Abstract
Suberoylanilide hydroxamic acid (SAHA) is a potent inhibitor of histone deacetylases (HDACs) that causes growth arrest, differentiation, and/or apoptosis of many tumor types in vitro and in vivo. SAHA is in clinical trials for the treatment of cancer. HDAC inhibitors induce the expression of less than 2% of genes in cultured cells. In this study we show that SAHA induces the expression of vitamin D-up-regulated protein 1/thioredoxin-binding protein-2 (TBP-2) in transformed cells. As the expression of TBP-2 mRNA is increased, the expression of a second gene, thioredoxin, is decreased. In transient transfection assays, HDAC inhibitors induce TBP-2 promoter constructs, and this induction requires an NF-Y binding site. We report here that TBP-2 expression is reduced in human primary breast and colon tumors compared with adjacent tissue. These results support a model in which the expression of a subset of genes (i.e., including TBP-2) is repressed in transformed cells, leading to a block in differentiation, and culture of transformed cells with SAHA causes re-expression of these genes, leading to induction of growth arrest, differentiation, and/or apoptosis.
Histone acetyl transferases and histone deacetylases (HDACs) regulate the acetylation of histones (1). Histone acetylation affects gene expression (2), and inhibitors of HDACs, such as the hydroxamic acid-based hybrid polar compound suberoylanilide hydroxamic acid (SAHA) (3), induce growth arrest, differentiation, and/or apoptosis of transformed cells in vitro (4–6) and inhibit tumor growth in vivo (7–12). SAHA is in phase I clinical trials for the treatment of solid and hematological tumors.
HDACs catalyze the removal of acetyl groups from lysine residues in the amino-terminal tails of the nucleosomal core histones (2, 13). HDACs can be divided into three classes. Class I HDACs (HDACs 1, 2, 3, and 8) are similar to the yeast RPD3 protein, are located in the nucleus, and are found in complexes associated with transcriptional corepressors (13). Class II HDACs (HDACs 4, 5, 6, 7, 9, and 10) are similar to the yeast HDA1 protein and are found in both the nucleus and cytoplasm (13–18). Both class I and II HDACs are inhibited by SAHA and related hydroxamic acid-based HDAC inhibitors (3, 14). Class III HDACs form a structurally distant class of NAD-dependent enzymes that are related to the yeast SIR2 proteins and are not inhibited by hydroxamic acid-based HDAC inhibitors (19, 20).
Differential display analysis of transformed lymphoid cell lines revealed that the HDAC inhibitor trichostatin A alters the expression of less than 2% of transcribed genes (21). One gene whose expression is induced by HDAC inhibitors is the p21WAF1 gene (4, 22, 23). The p21WAF1 protein is an inhibitor of cyclin-dependent kinases and inhibits cell cycle progression in the G1 phase of the cell cycle. p21WAF1 is induced within 2 h of culture of transformed cells with HDAC inhibitors. Induction by HDAC inhibitors requires an Sp1 site (23) and is associated with accumulation of acetylated histones in the promoter region of the gene (24, 25). Expression of the multidrug resistance 1 gene (MDR1) (26) and the SHP-1 gene (27) is induced by the HDAC inhibitors and requires the presence of a functional NF-Y binding site. These studies suggest that there are at least two classes of genes whose expression is induced by HDAC inhibitors: one class whose induction requires Sp1 sites in the promoter (e.g., p21WAF1) and a second that requires NF-Y sites (e.g., MDR1 and SHP-1).
The current study was designed to identify and characterize additional target genes of HDAC inhibitors. Using microarray analysis, we found that SAHA induces the expression of the thioredoxin-binding protein-2 (TBP-2) gene in LNCaP prostate cells. The induction of TBP-2 was associated with a decrease in thioredoxin (TRX) mRNA levels in these cells. We report here that TBP-2 mRNA levels are reduced in human breast and colon tumor tissue compared with matched samples of normal tissues. We cloned the promoter region of the TBP-2 gene and found that it is directly induced by SAHA and requires a NF-Y site for this induction. Induction of TBP-2 followed by decreased levels of TRX may play a critical role in SAHA-induced growth arrest and/or apoptosis in transformed cells.
Experimental Procedures
Cell Culture.
LNCaP prostate carcinoma, T24 bladder carcinoma, and MCF7 breast carcinoma cells were obtained from the American Type Culture Collection and cultured as suggested. SAHA was synthesized as described (4) and was dissolved and diluted in DMSO.
Microarray Analysis.
LNCaP human prostate carcinoma cells were cultured in the presence of vehicle alone (DMSO) or SAHA (7.5 μM) for 0.5, 2, 6, or 24 h, and total RNA was isolated from the cells by using Trizol reagent (GIBCO/BRL). Poly(A)+ mRNA was isolated from the total RNA by using Oligotex columns (Qiagen, Valencia, CA). Poly(A)+ mRNA from cells cultured with SAHA was compared with mRNA from cells cultured without SAHA, using the UniGEM human cDNA array (Incyte, St. Louis). A 2-fold change was considered as a threshold for real differences in gene expression.
