization trials. (C) Proteins that crystallized in 480-condition coarse-screen trials.
Protein crystal growth is largely an empirical process typically achieved by using a screen of conditions historically shown to produce crystals. Our pipeline allows us to evaluate the relative success rate for an unbiased set of proteins. Our screens consisted of 480 commercial solutions as described in Materials and Methods. Each condition was evaluated based on the 542 proteins taken through crystallographic trials. Conditions then were ranked based on the number of unique proteins that crystallized under that condition. Fig. 4 shows a summary of the conditions relative to the number of proteins crystallized. In retrospect, 94% of the proteins could have been crystallized by using only 192 of the best conditions of the 480 tested. Refining the process to use these conditions would reduce the protein requirement by 60% and would allow more fine screening, novel coarse-screen conditions, or the inclusion of lower-expressing proteins. Various precipitants such as polyethylene glycols (PEGs), alcohols, and ammonium sulfate are used in protein crystallization trials. High-molecular-weight PEGs seem to be the most efficient, enabling crystallization of 80% of the targets tested. Low-molecular weight PEGs, alcohols, and ammonium sulfate are less efficient. Further detailed analysis of the relationship between crystallization success, screen conditions, and protein properties may improve predictive selection of reagents.
Figure 4.
Comparison of major protein precipitants used for crystallization. Crystallization conditions for each T. maritima protein tested were compiled. The number of proteins crystallizing under each condition was determined, and conditions were rank-ordered by frequency (All Conditions). Each of the major protein precipitants was rank-ordered in the same manner for comparison.
Although first-tier experimentation is aimed at identifying which proteins readily crystallize, many crystals obtained in these screens were of sufficient size and quality to permit data collection and structure determination. To date, over 1,000 T. maritima protein crystals representing 122 proteins have been examined by x-ray diffraction (Table 2). Because of the small volume of protein (50 nl) used in these crystal trials, most crystals were very small (approximately 20–50 μm on edge). The majority (≈70%) of protein crystals diffracted, with 21% diffracting to 3 Å or better resolution. We have chosen 3 Å resolution as a practical minimum for routine structure determination, below which automated data analysis becomes difficult. The large number of proteins with such resolution from limited crystallization trials is remarkable, given that generalized protocols for purification were used and all proteins retained their 12-amino acid purification tag. These results affirm that many proteins with a propensity to crystallize will do so even under nonoptimal conditions.
Structure Determination on T. maritima.
The second tier of our approach focuses on structure determination from the successful first tier targets. As described earlier, the entire T. maritima proteome was analyzed for similarity to known structures and prioritized for novelty. Based on these predictions and the results of the proteome screen, targets were prioritized. Because the high-priority targets are not likely to have molecular replacement solutions, experimentally determined phases are required. Selenomethionine (SeMet) incorporation provides a fairly generic and robust means of phase determination (5). Second-tier targets are re-expressed as SeMet-derivatives and are processed with additional effort expended on secondary purification and fine screening of crystallization conditions. This approach provides a cost-effective and productive route to providing optimal crystals for structure determination. Two example structures that have passed through the JCSG HT pipeline are described below. TM0423 demonstrates the potential for a fully automated structure determination strategy, whereas TM0449 demonstrates the utility of the structural genomics approach for determining a previously uncharacterized protein fold.
