Abstract
Overexpression of the TCL1 oncogene has been shown to play a causative role in T cell leukemias of humans and mice. The characterization of Tcl1-deficient mice in these studies indicates an important developmental role for Tcl1 in early embryogenesis. In wild-type embryos, Tcl1 is abundant in the first three mitotic cycles, during which it shuttles between nuclei and the embryo cortical regions in a cell-cycle-dependent fashion. The absence of this protein in early embryogenesis results in reduced fertility of female mice. The present studies elucidate the mechanism responsible for the reduced female fertility through analysis of the oogenesis stages and early embryo development in Tcl1-deficient mice. Even though Tcl1−/− females display normal oogenesis and rates of oocyte maturation/ovulation and fertilization, the lack of maternally derived Tcl1 impairs the embryo's ability to undergo normal cleavage and develop to the morula stage, especially under in vitro culture conditions. Beyond this crisis point, differentiative traits of zygotic genome activation and embryo compaction can take place normally. In contrast with this unanticipated role in early embryogenesis, we observed an overexpression of TCL1 in human seminomas. This finding suggests that TCL1 dysregulation could contribute to the development of this germinal cell cancer as well as lymphoid malignancies.
The T cell leukemia/lymphoma 1 gene, TCL1, was initially identified as a gene involved in recurrent chromosomal translocations in human prolymphocytic leukemia (T-PLL), a neoplasia often seen in ataxia-telangiectasia patients (1, 2). In T-PLL, reciprocal translocation of the TCL1 locus at 14q32.1 with one of the T cell receptor (TCR) loci can result in inappropriate overexpression of the TCL1 gene because of the juxtaposition to TCR enhancer(s) (2). The structure of the encoded protein is unchanged and the overexpression of Tcl1 may contribute to T cell tumorigenesis by deregulating cell proliferation and/or cell survival. This interpretation is supported by the occurrence of mature T cell leukemia in mice carrying a TCL1 transgene under the control of a lck promoter ensuring T lineage expression (3).
The Tcl1 protein has recently been shown to interact with Akt, the product of an oncogene that is a key participant in transduction of antiapoptotic and proliferative signals in T cells. Tcl1 can enhance Akt kinase activity and induce its nuclear translocation (4, 5). In addition to its involvement in T cell malignancies, TCL1 is constitutively transcribed in the peripheral lymphoid tissues, predominantly in B cells (6–8). TCL1 is expressed throughout B cell development, from the preB to the mature B cell stages, but is extinguished at the mature plasma cell stage (2). The TCL1 gene shares significant homology with MTCP1 (mature T-cell proliferation-1), a gene identified by its chromosomal localization near the Xq28 breakpoint in T-PLL (9). Other genes of the TCL1 family have now been identified, the TCL1b gene in humans and the Tcl1b1-b5 genes in the mouse (10–12). All of these genes are located in close proximity to the human and murine TCL1 genes.
The developmentally regulated Tcl1 gene is normally expressed in ovary, testis, preimplantation embryos, fetal thymus, and bone marrow (12, 13). To gain insight into the biological role(s) of Tcl1, we have generated Tcl1 null mutant mice. The present report describes the Tcl1 loss-of-function phenotype, which indicates that Tcl1 is important for in vitro preimplantation embryo development. Moreover, aberrant TCL1 expression was observed in a survey of human germinal cell cancers.
Materials and Methods
Construction of Tcl1-Deficient Mice.