Northern Blotting.
Total RNA (10 μg) was analyzed by Northern blotting using a 32P-labeled 1.1-kb TBP-2 coding region cDNA probe or a 500-bp cDNA probe for human TRX according to Ausubel et al. (28).
Expression of TBP-2 in Normal and Tumor Tissues.
Northern blots containing poly(A)+ mRNA extracted from a panel of different normal human tissues was obtained from CLONTECH. Blots were hybridized first with a 32P-labeled 1.1-kb TBP-2 cDNA probe, then with a 32P-labeled 2.0-kb cDNA probe for β-actin, as a loading control.
Dot blots of cDNAs from matched pairs of normal and tumor tissues from human patients were obtained from CLONTECH. The manufacturer (CLONTECH) normalized the quantities of cDNA on the blot by using three housekeeping genes: ubiquitin, 23-kDa highly basic protein, and ribosomal protein S9. The blot was hybridized with the 32P-labeled 1.1-kb TBP-2 cDNA probe according to the manufacturer's instructions.
Cloning of the 5′ Regulatory Region of the TBP-2 Gene.
Rapid amplification of cDNA ends was performed to determine the transcriptional start site of the TBP-2 gene, using TBP-2-specific primer 1 (5′-GTTGGTTTTAAGAGTTAGAAATGACGG-3′) and nested primer 2 (5′-TAAGGTATTCTTAAGCAGTTTGAGC-3′) with Marathon-ready human brain cDNA (CLONTECH), according to the manufacturer's instructions. A product of approximately 200 bp was amplified, gel-purified, and subcloned, and nine independent clones were sequenced. From this sequence, two additional primers (5′-CCAATTGCTGGAGAAAAGATCCG-3′ and 5′-AAGATCCGATCTCCACAAGCACTCC-3′) were designed. These two primers were used to clone the promoter of the TBP-2 gene by genome walking with the GenomeWalker kit (CLONTECH). Products ranging from approximately 1,200 to 1,800 bp were amplified from three of the genomic libraries (SspI, PvuII, and DraI), gel-purified, and subcloned for sequencing. Sequencing was performed at the DNA Sequencing Facility at Cornell University (Ithaca, NY). At least two clones obtained from each of the three libraries were sequenced, and all were found to be virtually identical in the region directly upstream of the 5′ untranslated region of the TBP-2 gene. The sequence for the 1,763-bp SspI fragment was deposited in GenBank (accession no. AF408392). During the preparation of this manuscript, the sequence of the TBP-2 gene was deposited into the GenBank database (accession nos. AB051901 and AF333001) and the Human Genome database (accession no. NT_004883).
Luciferase Assays for Analysis of TBP-2 Promoter Activity.
Construction of TBP-2/pGL2-Luc vectors.
The 1,763-bp SspI fragment was subcloned into the multiple cloning site (SacI and NheI sites) of the pGL-2 luciferase reporter vector (Promega) to make the pTBP-2-(−2026)-Luciferase construct.
Deletion constructs of the TBP-2 promoter.
Deletion constructs of the TBP-2 promoter sequence were generated by PCR cloning. The following primers with the original nested TBP-2-specific primer end at −264 bp from the translation initiation site were used to amplify the TBP-2 promoter from the −2026 (1,736 bp) construct to generate 5′ deletion mutants: −1049: 5′-TGAGCTCAACAGCACAGGCACGCAGCC-3′; −901: 5′-TGAGCTCCAAGAGAAGGACAAAGGGC-3′; −784: 5′-TGAGCTCGCCAGGAATAACGACAGGC-3′; −679: 5′-TGAGCTCCAGAAACGTCCACACCCG-3′; −604: 5′-TGAGCTCCTGGACCCGGGAGAAGACG-3′; −530: 5′-TGAGCTCTGCGCCGCTCCAGAGCGC-3′; −482: 5′-TGAGCTCGTGTCCACGCGCCACAGCG-3′; −440: 5′-TGAGCTCTGGTAAACAAGGACCGGG-3′; −395: 5′-TGAGCTCGCAGCACGAGCCTCCGGG-3′; and −349: 5′-TGAGCTCGGCTACTATATAGAGACG-3′.
All of the above primers contained NheI sites (underlined) to facilitate cloning, and all clones were sequenced before analysis.
Generation of a TBP-2 promoter construct containing mutations in the inverted CCAAT box.
Point mutations that abolish the inverted CCAAT box site (ATTGG → AGTAG) were introduced by PCR-directed mutagenesis (28) using primers 5′-ACTGAGTAGTCGGGCTCCTGGTAAAC-3′ and 5′-CCGACTACTCAGTGAGATCGCTGTGG-3′. The mutations were confirmed by DNA sequencing.