TM0423 is annotated as a glycerol dehydrogenase. The fold of TM0423 was predicted by FFAS (24) to be similar to that of dehydroquinate synthase from Aspergillus nidulans (25) with a Z score of 52. The recently published structure of the Bacillus stearothermophilus glycerol dehydrogenase (26) provided an even better model for TM0423, but was not available for molecular replacement at the time of data collection. In all, five native TM0423 crystals were screened. The best crystals contained a single molecule in the asymmetric unit and diffracted beyond 1.5 Å resolution. Four selenomethionine crystals also diffracted strongly, and all of the structure determination steps proceeded in real-time concurrent with the 2.0-Å resolution MAD experiment. The standard BLU-ICE/distribution control system (DCSS; see http://smb.slac.stanford.edu/blu-ice/) provided a streamlined interface for the setup and collection of MAD data. The automated execution and analysis of a selenium x-ray fluorescence scan resulted in the transparent selection of optimal wavelengths. During the experiment, the DCSS automatically deals with wavelength changes and beam optimizations. Therefore, all of the human effort was devoted to the crystallographic analysis so that direct feedback and adjustment of the diffraction experiment was possible. Automated model-building with ARP/wARP (27) rapidly converged to an almost complete peptide chain-trace. At this point, it was possible to interrupt the MAD data collection and proceed instead with a high-resolution (1.5 Å) dataset from the same crystal. In this way, the quality of the final resulting structure was improved, and the beamline efficiency was increased. The final steps of the crystallographic analysis, including refinement and model building, required some human intervention and manual effort. The final model (Fig. 5A) includes a zinc ion and a Tris buffer molecule bound in the enzyme's active site. The latter seems to be mimicking a glycerol substrate, which helped confirm the annotation that TM0423 is a glycerol dehydrogenase. Atomic coordinates and structure factors for TM0423 have been deposited with the PDB with accession code 1KQ3.
Figure 5.
Example structures from the pipeline process. (A) Ribbon diagram of T. maritima glycerol dehydrogenase. Strands are shown in blue and helices are shown in red. Tris is shown in a stick representation. The van der Waals volume of the Zn2+ ion is also shown. (B) Ribbon diagram of T. maritima thy1 thymidylate synthase-complementing protein (TM0449). Four FAD molecules are shown in a stick representation.
TM0449 is annotated as a thymidylate synthase-complementing protein with no significant homology to any known protein structure and, therefore, was likely to have a previously uncharacterized fold. Crystals of TM0449 diffracted to 2.25-Å resolution and crystallized with four molecules in the asymmetric unit. The crystallographic analysis was again conducted in real time during the diffraction experiment; in this case, it was not possible to obtain a fully interpretable electron density map during the time-course of the data collection. Nevertheless, the location of the selenium substructure was relatively straightforward. Subsequent heavy atom refinement and phase-determination steps, however, were not stable until all three wavelengths of the MAD experiment were incorporated. During the first pass of automated model building, only 220 of the 880 residues in the asymmetric unit could be traced. Subsequently, the experimental phases were improved by incorporating non-crystallographic symmetry (NCS) averaging, which leads to a more complete trace, including 578 of 880 residues. This model included 28 chain fragments, which were manually pieced together to reveal a complete monomer of TM0449 and then the entire tetramer. Clearly, automated methods for tasks like NCS averaging will significantly impact the quality and completeness of automatically built models, especially when the resolution of the diffraction data are limited. In turn, these improvements will reduce the amount of manual intervention in the subsequent model completion and refinement steps. The final model for TM0449 is shown in Fig. 5B. The coordinates and structure factors have been deposited with the PDB with the accession 1KQ4.
Thymidylate synthase-complementing proteins have been implicated in cell survival in the absence of external sources of thymidylate. A large active site pocket is formed in the center of the tetramer, with four channels running along the molecular interfaces. Four FAD molecules are present in the active site, with the AMP moieties pointing toward the central cavity and the riboflavin moieties of each FAD molecule pointing toward the surface of the protein. A structural similarity search performed by using the DALI server (28) failed to detect significant similarities to any other protein structure, indicating that TM0449 indeed exhibits a previously uncharacterized fold (11). This structural information was recently used to predict an alternate flavin-dependant mechanism for thymidylate synthesis (29) and clearly demonstrates the value of structural genomics in determining protein function.
The need to determine protein structures rapidly and cost-effectively has greatly expanded because of genomic and proteomic studies. Beyond exploring protein structure space, such structure determinations are essential for identifying and understanding protein function. Traditional approaches to structure determination are labor-intensive, costly, and slow. Our approach has been to establish instrumentation and methods to permit an HT processing of proteins for crystallography and structure determination. To demonstrate this potential, we have used this pipeline for the entire proteome of T. maritima and can use these results for processing of the subset of important structural genomics targets. By using these instrumentation and methods, we were able to process the majority of the T. maritima proteome through expression, purification, and crystallization in a matter of months. This success allowed us to identify and prioritize the 432 protein targets that crystallized under the conditions tested. The success of these crystallization trials has a high likelihood of translating into a substantial number of structures from the T. maritima proteome. Our subsequent efforts at structure determination from this pipeline have confirmed the productivity of this approach. A future key goal is to integrate the individual automated process steps into a seamless structure determination pipeline. Completion of this process integration will provide yet another significant increase in the capability and capacity to process an even higher number of samples more cost-effectively and to be able to tackle substantially more complex systems.