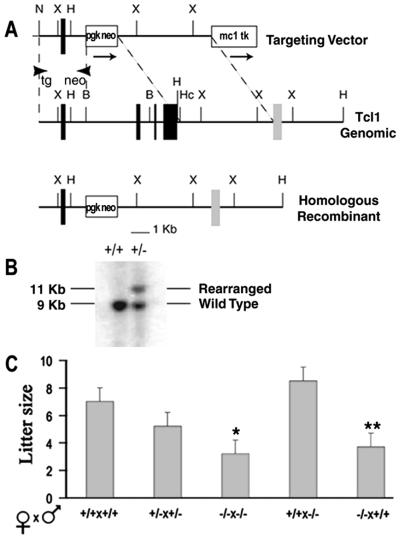
The Tcl1 genomic DNA was cloned from a 129/SVJ mouse genomic library, as described (13), and subcloned into pBluescript SK vector (Stratagene). A targeting vector was designed to replace a 5-kb BamHI–HincII fragment containing exons 2, 3, and 4 by phosphoglycerate kinase (PGK)-neo in the same transcriptional orientation as the Tcl1 gene. The targeting vector was flanked at the 5′-end by a 2.5-kb fragment, including Tcl1 exon I and 5′-untranslated region and, at the 3′-end, by a 5-kb of genomic DNA containing polyoma enhancer fragment–herpes simplex thymidine kinase (MC1-TK), as a counterselectable marker against random integration events. The targeting vector was linearized by digestion at the unique NotI site and electroporated into 107 RW-4 embryonic stem cells (Genome Systems, St. Louis) (passage 6) with 230 Volts and 500 μF. Positive–negative selection was initiated 24 hr after transfection using 180 μg/ml of active G418 (GIBCO/BRL) and ganciclovir (2 μM). Clones resistant to G418-ganciclovir were screened for homologous recombination by Southern blot hybridization using a 0.4-kb HincII fragment as 3′ external probe located outside the targeting construct and confirmed by long PCR using the following primers (see Fig. 1): Tg, 5′-CGGTGGATGTGGAATGTGTGC-3′; neo, 5′-ACCACCAACGGCTTCCTCCACT-3′. Chimeric mice and F1 heterozygotes were generated from Tcl1 mutant embryonic stem cells by standard methods (14). Wild-type and mutant C57BL/6Jx123XSvS F2 hybrids were used in all of the experiments. Tcl1−/− mice obtained in crosses were genotyped by PCR analysis performed in 2 separate reactions on genomic tail DNA using wild-type primers designed to detect the exon 2 of Tcl1 genomic locus: ex2for, 5′-GAAGCTATGTCCCCCAGTCA-3′; ex2rev, 5′-CAGGATCTGCCAATACATCG-3′; and G418 primers to detect the mutant null allele: G418for, 5′-ATTGTCTTCCCAATCCTCCC-3′; G418rev, 5′-CGACTGTGCCTTCTAGTTGC-3′.
Figure 1.
Gene targeting of the Tcl1 locus and reduced female fertility. (A) Configuration of the targeted genomic Tcl1 gene and the PGK-neo targeting vector. PGK-neo cassette replaces a BamHI (B) to HincII (Hc) fragment containing exons 2, 3, and 4. Arrows show transcriptional orientation of neo and TK genes. Restriction endonuclease cleavage sites are indicated. X, XbaI; H, HindIII; N, NotI; PGK, phosphoglycerate kinase; MC1, polyoma enhancer fragment; TK, herpes simplex thymidine kinase. (B) Representative genomic blot probed with an external probe recognizing a 9-kb wild type HindIII (H) fragment. The 11-kb fragment shows the rearranged homologous recombinant allele. (C) Litter size of wild-type and Tcl1 knockout female mice. The number of pups (mean ± SEM) in the litters produced by various mating pair was determined the day of birth. Mice (2–3 months old) were bred for a period of 3 months. Mating pairs analyzed in each group were (from left to right): 11, 12, 11, 5, and 4. Difference from wild type litters: *, P = 0.0001; **, P = 0.006, calculated by ANOVA.
Antibodies.
A rat anti-murine Tcl1 monoclonal antibody, 5A4, and a rabbit affinity-purified antimurine Tcl1 polyclonal IgG were prepared (S.-M.K., unpublished data) and used for immunohistochemistry and microinjection, respectively. For microinjection of mouse embryos, rabbit polyclonal IgG reacting with murine and human Sp2 (Santa Cruz Biotechnology) and preimmune rabbit IgG (Sigma) were used as controls.
Embryo Isolation, in Vitro Culture, and Microinjection.
Hormonally primed, 40- to 60-day-old female Tcl1−/−, Tcl1+/−, and Tcl1+/+ mice were mated with adult Tcl1+/+ or Tcl1−/− males taking the midnight after mating as fertilization time. Wild-type eggs were obtained from 40- to 60-day-old B6D2F1 mice (Charles River Italia, Calco, Italy). Routinely, 1-cell embryos were collected 10–12 hr after fertilization, and cultured in vitro for a total of 5 days as described (15). Cultured embryos were scored daily (between 11:00 a.m. and 1:00 p.m.) for the presence of pronuclei, progression through cleavage stages, and morphology. For microinjection, antibodies were microdialyzed against 10 mM Tris/0.1 mM EDTA, pH 7.4 (TE) through MF filters (Millipore, Rome) and diluted to a final concentration of approximately 125–250 ng/μl in TE. Cytoplasmic microinjection of anti-Tcl1 antibody (≈5 pL per egg) was performed at 14–16 hr after fertilization in 1-cell embryos.
Tcl1 Expression in Mouse Embryo.