Reporter gene assay.
293T cells were plated in 24-well plates in triplicate and transfected with 100 ng of either pGL-2 vector, pGL-2-SV40 (simian virus 40) promoter vector (positive control containing the SV40 promoter), or the pGL2-TBP-2 promoter constructs, using FuGENE 6 transfection reagent (Roche Applied Sciences) according to the manufacturer's instructions. Cells were harvested after 24–48 h, and luciferase activity was measured by using the Dual Luciferase Assay System (Promega), according to the manufacturer's instructions. For experiments in which SAHA or other HDAC inhibitors were used, the transfection medium was replaced with fresh medium containing either vehicle control (DMSO) or SAHA (0.5, 1, or 2 μM), 12 h after transfection. After another 12–24 h, the cells were harvested and the lysates were analyzed for luciferase activity as described above. For the experiment with dominant negative NF-Y mutant, 293T cells were cotransfected with 100 ng pGL2-TBP-2 −2026 promoter construct and 6.75 and 12.5 ng NF-YA29 expression vector (29). The total amount of DNA was adjusted to 200 ng by addition of sonicated salmon sperm DNA. The treatment with 2 μM SAHA and luciferase assays was performed as described above.
Gel Mobility-Shift Assays.
Nuclear extracts were prepared from T24 cell with or without treatment of 7.5 μM SAHA for 12 h as described (30). The double-stranded DNA oligonucleotide containing the inverted CCAAT box (5′-CGATCTCACTGAttgGTCGGGCTCCTG-3′) was 5′ end-labeled with [γ-32P]ATP (ICN) using T4 polynucleotide kinase (New England Biolabs), and purified by using a G-25 sephadex column (Roche Applied Sciences). Gel mobility-shift assays were performed as described (26). For the competition studies, both the wild-type double-stranded oligonucleotide and a mutant double-stranded oligonucleotide containing the inverted CCAAT box mutation as described above for reporter gene assay were used. For the gel mobility-supershift assays, nuclear extracts were preincubated with either rabbit anti-human NF-YA polyclonal antibody, goat anti-human C/EBP antibody, or preimmune rabbit IgG (Santa Cruz Biotechnology) at 4°C for 3 h before the addition of the labeled oligonucleotide.
Results
SAHA Induces Expression of TBP-2 in Transformed Cells.
To identify genes whose expression is modified by SAHA, LNCaP human prostate carcinoma cells were cultured in the presence of either DMSO (vehicle control) or 7.5 μM SAHA for 0.5, 2, 6 or 24 h, poly(A)+ mRNA was isolated, and cDNA microarray (Incyte) analysis was performed. TBP-2 was the only gene detected that was induced by more than 2.0-fold at early times after culture with SAHA (30 min). The level of expression of this gene remained increased in LNCaP cells cultured with SAHA for at least 24 h.
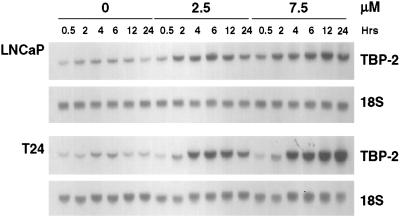
To confirm the microarray results, TBP-2 mRNA levels were analyzed by Northern analysis in several transformed cell lines cultured with SAHA. SAHA (2.5 or 7.5 μM) induces TBP-2 mRNA levels within 2 h in LNCaP cells (Fig. 1). The maximum induction (4-fold) was observed after culture with SAHA (7.5 μM) for 12 h. At 2.5 μM, SAHA causes complete growth suppression and at 7.5 μM causes death in these cells (12). SAHA (2.5 or 7.5 μM) induced TBP-2 mRNA within 2 h in each transformed cell line examined: T24 bladder carcinoma (Fig. 1), ARP-1 human myeloma, murine erythroleukemia, 293T kidney carcinoma, and MCF7 breast carcinoma cell lines (data not shown). A 20-fold induction of TBP-2 mRNA was observed after 24 h of culture with SAHA (7.5 μM) in T24 bladder carcinoma cells.
Figure 1.
Induction of TBP-2 mRNA levels in transformed cells by SAHA. LNCaP prostate carcinoma and T24 bladder carcinoma cells were cultured with vehicle alone (0) or SAHA (2.5 or 7.5 μM) for the indicated times. Total RNA was extracted from the cells, and the levels of TBP-2 were determined by Northern blotting using a 1.1-kb 32P-labeled TBP-2 cDNA probe (Upper for each cell line). Blots were rehybridized with a γ-32P-labeled 18S oligonucleotide probe to indicate RNA loading and are shown (Lower for each cell line). Similar results were obtained for a total of six transformed cell lines.
Expression of TBP-2 in Normal and Tumor Tissues.
