Abstract
We demonstrated that a subterranean, visually blind mammal has a functional set of three Per genes that are important components of the circadian clockwork in mammals. The mole rat superspecies Spalax ehrenbergi is a blind subterranean animal that lives its entire life underground in darkness. It has degenerated eyes, but the retina and highly hypertrophic harderian gland are involved in photoperiodic perception. All three Per genes oscillate with a periodicity of 24 h in the suprachiasmatic nuclei, eye, and harderian gland and are expressed in peripheral organs. This oscillation is maintained under constant conditions. The light inducibility of sPer1 and sPer2, which are similar in structure to those of other mammals, indicates the role of these genes in clock resetting. However, sPer3 is unique in mammals and has two truncated isoforms, and its expressional analysis leaves its function unresolved. Per's expression analysis in the harderian gland suggests an important participation of this organ in the stabilization and resetting mechanism of the central pacemaker in the suprachiasmatic nuclei and in unique adaptation to life underground.
Life on Earth is adapted to cyclical phenomena imposed by the external environment (1). Most organisms have circadian systems that synchronize physiological events to the external 24-h cycle (2). The underlying molecular-genetic mechanisms of these clocks exhibit an extraordinary evolutionary conservation from cyanobacteria through plants, fruit flies, and mammals. All of these clock systems consist of autoregulatory transcriptional/translational feedback loops with positive/negative regulatory elements and similar genetic machinery (3, 4).
Two basic helix–loop–helix PAS (PER-ARNT-SIM) transcription factors, CLOCK and MOP3 (BMAL1), form the positive elements of the system and drive transcription of three Period (Per 1, 2, 3) and two Cryptochrome (Cry 1, 2) genes. The protein products of these genes are thought to be components of a negative feedback complex that inhibits the CLOCK/MOP3 heterodimer, thereby closing the circadian loop.
The enigma of circadian rhythms in a blind subterranean mammal is intriguing (5–7). We have already shown that a CLOCK/MOP3-driven clock exists in Spalax (8). Here we continue to decipher its circadian machinery.
The Evolutionary Model of Blind Subterranean Mammals
The blind subterranean mammals, mole rats of the Spalax ehrenbergi superspecies in Israel, consist of four species that have been studied multidisciplinarily as an evolutionary model of speciation and adaptation (5–7). Spalax lives in total darkness, yet, it perceives the daily and seasonal temporal cycles underground (9). Behaviorally, Spalax displays polyphasic and polytypic day-night activity patterns (10, 11) coupled with polymorphic (12) and seasonal‖ variation, supported by a unique photoperiodic perception mechanism (9). Spalax has a degenerated s.c. functional eye (13, 14), which, together with the harderian gland, participates in photoperiodic perception (9, 15–18). The retina harbors Rhodopsin (19, 20) and Coneopsin (21), adaptively effective in photoperiodic perception (22, 23), and expresses alfa-B-crystallin (24). The photic signals entrain c-fos in the suprachiasmatic nuclei (SCN) Zeitgeber (25) and can possibly activate circadian genes.
Evolutionarily, Spalax's perceptive brain structures (SCN and striatum) were expanded and sight pathways were drastically (>90%) reduced. The visual cortex was replaced by somatosensory cortex (26–28), Per homologous ACNGGN-repeats cycle in the hypothalamus (29) and melatonin precursors occur in the harderian and pineal glands and retina (30).
What is the genetic basis of circadian rhythmicity in Spalax? We cloned, sequenced, and unraveled the expression of the circadian Clock and MOP3 cDNAs of three species of the S. ehrenbergi superspecies in Israel (8). Both genes are relatively conserved, yet Clock displays a unique Q-rich area as compared with other mammals, assumed to function in circadian rhythmicity, and Spalax CLOCK/MOP3 dimer is less potent than its human counterpart in driving transcription.
Here we describe the cloning, sequencing, and expression of the three Period cDNAs of Spalax. Its three Per cDNAs are conserved, yet they show features unique to Spalax especially in Per3, Per1, and Per2 cycles in the SCN, eye, and harderian gland. Per3 is structurally unique among studied sighted mammals and awaits functional elucidation.