Mouse MetII oocytes and preimplantation embryos at the appropriate developmental stages were fixed in 2.4% paraformaldehyde in M2 medium (14) for 1 hr at room temperature (RT). Fixed embryos were then carefully washed and incubated for 3 hr in PBS containing 0.1 M glycine and 0.3 mg/ml BSA, and then permeabilized in PBS containing 0.1% Triton X-100 for 15 min at RT. Permeabilized embryos were washed in PBS containing 1 mg/ml BSA (PBS-BSA) and then processed for immunostaining. An overnight incubation at 4°C in the presence of the first antibody was followed by a washing in PBS–BSA and 1 hr of incubation with the secondary antibody at RT. The 5A4 antimurine Tcl1 monoclonal rat antibody (1:75) and a FITC-conjugated anti-rat IgG (1:400) were used to detect Tcl1, whereas a rabbit polyclonal (1:200) and a FITC-conjugated anti-rabbit IgG (1:200) were used to detect β-catenin. Specimens were mounted on slides and observed by laser scanning confocal microscopy. For semiquantitative analysis of fluorescence, embryos at various developmental stages were pooled in the same drop and immunostained with the anti-Tcl1 antibody. Fluorescence emission was collected under similar excitation conditions and then quantitatively analyzed by using the metamorph imaging system (Universal Imaging, Media, PA) software.
Semiquantitative reverse transcription–PCR reactions were performed with rTth Reverse Transcriptase on groups of 5 1-cell or 2-cell embryos for 35 cycles and by using [α-P32]dCTP (DuPont Italiana, Cologno Milanese, Italy) as tracer, as described (16). Tcl1 mRNA was quantitated by using the ribosomal protein S16 mRNA as internal standard. Primer pairs were: Tcl1 (amplification fragment, 264 bp), 5′-GATCTGGGAGAAGCACGTGTA-3′ [118–138 nt, sense] and 5′-TTCAAGCAACATGTCCTCCA-3′ (363–382 nt, antisense); and S16 (amplification fragment, 103 bp), 5′-AGGAGCGATTTCCTGGTGTGC-3′ (1451–1471 nt, sense) and 5′-GCTACCAGGGCCTTTGAGATGGA-3′ (1621–1641 nt, antisense).
Immunohistochemistry.
A monoclonal antihuman Tcl1 antibody (17) and two polyclonal anti-Akt and anti-phosphoAkt (Ser-473) antibody preparations (Cell Signaling, Beverly, MA; catalog nos. 9272 and 9277), were used for immunohistochemical analysis of germ cell tumors according to the manufacturer's instructions.
Results
Tc11 Knockout Mice Display a Maternal Fertility Defect.
A vector replacing exons 2, 3, and 4 of the Tcl1 gene by a PGK-neo cassette (Fig. 1A) was used to target embryonic stem cells (Fig. 1B). Subsequent germ-line transmission was obtained and null mice were generated as described in Materials and Methods. Tcl1−/− mice appeared normal at birth and had no discernible histologic abnormalities other than a modest impairment in the development and function of the immune system that is described elsewhere (S.-M.K., C.M.C., M.D.C., and G.R., unpublished data). In addition, a female fertility defect attracted our attention when we noted that litters of the Tcl1−/− females contained fewer pups than those of Tcl1+/+ and Tcl1+/− females. The reduced litter size was related to the dose of the null allele in the female rather than in the male parent, as evidenced by similar pup number/litter produced by Tcl1−/− and Tcl1+/+ males (Fig. 1C). This reduction in litter size became more dramatic with increasing age of the knockout mice (data not shown). Resembling a typical maternal effect, the pup number reduction in litters of Tcl1−/− females suggested these mice may have impaired oogenesis, oocyte maturation/ovulation, fertilization, preimplantation embryo development, implantation, and/or postimplantation embryo development.
Reduced Fertility in Tcl1−/− Females Caused by Impairment of Blastomere Proliferation in the Early Preimplantation Embryo.
In an initial evaluation of possible cause(s) for the fertility defect, the follicle/oocyte growth processes appeared to be unaffected by the absence of Tcl11. Ovaries of juvenile (4–15 days old) and adult (8–12 weeks old) Tcl1−/− mice were histologically normal and impaired ovarian follicle function was rendered unlikely by the observation that hormonally primed Tcl1+/+, Tcl1+/−, and Tcl1−/− females produced similar numbers of ovulated metaphase II (Met II) oocytes with normal fertilization capacities (Table 1). Tcl1 therefore does not appear to affect ovarian oocyte and follicle development.
Table 1.
Ovulation and fertilization rates of Tcl1 wild-type and knockout mice
| Cross
|
Ovulated eggs*
|
Eggs with pronuclei†
|
||||
|---|---|---|---|---|---|---|
| N‡ | Female × male | N§ | Eggs per mouse | P¶ | N§ | Fertilization, %‖ |
| 6 | +/+ × +/+ | 125 | 20.8 + 3.6 | 113 | 90.4 | |
| 4 | +/− × +/+ | 96 | 24.0 + 6.8 | >0.5 | 88 | 91.7 |
| 5 | −/− × +/+ | 117 | 23.4 + 4.2 | >0.8 | 101 | 86.3 |
| 4 | −/− × −/− | 78 | 19.5 + 1.0 | >0.9 | 64 | 82.0 |
Ovulated eggs (not including fragmented or degenerated eggs) were scored at 12 p.m. of day 0.