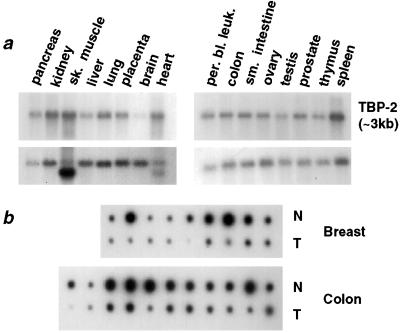
The pattern of expression of TBP-2 mRNA was examined in a panel of 16 normal human tissues. TBP-2 mRNA was detected in all of the tissues examined with the highest levels of expression in skeletal muscle, kidney, and spleen, and the lowest levels of expression in the brain (Fig. 2a).
Figure 2.
Expression of TBP-2 in normal and tumor tissues. (a) Multiple tissue Northern blots, containing poly(A)+ RNA from the indicated tissues (CLONTECH), were hybridized with a 1.1-kb 32P-labeled TBP-2 cDNA probe (Upper). The bolts were rehybridized with a 2.0-kb probe for β-actin, as a control for loading (Lower). sk. muscle, skeletal muscle; per. bl. leuk., peripheral blood leukocytes. (b) A dot blot containing matched samples of cDNA extracted from normal human tissues and tumors (CLONTECH) was hybridized with a 1.1-kb 32P-labeled TBP-2 cDNA probe. Samples of colon and breast tumors (T) are shown, with the cDNA from the normal tissue (N) shown directly above each corresponding tumor sample.
To address whether genes whose expression is induced in transformed cells by SAHA are down-regulated during tumorigenesis, the expression of TBP-2 was analyzed in normal and tumor tissues. Hybridization of the TBP-2 coding region cDNA probe to a dot-blot array of cDNAs from matched normal tissues and primary tumor specimens of patients showed a consistent reduction in the levels of TBP-2 mRNA expression in colon and breast carcinomas (Fig. 2b).
SAHA Reduces TRX mRNA Levels in Transformed Cells.
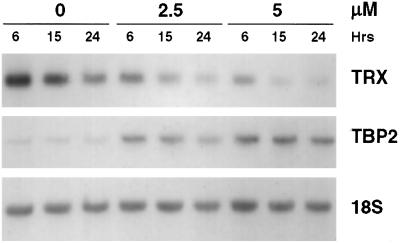
TBP-2 has been identified as a protein that associates with the active (reduced) form of TRX, a dithiol-reducing redox regulatory protein (31). Binding of TBP-2 to TRX both inhibits the thiol-reducing activity and reduces the level of expression of TRX. To determine whether induction of TBP-2 by SAHA is associated with reduced TRX levels, Northern blot analysis of RNA prepared from T24 cells cultured with SAHA (2.5 and 5 μM), using a full-length TRX cDNA probe, was performed. A decrease in the levels of TRX mRNA was observed within 15 h of culture with SAHA accompanied by a concomitant increase in the level of TBP-2 mRNA (Fig. 3). A similar SAHA-induced decrease in TRX mRNA and increase in TBP-2 mRNA was detected in ARP-1 and MCF7 cells (data not shown).
Figure 3.
Expression of TRX in transformed cells cultured with SAHA. T24 bladder carcinoma cells were cultured with vehicle alone (0) or 2.5 or 5 μM SAHA for 6, 15, or 24 h. RNA was extracted and analyzed by Northern blotting for levels of TRX, using a 500-bp 32P-labeled cDNA probe (Top). The blots were subsequently rehybridized with the 1.1-kb 32P-labeled TBP-2 cDNA probe to confirm induction of TBP-2 (Middle) and a γ-32P-labeled 18S oligonucleotide probe to indicate RNA loading (Bottom).
Cloning and Characterization of the TBP-2 Promoter.
To investigate the mechanism by which SAHA induces the expression of TBP-2 mRNA, we cloned the TBP-2 promoter region with a combination of rapid amplification of 5′ cDNA ends and genome walking. The promoter sequence was analyzed with the matinspector professional program (http://transfac.gbf.de/cgi-bin/matSearch/matsearch.pl) for the presence of classical promoter features. The presence of a putative TATA box as well as putative binding sites for the transcription factors, such as, NF-Y, Myc-Max, E2F, vitamin D receptor/retinoid X receptor, and NF-κB were identified. The proscan computer program (version 1.7, http://bimas.dcrt.nih.gov/molbio/proscan/) predicted that a TATA box existed at the same location predicted by matinspector.