Acknowledgments
We thank Mike Hornsby, Kevin Rodrigues, Teresa Lesley, and the Genomics Institute of the Novartis Research Foundation Engineering Department and Syrrx staffs for their contributions. This work was supported in part by National Institutes of Health Protein Structure Initiative Grant GM62411 from the National Institute of General Medical Sciences (www.nigms.nih.gov). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, the National Institutes of Health, National Center for Research Resources, the Biomedical Technology Program, and the National Institutes of General Medical Sciences. This paper is manuscript no. 15073-CH of The Scripps Research Institute.
Abbreviations
- JCSG
Joint Center for Structural Genomics
- PDB
Protein Data Bank
- HT
high-throughput
- TCEP
tri(2-carboxyethyl)phosphine hydrochloride
- TM
T. maritima
- MAD
multiwavelength anomalous dispersion
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1KQ3 and 1KQ4).
References
- 1.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens R C. Curr Opin Struct Biol. 2000;10:558–563. doi: 10.1016/s0959-440x(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 3.Abola E, Kuhn P, Earnest T, Stevens R C. Nat Struct Biol Suppl. 2000;7:973–977. doi: 10.1038/80754. [DOI] [PubMed] [Google Scholar]
- 4.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, et al. Nature (London) 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spraggon, G., Lesley, S. A., Kreusch, A. & Priestle, J. P. (2002) Acta Crystallogr. D, in press. [DOI] [PubMed]
- 7.Leslie A G. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. Warrington, U.K.: Daresbury Lab.; 1992. , No. 26. [Google Scholar]
- 8.Weeks C M, Miller R. Acta Crystallogr D. 1999;55:492–500. doi: 10.1107/s0907444998012633. [DOI] [PubMed] [Google Scholar]
- 9.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 11. Kuhn, P., Lesley, S. A., Mathews, I. I., Canaves, J. M., Brinen, L. S., Dai, X., Deacon, A. M., Elsliger, M. A., Eshaghi, S., Floyd, R., et al. (2002) Proteins Struct. Funct. Genet., in press.
- 12.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 13.Vaguine A A, Richelle J, Wodak S J. Acta Crystallogr D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 14.Colovos C, Yeates T O. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pontius J, Richelle J, Wodak S J. J Mol Biol. 1996;264:121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- 16.Hooft R W W, Vriend G, Sander C, Abola E E. Nature (London) 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaroszewski L, Rychlewski L, Godzik A. Protein Sci. 2000;9:1487–1496. doi: 10.1110/ps.9.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesley S A. Protein Expression Purif. 2001;22:159–164. doi: 10.1006/prep.2001.1465. [DOI] [PubMed] [Google Scholar]
- 20.Jancarik J, Kim S H. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 21.Santarsiero B D, Yegian D T, Lee C C, Spraggon G, Gu J, Scheibe D, Uber D C, Cornell E W, Nordmeyer R A, Kolbe W F, et al. J Appl Crystallogr. 2002;35:278–281. [Google Scholar]
- 22.Slonczewski J L, Rosen B P, Alger J R, Macnab R M. Proc Natl Acad Sci USA. 1981;78:6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Rychlewski L, Jaroszewski L, Li W, Godzik A. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter E P, Hawkins A R, Frost J W, Brown K A. Nature (London) 1998;394:299–302. doi: 10.1038/28431. [DOI] [PubMed] [Google Scholar]
- 26.Ruzheinikov S N, Burke J, Sedelnikova S, Baker P J, Taylor R, Bullough P A, Muir N M, Gore M G, Rice D W. Structure (London) 2001;9:789–802. doi: 10.1016/s0969-2126(01)00645-1. [DOI] [PubMed] [Google Scholar]
- 27.Perrakis A, Morris R M, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]