Fertilization rate was assessed by scoring the eggs for the presence of two pronuclei at 3 p.m. of day 0.
Total number of mated females analyzed.
Total number of eggs recovered.
Difference from +/+ × +/+ cross was calculated by multiple ANOVA.
Difference between crosses: P > 0.1, calculated by χ2.
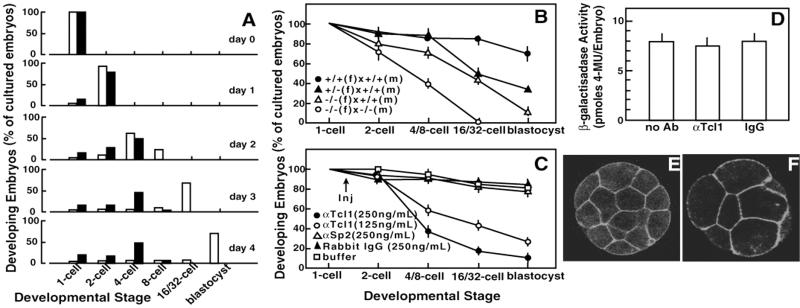
Tcl1 relevance to preimplantation embryo development was investigated by comparing the in vitro ability of Tcl1−/− embryos to develop from the 1-cell stage to the blastocyst stage (Fig. 2A). A majority of the Tcl1−/− embryos developed more slowly than wild-type embryos and failed to proceed beyond the 4- to 8-cell stage. This delay or blockage of blastomere proliferation of Tcl1-deficient embryos in vitro was examined further by comparing the preimplantation development in embryos obtained from crosses between wild-type and/or Tcl1-deficient parents (Fig. 2B). Embryos derived from wild-type females and Tcl1−/− males developed normally to the blastocyst stage (data not shown) in agreement with the normal litter size observed for their offsprings. In contrast, about 50% of the embryos produced by crossing Tcl1+/− females with wild-type fathers did not develop beyond the 8-cell stage. The impaired development was more pronounced in heterozygous embryos derived from a cross between Tcl1−/− females and wild-type males, and none of the Tcl1−/− embryos progressed beyond the 8-cell stage (Fig. 2B). These results were confirmed by experiments in which wild-type embryos received a single cytoplasmic injection of anti-Tcl1 antibodies at the late 1-cell stage and then were allowed to develop in vitro for 5 days (Fig. 2C).
Figure 2.
Preimplantation development of in vitro cultured Tcl1-deficient embryos. (A) Comparison of developmental abilities of wild-type and Tcl1 knockout embryos. One-cell embryos derived from the wild-type (open bars) and knockout (solid bars) were cultured in vitro and scored daily for 5 days in three independent experiments having 20–40 embryos in each experimental group. (B and C) Developmental abilities of Tcl1-defective embryos (B) and of wild-type embryos made defective in Tcl1 by cytoplasmic injection of anti-Tcl1 antibody (C). The arrow indicates the time of injection. Data represent the mean ± SEM of percentages obtained in at least three independent experiments having 20–40 embryos in each experimental group. Symbols indicate the embryos obtained in different crosses that progressed to the appropriate developmental stage at the time of the score. (D) Effect of anti-Tcl1 antibody on the expression of the DNA construct phsplacZ in wild-type embryos at ZGA. Histograms represents the mean β-galactosidase activity ± SEM of 25–35 single injected embryos, pooled from three independent experiments. Differences between treatments: P > 0.75, calculated by ANOVA. (E and F) Immunofluorescence detection of β-catenin by confocal microscopy in compacted Tcl1+/+ (E) and Tcl1−/− (F) embryos. One-cell embryos were in vitro cultured for 3 days. Note the reduced number of blastomeres in the presence of normal blastomere flattening in Tcl1−/− embryos.
To determine whether the compromised blastomere proliferation of Tcl1−/− embryos was accompanied by defective embryo acquisition of differentiative traits, two major steps of preimplantation development, zygotic gene activation (ZGA) at the early 2-cell stage and embryo compaction at the 8-cell stage, were analyzed in embryos depleted of Tcl1 by injection of anti-Tcl1 antibodies. Normal ZGA in these embryos was evidenced by the expression of phspLacZ, a DNA construct directed by the hsp70.1 gene promoter (18), which is spontaneously activated during ZGA at the early 2-cell stage in the mouse (19, 20) (Fig. 2D). Embryo compaction was evaluated by outlining blastomere boundaries with β-catenin immunostaining (21) (Fig. 2 E and F). Tcl1-deficient embryos displayed both of these differentiative traits normally and with appropriate timing, despite their block in blastomere proliferation. Apoptosis was not evident in the cleavage-blocked embryos when analyzed by the terminal deoxynucleotidyltransferase-mediated dUTP end labeling (TUNEL) assay (data not shown).