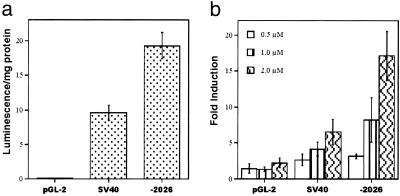
To confirm that the sequence identified by genome walking has promoter activity, the obtained sequence was cloned into a promoter-less pGL-2 luciferase reporter vector, and luciferase reporter assays were performed. Transfection of the pGL-2-TBP-2 construct, −2026, into 293T cells yielded reporter gene activity equivalent to or greater than the SV40-positive control promoter whereas transfection with pGL-2 vector yielded barely detectable reporter gene activity (Fig. 4a), indicating that the cloned TBP-2 promoter is functional.
Figure 4.
The cloned TBP-2 promoter is functional and induced by SAHA. 293T cells were transfected with 100 ng of either an empty pGL2 vector, a pGL2-SV40 positive control vector, or the TBP-2 construct (−2026). (a) Firefly luminescence was measured 24 h after transfection. (b) The transfection medium was removed 12 h after transfection and replaced with medium containing DMSO alone or SAHA (0.5, 1, or 2 μM). Luminescence was measured 24 h later and normalized for total protein concentration of each sample. Fold induction is obtained by normalizing the luciferase value in the presence of SAHA against luciferase value in the absence of SAHA.
To test whether SAHA induces the activity of the cloned TBP-2 promoter, 293T cells were transfected with reporter constructs and cultured with SAHA. The activity of the TBP-2 promoter fragment was induced in a dose-dependent manner by SAHA (Fig. 4b). The activity of the SV40 control promoter was induced by SAHA, but not to the same extent as the TBP-2 promoter (Fig. 4b). The activity of the TBP-2 promoter was also induced by m-carboxycinnamic acid bishydroxamic acid (CBHA) a related hydroxamic acid-based hybrid polar inhibitor of HDAC activity (data not shown).
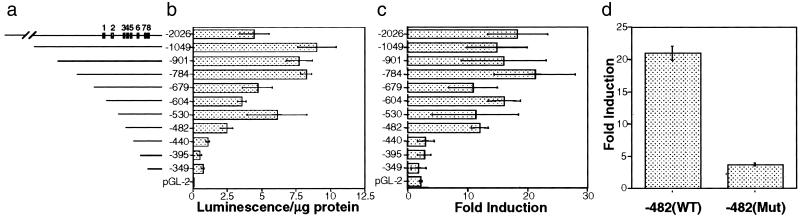
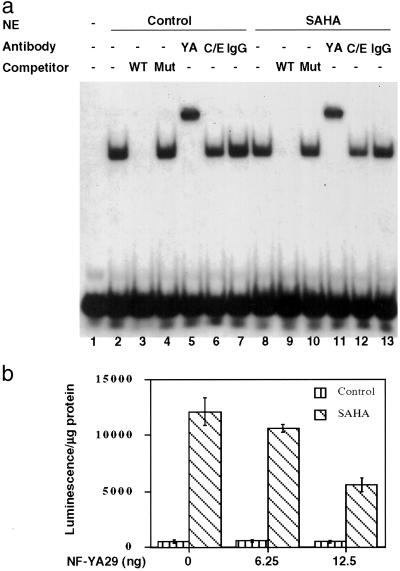
To determine which potential transcription factor binding sites are important for TBP-2 gene transcription and induction by SAHA, a series of deletion constructs was generated (Fig. 5a). Transient transfection assays showed that promoter constructs −2026 to −482 varied in luciferase activity by 4-fold (Fig. 5b), most likely representing loss of transcription factor activator or repressor binding sites. Deletions beyond −482 resulted in a consistent drop in transcriptional activity. Addition of SAHA (2 μM) to transfected cells caused an induction of luciferase activity of 12- to 20-fold after 24 h of culture for promoter constructs −2026 to −482 (Fig. 5c). However, promoter constructs −440 to −349 showed reduced levels of induction (2- to 3-fold) in response to SAHA (Fig. 5c). This finding suggested the presence of a site between promoter constructs −482 and −440 that is critical for optimal induction of TBP-2 by HDAC inhibitors. This region of the promoter contains putative E box and inverted CCAAT box sites. Several transcription factors, including NF-Y (32), bind to the inverted CCAAT box. We introduced two point mutations into the inverted CCAAT box (ATTGG → AGTAG) and generated pGL-2-luciferase constructs to test the activity and SAHA inducibility of the inverted CCAAT box mutant promoter. The −482 construct consisting of the mutated inverted CCAAT box showed a lower level of induction (3.7-fold) by SAHA than the wild-type −482 construct, which was induced 21-fold (Fig. 5d). These results indicate that the inverted CCAAT box in the TBP-2 promoter is critical for the optimal induction of the TBP-2 promoter by SAHA. Electrophoretic mobility-shift assays using nuclear extracts prepared from control and SAHA-treated T24 cells were performed to determine whether NF-Y binds this inverted CCAAT box in the TBP-2 promoter. A specific protein-DNA complex was detected (Fig. 6a, lanes 2 and 8). The wild-type unlabeled competitor blocked the formation of the complex (Fig. 6a, lanes 3 and 9), but the inverted CCAAT box mutant competitor had no effect (Fig. 6a, lanes 4 and 10). Supershift analysis was then performed to identify the proteins bound to the inverted CCAAT box. The polyclonal antibody against NF-YA resulted in a supershift of the protein–DNA complex, whereas an antibody against another CCAAT box binding protein C/EBP did not alter the mobility of the complex. These results indicate that the inverted CCAAT box site in the TBP-2 promoter is capable of binding NF-Y. Similar results were observed from the nuclear extracts of untreated (Fig. 6a, lanes 2–7) and cells cultured with SAHA (Fig. 6a, lanes 8–13).