Tcl1 Expression in Wild-Type Embryos.
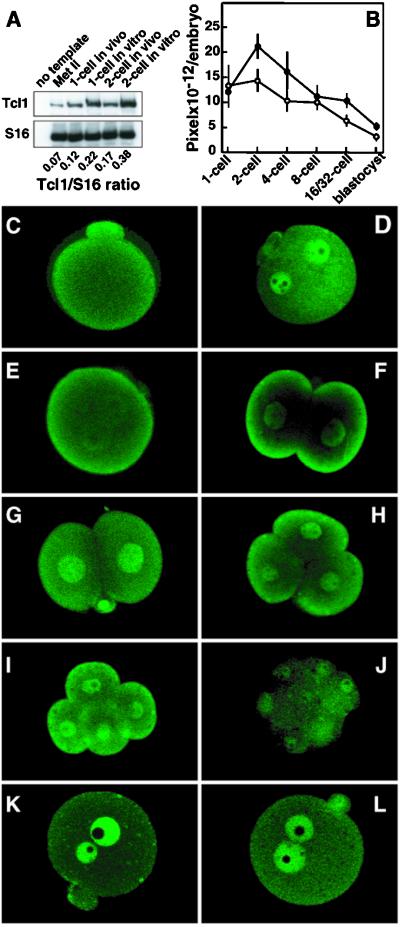
The indication that Tcl1 is required for preimplantation development led us to examine normal Tcl1 expression and cytological localization in wild-type embryos. Semiquantitative reverse transcription–PCR and immunofluorescence analysis indicated that Tcl1 mRNA and protein are present in unfertilized eggs, 1-cell embryos and 2-cell embryos, and the Tcl1 levels increase significantly under in vitro culture conditions (Fig. 3A) (22). At the late preimplantation stages, however, the embryonic Tcl1 content progressively declines irrespectively of the culture conditions. When intracellular Tcl1 distribution was analyzed in wild-type eggs and preimplantation embryos by confocal microscopy (Fig. 3 C–J), Tcl1 was found to be sharply localized to the cortex of unfertilized Met II oocytes (Fig. 3C). In 1-cell embryos, Tcl1 loses its cortical localization and is translocated to both male and female pronuclei during the S and early G2 phases of the first cell cycle (Fig. 3D). When the embryos approach cleavage, however, Tcl1 appears to relocate to the cortical embryo regions (Fig. 3E) that directly face the external environment (Fig. 3F). This cortical-nuclear-cortical Tcl1 shuttle pattern was also observed in two and 4-cell embryos (Fig. 3 G–I). When 1-cell embryos with newly formed pronuclei were cultured with either the DNA-polymerase inhibitor aphidicolin (Fig. 3K) or the cytokinesis inhibitor cytochalasin D (Fig. 3L) until the late 1-cell stage, the Tcl1 translocation from pronuclei to cortical region was inhibited completely, thereby indicating the dependency of this process on both cell cycle progression to the G2 phase and microfilament integrity. Notably, this Tcl1 shuttling pattern is lost at the 8-cell stage (Fig. 3J), a finding that correlates with the Tcl1 protein down-regulation observed in the wild-type embryos and the developmental blockade in Tcl1−/− embryos.
Figure 3.
Expression of Tcl1 protein in wild-type preimplantation embryos. (A) Semiquantitative reverse transcription–PCR amplification of Tcl1 and ribosomal protein S16 mRNAs of oocytes and early wild-type embryos. Numbers below lanes indicate Tcl1/S16 incorporation ratios. (B) Quantitative immunofluorescence analysis of Tcl1 protein in preimplantation wild-type embryos using anti-Tcl1 antibodies. (○) One-cell embryos collected 15 hr after hCG and allowed to develop in vitro to the appropriate developmental stage; (●) embryos collected at the appropriate developmental stage directly from the tubes of pregnant animals and immediately processed for the assay. (C–L) Tcl1 immunolocalization in Met II oocytes and preimplantation embryos by confocal microscopy. Representative embryos are shown in the panel. (C) Met II oocyte. (D) Mid 1-cell embryo at the S phase. (E) Late 1-cell embryo at the late G2 phase. (F) Early 2-cell embryo at the early G1 phase. (G) Mid 2-cell embryo at the S phase. (H) Three- to 4-cell embryo at the early G1 phase. (I) Four-cell embryo at the late G2 phase. (J) Eight-cell embryo. (K) Late 1-cell embryo cultured in vitro in the presence of 2 μg/ml aphidicolin. (L) Late 1-cell embryo cultured in vitro in the presence of 5 μg/ml cytochalasin D.
Tcl1 Expression in Germinal Cell Cancers.
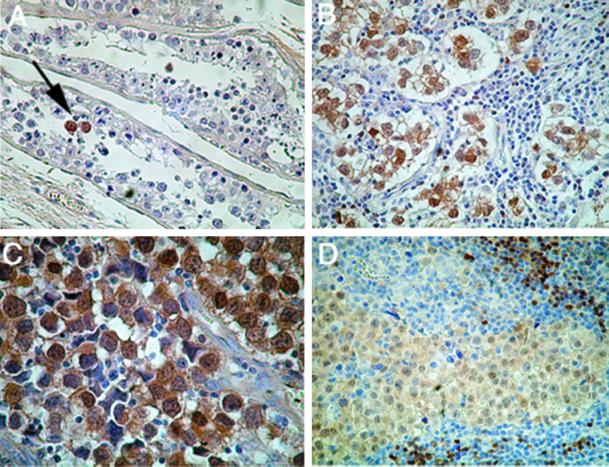
The finding that Tcl1 expression is tightly regulated during early embryo development, where it functions to ensure blastomere proliferation, prompted us to examine the possibility that this gene could be involved in human cancers other than T-PLL and B cell neoplasms. With a focus on early embryonic or germ cell tissues, 29 tumors of the human gonads, including 13 ovarian carcinomas, 13 testicular seminomas, one mediastinal dysgerminoma, and two teratocarcinomas were examined for Tcl1 expression by immunohistochemistry analysis with an anti-human Tcl1 monoclonal antibody (17). This analysis revealed the expression of Tcl1 in 10/13 seminomas and in the mediastinal dysgerminoma (Fig. 4), but not in the other types of cancer. Tcl1 expression was confined to the nuclear and cytoplasmic regions of the seminoma cells and was not seen in normal neighboring cells.(Fig. 4A). Because Tcl1 expression is normally detectable in the human testis only by reverse transcription–PCR analysis, and not by immunohistochemical analysis (not shown), the selective immunodetection of Tcl1 in neoplastic cells indicates the overexpression of this gene in this type of germinal cell cancer.
Figure 4.
TCL1 expression in human germ cell neoplasia. (A) Testicular tubules showing normal spermatogenesis in adult testis with focal Tcl1 positive cells (arrow) of intratubular germ cell neoplasia. Nonneoplastic germ cells are Tcl1 negative (original magnification, ×250). (B) Testicular seminoma, classical type of neoplastic testis cells show strong nuclear immunoreactivity (original magnification, ×400). (C) Testicular seminoma: neoplastic cells show strong nuclear and cytoplasmic immunoreactivity (original magnification, ×630). (D) Mediastinal (thymic) dysgerminoma: neoplastic cells show weak to medium level positivity for Tcl1, both in the nucleus and in the cytoplasm. Clusters of B lymphocytes display the normal Tcl1 pattern of immunoreactivity (Upper Right) (original magnification, ×400).
Because of the functionally relevant interaction that may occur between Tcl1 and Akt in vivo, we also used a set of activation specific AKT antibodies in our immunohistochemical analysis of seminomas and normal testis. Although Akt immunopositivity was detected for normal adult testis, with a stronger signal being seen with the anti-phosphoAkt antibody, Akt was undetectable in any of the Tcl1 positive tumors, with the single exception of an intratubular germ cell component present in one of the seminomas (data not shown).
Discussion
The present observation of impaired fertility in Tcl1-deficient females prompted the analysis of Tcl1 intracellular distribution and function in the early mouse embryo. The mouse zygotic genome is transcriptionally activated soon after the first embryo cleavage (15, 20, 23). Two additional mitotic divisions then lead the embryo to the 8-cell stage at which its peripheral blastomeres compact and establish tight and gap junctional communications with each other (21, 24), eventually resulting in blastocoel cavity formation. Our results indicate that Tcl1-deficient embryos acquire major differentiative traits normally and with the appropriate developmental timing. At the time of compaction, however, these embryos consistently display a reduced number of blastomeres, indicating a specific defect(s) in blastomere proliferation per se. As a consequence of the impaired blastomere proliferation, the in vitro cultured Tcl1−/− embryos are unable to reach the blastocyst stage and, likewise, neither are the heterozygote embryos derived from Tcl1−/− mothers. The presence of a paternal Tcl1 allele can only partially rescue the defect in initial blastomere proliferation, but not the eventual embryo development to the blastocyst stage, as a further indication that maternally derived Tcl1 is essential to early mouse embryo development. The Tcl1 dependency of initial embryo cleavages thus represents one of the earliest phenotypes so far elucidated in mouse gene targeted models.