Figure 5.
Deletion and mutation analyses of TBP-2 promoter. (a) Schematic representation of the putative TBP-2 promoter region and the deletion mutants. The positions of putative transcription factors binding sites in the promoter are shown: 1, NF-κB binding site; 2, vitamin D receptor/retinoid X receptor responsive element; 3, E2F binding site; 4, E box; 5, inverted CCAAT box; 6, CCAAT box; 7, E box; 8, TATA box. (b) Activity of deletion mutants. Different lengths of the 5′ flanking region of human TBP-2 gene were amplified by PCR and cloned upstream of the luciferase gene in the pGL-2 vector. The constructs were transfected into 293T cells and luciferase activity was measured. (c) SAHA induction of deletion mutants. Parallel experiments were carried out as in b except SAHA was added 12 h after transfection. Luciferase activity was normalized against total protein. (d) The inverted CCAAT box is critical for SAHA inducibility. PCR-based mutagenesis was performed to mutate the inverted CCAAT box in the isolated TBP-2 promoter. The mutant promoter was then cloned into pGL-2 and transfected into 293T cells with or without SAHA (2 μM) treatment. The TBP-2 promoter construct with the mutations in the inverted CCAAT box has about 8% of wild-type promoter activity (not shown). Fold induction was calculated as in b and results shown are the average of three independent transfections ± SD.
Figure 6.
Role of NF-Y in induction of TBP-2. (a) Binding of NF-Y to the inverted CCAAT box in TBP-2 promoter. Electrophoretic mobility-shift assay (lanes 1–4 and 8–10) detects specific complex formation at the inverted CCAAT box. 32P-labeled wild-type probe (20,000 cpm, ≈0.5 ng; lane 1) was incubated with 10 μg nuclear extracts prepared from untreated (lanes 2–7) or 7.5 μM SAHA-treated (12 h) (lanes 8–13) T24 cells, in the absence (lanes 2 and 8) or presence of 25 ng (×50) wild-type (lanes 3 and 9) or mutant (lanes 4 and 10) oligonucleotide competitors. For supershift assays, nuclear extracts were incubated with 2 μg rabbit anti-NF-YA (lanes 5 and 11), 2 μg goat anti-C/EBP (lanes 6 and 12), or 2 μg normal rabbit IgG (lanes 7 and 13). WT, wild-type probe competitor; Mut, mutant probe competitor; YA, anti-NF-YA; C/E, anti-C/EBP. (b) Dominant negative NF-Y mutant (NF-YA29) decreases the promoter induction by SAHA. The pGL2-TBP-2 −2026 promoter construct (100 ng) was cotransfected with NF-YA29 expression vector as indicated, and then treated with or without SAHA (2 μM) for 24 h.
To further investigate the role of NF-Y in SAHA induction of the TBP-2 promoter a dominant negative NF-YA mutant expression vector, NF-YA29, was cotransfected with −2026 pGL2-TBP-2 promoter construct into 293T cells. NF-Y29 is a dominant negative form of NF-YA with a mutation of 3 aa in the DNA binding domain. NF-Y29 forms a complex with NF-YB (and NF-YC), but fails to bind the CCAAT box (29). NF-YA29 decreased the TBP-2 promoter induction by SAHA (Fig. 6b). Taken together, these results support a role for NF-Y in the induction of TBP-2 transcription by SAHA.
Discussion
In this study we show that inhibition of HDAC activity in transformed cells with SAHA increases the level of TBP-2 mRNA. TBP-2 is a TRX binding protein. The expression of the TBP-2 gene was induced by SAHA in several human transformed cell lines, including prostate carcinoma, bladder carcinoma, myeloma, breast carcinoma cell lines, and murine erythroleukemia cells. All of these cell lines are growth inhibited by SAHA (4, 12, 33) at concentrations that cause induction of TBP-2 mRNA.