Our observation that Tcl1 expression is higher under in vitro versus in vivo conditions is concordant with the findings of Minami et al. (22) who have described enhanced Tcl1 gene expression in embryos cultured without oviductal tissues. This higher expression of Tcl1 might account also for the more dramatic in vitro phenotype observed for Tcl1−/− embryos compared with the in vivo phenotype, wherein the absence of Tcl1 in females reduces their litter size and significantly shortens their reproductive life. This suggests that alternative mechanisms may counteract the lack of Tcl1 in vivo. Tcl1 has been shown to interact with Akt to increase Akt kinase activity (4, 5) through the formation of Akt-Tcl1 hetero-oligomers (25, 26). Tcl1 may thus act as a structural amplification loop in the phosphatidylinositol 3-kinase (PI3-kinase) Akt pathway (25). Although the mechanism of Tcl1 action in early embryo development is not yet fully elucidated, the Tcl1 cofactor role in the PI3-kinase Akt pathway may be essential under conditions where growth factors are minimal, as for the in vitro conditions used in this study or in the absence of oviduct factors (22).
The nuclear translocation of Tcl1 is assumed to have functional significance on the basis of the Tc11 β-barrel structure (27, 28), TCL1 expression patterns in normal lymphoid and cancerous tissues (8, 17), and the enhancing effects of TCL1 and AKT1 cotransfection (4). The present analysis of early mouse embryos unambiguously demonstrates that Tcl1 is repetitively translocated from the extranuclear cortical region to the nucleus in a cell-cycle-dependent manner during the first three mitotic divisions. The initial localization of Tcl1 in the outmost region of embryo cortex may suggest a role for this factor in an external milieu sensing capacity. Interestingly, the abridgement of the Tcl1 shuttling by the 8-cell stage and the concordant progressive decrease in Tcl1 expression coincides precisely with the developmental blockade observed in Tcl1−/− embryos.
A definitive interpretation of the pathogenetic significance of the elevated Tcl1 expression that we observed in seminomas will require further genetic analysis. Nevertheless, Tcl1 overexpression in human seminomas is highly suggestive in view of the fact that ectopic TCL1 overexpression is associated with the development of lymphoid tumors (3, 29, 30). The Tcl1 associate, Akt1, may be linked to the spermatogenesis process in that Akt1−/− mice have increased testicular cell apoptosis and attenuated spermatogenesis (31). Although an increase in phosphorylated Akt apparently does not accompany the elevated Tcl1 expression in human seminomas, the most well-documented effect of Tcl1 is to increase Akt kinase activity. Tcl1 could thus act to maximize kinase activity of the Akt, which is present in very low levels in the seminoma cells. In this regard, transfection experiments have indicated that whereas Tcl1 is unable to directly induce Akt phosphorylation, it can effectively enhance Akt kinase activity (4). In conclusion, the present studies indicate that TCL1 plays a significant role in early embryo development and suggest that it may also contribute to the pathophysiology of male germ cells.
Acknowledgments
We thank Mauro Helmer Citterich and Maria Teresa Taffuri for technical assistance, Dr. Simona Torcia for help in quantitative immunofluorescence assays, Dr. Alessandra Minasi for initial embryo analysis, and Jay Rothstein for valuable comments and animal construction. This work has been supported by Associazione Italiana Ricerca sul Cancro, Telethon Grant D.102, Istituto Pasteur–Fondazione Cenci Bolognetti, COFIN 2000, PNR Tema 2 Oncologia, European Community Grant QLG2-CT-1999-0786, and National Institutes of Health Grants CA76259 and AI39816. M.D.C. is an Howard Hughes Medical Investigator, and S.-M.K. was a Howard Hughes Medical Institute Research Associate.
Abbreviations
- T-PLL
human prolymphocytic leukemia
- Met II
metaphase II
References
- 1.Russo G, Isobe M, Pegoraro L, Finan J, Nowell P C, Croce C M. Cell. 1988;53:137–144. doi: 10.1016/0092-8674(88)90495-3. [DOI] [PubMed] [Google Scholar]
- 2.Virgilio L, Narducci M G, Isobe M, Billips L G, Cooper M D, Croce C M, Russo G. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virgilio L, Lazzeri C, Bichi R, Nibu K, Narducci M G, Russo G, Rothstein J L, Croce C M. Proc Natl Acad Sci USA. 1998;95:3885–3889. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, Russo G, Tsichlis P, Croce C M. Proc Natl Acad Sci USA. 2000;97:3028–3033. doi: 10.1073/pnas.040557697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 6.Narducci M G, Stoppacciaro A, Imada K, Uchiyama T, Virgilio L, Lazzeri C, Croce C M, Russo G. Cancer Res. 1997;57:5452–5456. [PubMed] [Google Scholar]
- 7.Takizawa J, Suzuki R, Kuroda H, Utsunomiya A, Kagami Y, Joh T, Aizawa Y, Ueda R, Seto M. Jpn J Cancer Res. 1998;89:712–718. doi: 10.1111/j.1349-7006.1998.tb03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teitell M, Damore M A, Sulur G G, Turner D E, Stern M H, Said J W, Denny C T, Wall R. Proc Natl Acad Sci USA. 1999;96:9809–9814. doi: 10.1073/pnas.96.17.9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern M H, Soulier J, Rosenzwajg M, Nakahara K, Canki-Klain N, Aurias A, Sigaux F, Kirsch I R. Oncogene. 1993;8:2475–2483. [PubMed] [Google Scholar]
- 10.Pekarsky Y, Hallas C, Isobe M, Russo G, Croce C M. Proc Natl Acad Sci USA. 1999;96:2949–2951. doi: 10.1073/pnas.96.6.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugimoto J, Hatakeyama T, Narducci M G, Russo G, Isobe M. Cancer Res. 1999;59:2313–2317. [PubMed] [Google Scholar]
- 12.Hallas C, Pekarsky Y, Itoyama T, Varnum J, Bichi R, Rothstein J L, Croce C M. Proc Natl Acad Sci USA. 1999;96:14418–14423. doi: 10.1073/pnas.96.25.14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narducci M G, Virgilio L, Engiles J B, Buchberg A M, Billips L, Facchiano A, Croce C M, Russo G, Rothstein J L. Oncogene. 1997;15:919–926. doi: 10.1038/sj.onc.1201246. [DOI] [PubMed] [Google Scholar]
- 14.Hogan B, Bedginton R, Constantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 15.Bevilacqua A, Fiorenza M T, Mangia F. Development (Cambridge, UK) 2000;127:1541–1551. doi: 10.1242/dev.127.7.1541. [DOI] [PubMed] [Google Scholar]
- 16.Fiorenza M T, Mangia F. BioTechniques. 1998;24:618–623. doi: 10.2144/98244st02. [DOI] [PubMed] [Google Scholar]
- 17.Narducci M G, Pescarmona E, Lazzeri C, Signoretti S, Lavinia A M, Remotti D, Scala E, Baroni C D, Stoppacciaro A, Croce C M, Russo G. Cancer Res. 2000;60:2095–2100. [PubMed] [Google Scholar]
- 18.Bevilacqua A, Kinnunen L H, Bevilacqua S, Mangia F. Dev Biol. 1995;170:467–478. doi: 10.1006/dbio.1995.1230. [DOI] [PubMed] [Google Scholar]
- 19.Bensaude O, Babinet C, Morange M, Jacob F. Nature (London) 1983;305:331–333. doi: 10.1038/305331a0. [DOI] [PubMed] [Google Scholar]
- 20.Christians E, Campion E, Thompson E M, Renard J P. Development (Cambridge, UK) 1995;121:113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- 21.Pauken C M, Capco D G. Mol Reprod Dev. 1999;54:135–144. doi: 10.1002/(SICI)1098-2795(199910)54:2<135::AID-MRD5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 22.Minami N, Sasaki K, Aizawa A, Miyamoto M, Imai H. Biol Reprod. 2001;64:30–35. doi: 10.1095/biolreprod64.1.30. [DOI] [PubMed] [Google Scholar]
- 23.Conover J C, Temeles G L, Zimmermann J W, Burke B, Schultz R M. Dev Biol. 1991;144:392–404. doi: 10.1016/0012-1606(91)90431-2. [DOI] [PubMed] [Google Scholar]
- 24.Torres M, Stoykova A, Huber O, Chowdhury K, Bonaldo P, Mansouri A, Butz S, Kemler R, Gruss P. Proc Natl Acad Sci USA. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunstle G, Laine J, Pierron G, Kagami Si S, Nakajima H, Hoh F, Roumestand C, Stern M H, Noguchi M. Mol Cell Biol. 2002;22:1513–1525. doi: 10.1128/mcb.22.5.1513-1525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laine J, Kunstle G, Obata T, Noguchi M. J Biol Chem. 2002;277:3743–3751. doi: 10.1074/jbc.M107069200. [DOI] [PubMed] [Google Scholar]
- 27.Hoh F, Yang Y S, Guignard L, Padilla A, Stern M H, Lhoste J M, van Tilbeurgh H. Structure (London) 1998;6:147–155. doi: 10.1016/s0969-2126(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 28.Fu Z Q, Du Bois G C, Song S P, Kulikovskaya I, Virgilio L, Rothstein J L, Croce C M, Weber I T, Harrison R W. Proc Natl Acad Sci USA. 1998;95:3413–3418. doi: 10.1073/pnas.95.7.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gritti C, Dastot H, Soulier J, Janin A, Daniel M T, Madani A, Grimber G, Briand P, Sigaux F, Stern M H. Blood. 1998;92:368–373. [PubMed] [Google Scholar]
- 30.Bichi R, Shinton S A, Martin E S, Koval A, Calin G A, Cesari R, Russo G, Hardy R R, Croce C M. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W S, Xu P Z, Gottlob K, Chen M L, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]