TBP-2 was initially identified as a gene induced in HL-60 cells cultured with 1,2-dihydroxyvitamin D3 (34). TBP-2 associates with the active (reduced) form of TRX, a dithiol-reducing redox regulatory protein (31). Binding of TBP-2 to TRX inhibits the thiol-reducing activity and reduces the level of TRX (31). We found that T24, MCF-7, and ARP-1 cells cultured with SAHA for 15 h or longer have decreased levels of TRX mRNA, which coincides with the induction of TBP-2 mRNA. TRX and glutaredoxin are major cellular protein disulfide reductases (35) and serve as electron donors for a number of enzymes including ribonoucleotide reductases and TRX peroxidases. Several transcription factors including NF-κB, estrogen, and glucocorticoid receptors are substrates for reduced TRX and show altered activity in reduced versus oxidized states. TRX also functions in defense against oxidative stress, growth control, and apoptosis. TRX is secreted from cells and stimulates proliferation of a variety of human solid tumor cells (36). Additionally, secreted TRX has cytokine activity (35). TRX is overexpressed in many human tumors, most notably colon and gastric cancers (37, 38). Exogenous expression of TRX confers increased clonogenicity and tumorigenicity to transformed cells and inhibits cells from undergoing apoptosis in response to a variety of stimuli (39, 40). Conversely, overexpression of a dominant negative redox-inactive form of TRX reduces the clonogenicity and tumorigenicity of MCF-7 breast cancer cells (39) and WEHI7.2 mouse lymphocytic leukemia cells (41). Our results suggest that induced TBP-2 gene expression may be an important mechanism, at least in part, by which SAHA and related HDAC inhibitors cause growth arrest and apoptosis of transformed cells. The decreased levels of the active form of TRX could lead to an accumulation of reactive oxygen species and inactive forms of proteins required for DNA synthesis and cell cycle progression, causing growth arrest and/or enhancing the apoptotic pathway in cancer cells.
The promoter region of the TBP-2 gene was found to contain a TATA box and binding sites for a number of transcription factors, several of which are present in complexes containing HDACs. The activity of the TBP-2 promoter was directly induced by SAHA and the structurally related HDAC inhibitor CBHA (m-carboxycinnamic acid bishydroxamic acid) in transient transfection assays. Deletion analysis of the TBP-2 promoter revealed that a 42-bp region containing an E-box and an inverted CCAAT box is required for maximal induction of TBP-2 activity by SAHA. Mutations of the inverted CCAAT box markedly reduced the ability of SAHA to induce the TBP-2 promoter, supporting that this site is required for optimal induction by HDAC inhibitors. Electrophoretic mobility-shift assays showed that the transcription factor NF-Y forms a specific complex with the inverted CCAAT box in the TBP-2 promoter, whereas another inverted CCAAT box binding protein C/EBP does not. Additionally, we find that a dominant negative form of NF-YA blocks induction of TBP-2 by SAHA. Thus, our findings are similar to those reported for the multidrug resistance 1 gene (MDR1) (26) and the SHP-1 gene (27). NF-Y is a transcription factor consisting of three independent subunits that binds specifically to the CCAAT box in both orientations (42). Activation of promoter activity by NF-Y may be mediated by the P/CAF coactivator, which has histone acetyltransferase activity and is recruited by NF-Y to the MDR1 promoter (36).
We found that TBP-2 is a ubiquitously expressed gene in normal tissues and that the expression of TBP-2 is reduced in human breast and colon tumors. SAHA induces TBP-2 expression in six human transformed cell lines tested. Taken together, our results support a model in which the expression of a subset of genes in transformed cells is repressed, leading to a block in differentiation. Culture of transformed cells with SAHA results in re-expression of these genes, which in turn, could lead to alterations in the expression of other genes and the induction of growth arrest, differentiation, and/or apoptosis.
Acknowledgments
We are grateful to Gisela Perez and Lang Ngo for their technical expertise. These studies were supported, in part, by grants from the Burke Foundation (to L.M.B. and H.I.S.), the Japan Foundation for Cancer Research, the DeWitt Wallace Fund for Memorial Sloan-Kettering Cancer Center, National Institutes of Health Grants CA-0974823 and U01 CA-84292 (to V.M.R.), and funds granted by the Michael and Ethel Cohen Foundation (to X.Z.). X.Z. is a Cohen Fellow in Biomedical Research. Memorial Sloan-Kettering Cancer Center and Columbia University jointly hold the patents on the hydroxamic acid hybrid polar compounds, including SAHA, which are exclusively licensed to Aton Pharma, Inc. of which R.A.R., P.A.M., and V.M.R. are founders. Both institutions and the founders have an equity position in Aton Pharma, Inc.
Abbreviations
- HDAC
histone deacetylase
- SAHA
suberoylanilide hydroxamic acid
- SV40
simian virus 40
- TBP-2
thioredoxin-binding protein-2
- TRX
thioredoxin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF408392).
References
- 1.Marks P A, Rifkind R A, Richon V M, Breslow R, Miller T, Kelly W K. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 3.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richon V M, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1996;93:5705–5708. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffey D C, Kutko M C, Glick R D, Swendeman S L, Butler L, Rifkind R, Marks P A, Richon V M, LaQuaglia M P. Med Pediatr Oncol. 2000;35:577–581. doi: 10.1002/1096-911x(20001201)35:6<577::aid-mpo18>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Munster P N, Troso-Sandoval T, Rosen N, Rifkind R, Marks P A, Richon V M. Cancer Res. 2001;61:8492–8497. [PubMed] [Google Scholar]
- 7.Qiu L, Kelso M J, Hansen C, West M L, Fairlie D P, Parsons P G. Br J Cancer. 1999;80:1252–1258. doi: 10.1038/sj.bjc.6690493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen L A, Amin S, Marks P A, Rifkind R A, Desai D, Richon V M. Anticancer Res. 1999;19:4999–5005. [PubMed] [Google Scholar]
- 9.He L Z, Tolentino T, Grayson P, Zhong S, Warrell R P, Jr, Rifkind R A, Marks P A, Richon V M, Pandolfi P P. J Clin Invest. 2001;108:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffey D C, Kutko M C, Glick R D, Butler L M, Heller G, Rifkind R A, Marks P A, Richon V M, La Quaglia M P. Cancer Res. 2001;61:3591–3594. [PubMed] [Google Scholar]
- 11.Butler L M, Webb Y, Agus D B, Higgins B, Tolentino T R, Kutko M C, LaQuaglia M P, Drobnjak M, Cordon-Cardo C, Scher H I, et al. Clin Cancer Res. 2001;7:962–970. [PubMed] [Google Scholar]
- 12.Butler L M, Agus D B, Scher H I, Higgins B, Rose A, Cordon-Cardo C, Thaler H T, Rifkind R A, Marks P A, Richon V M. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 13.Gray S G, Ekstrom T J. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Marks P A, Rifkind R A, Richon V M. Proc Natl Acad Sci USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guardiola A R, Yao T P. J Biol Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 16.Tong J J, Liu J, Bertos N R, Yang X J. Nucleic Acids Res. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao H Y, Lee C H, Komarov A, Han C C, Evans R M. J Biol Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 18.Fischer D D, Cai R, Bhatia U, Asselbergs F A, Song C, Terry R, Trogani N, Widmer R, Atadja P, Cohen D. J Biol Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 19.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 21.Van Lint C, Emiliani S, Verdin E. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao H, Hasegawa T, Miyaishi O, Ohkusu K, Isobe K. Biochem Biophys Res Commun. 1997;237:457–460. doi: 10.1006/bbrc.1997.7158. [DOI] [PubMed] [Google Scholar]
- 23.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 24.Sambucetti L C, Fischer D D, Zabludoff S, Kwon P O, Chamberlin H, Trogani N, Xu H, Cohen D. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 25.Richon V M, Sandhoff T W, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Scotto K W. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Banville D, Zhao H F, Zhao X, Shen S H. Gene. 2001;269:141–153. doi: 10.1016/s0378-1119(01)00445-0. [DOI] [PubMed] [Google Scholar]
- 28.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 29.Mantovani R, Li X Y, Pessara U, van Huisjduijnen R H, Benoist C, Mathis D. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 30.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 32.Mantovani R. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang L, Pardee A B. Mol Med. 2000;6:849–866. [PMC free article] [PubMed] [Google Scholar]
- 34.Chen K S, DeLuca H F. Biochim Biophys Acta. 1994;1219:26–32. doi: 10.1016/0167-4781(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 35.Arner E S, Holmgren A. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 36.Gasdaska J R, Berggren M, Powis G. Cell Growth Differ. 1995;6:1643–1650. [PubMed] [Google Scholar]
- 37.Berggren M, Gallegos A, Gasdaska J R, Gasdaska P Y, Warneke J, Powis G. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 38.Grogan T M, Fenoglio-Prieser C, Zeheb R, Bellamy W, Frutiger Y, Vela E, Stemmerman G, Macdonald J, Richter L, Gallegos A, Powis G. Hum Pathol. 2000;31:475–481. doi: 10.1053/hp.2000.6546. [DOI] [PubMed] [Google Scholar]
- 39.Gallegos A, Gasdaska J R, Taylor C W, Paine-Murrieta G D, Goodman D, Gasdaska P Y, Berggren M, Briehl M M, Powis G. Cancer Res. 1996;56:5765–5770. [PubMed] [Google Scholar]
- 40.Baker A, Payne C M, Briehl M M, Powis G. Cancer Res. 1997;57:5162–5167. [PubMed] [Google Scholar]
- 41.Freemerman A J, Powis G. Biochem Biophys Res Commun. 2000;274:136–141. doi: 10.1006/bbrc.2000.3091. [DOI] [PubMed] [Google Scholar]
- 42.Mantovani R. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]