Materials and Methods
Animals.
We analyzed adults (100–150 g), belonging to Spalax judaei, (2n = 60) from Anza, Samaria (7). Field-trapped animals were kept at 22–24°C with seasonal photoperiod. We selected diurnal animals that were kept under a 12-h light/12-h dark cycle. For analysis of Per transcriptional activity in constant darkness, light was turned off at Zeitgeber time (ZT) 12, and animals were kept in the dark for at least 2 days before being killed under dim illumination (15-W safety red light). Light inducibility experiments were done on animals kept in light/dark for a week with a short light pulse (15 min, >200 Lx) at specified ZT followed by release into constant darkness. For gene induction analysis brains were taken 1 h after illumination. Each experiment was done on three sets of animals.
Cloning of Spalax Per cDNAs.
We cloned the three Spalax Per cDNAs by reverse transcription–PCR (RT-PCR) (31). Oligos were synthesized according to the ORF of the known human and mouse homologous sequences (GenBank accession nos. AF022991, AB002370, and AB047686 for human Per1, Per2, and Per3, respectively and AF022992, AF036893, and AB013605 for mouse Per1, Per2, and Per3, respectively). Whole brain total RNA was prepared by using the TriReagent RNA isolation reagent (Molecular Research Center, Cincinnati). First-strand cDNA was synthesized with oligo(dT) as a primer and SuperScript II reverse transcriptase enzyme (GIBCO/BRL). This cDNA product was taken for PCR by using TaqDNA polymerase (Appligene, Strasbourg, France). The annealing temperature, elongation time, and MgCl2 concentration were adjusted for each specific PCR. In the case of sPer3 isolation, we verified our RT-PCR results by also cloning through cDNA library screening (32). Spalax brain cDNA library in Lambda-TripleEx was screened by using a partial mPer3 cDNA as a probe. Sequencing was determined by thermocycling sequencing using di-deoxy nucleotide terminators (3700 DNA Analyzer, Perkin–Elmer/Applied Biosystems) at the sequencing unit of the Weizmann Institute of Science (Rehovot, Israel).
Evolutionary Analysis.
The evolutionary analysis of the Per cDNAs presented here is based on distances and divergence calculations (Wisconsin package version 10, GCG).
The distances program (33) calculates pairwise distances between aligned sequences expressed as substitutions per 100 bases or amino acids. To correct the distances for multiple substitutions at a site, we used Kimura's nucleic acid (33) and protein (34) methods.
The diverge program estimates the pairwise number of synonymous and nonsynonymous substitutions per site between two or more coding aligned nucleic acid sequences (35, 36).
In Situ Hybridization (ISH).
Tissues used for ISH were treated and examined as described in Albrecht et al. (37). The Spalax Per1 probe corresponded to nucleotides 615-1300, the sPer2 probe corresponded to nucleotides 85–605, and the probe of sPer3 corresponded to nucleotides 1751–2590.
Quantification was performed by densitometric analysis of hybridization signals on x-ray films with Scion (Frederick, MD) IMAGE 4.0.2 software. For silver grain images, tissue was visualized by fluorescence of Hoechst dye-stained nuclei, and silver grain signals were artificially colored for clarity. Quantitation of ISH results was analyzed with GraphPad (San Diego) prism software. Data sets were compared by ANOVA with subsequent Bonferroni correction for multiple comparisons, with P < 0.05 as the criterion of significance.
Quantitative RT-PCR.
For quantitation of the sPer genes expression in the harderian gland and the liver of Spalax, a quantitative RT-PCR was performed. Equal amounts of total RNA from animals killed at the relevant ZT points were taken for first-strand cDNA synthesis (see above). The cDNAs were synthesized by adding equal traces of [32P]dCTP to ensure equal amounts of cDNA templates in the PCRs. For the PCR we used oligos synthesized according to the sequence of the different sPer isolated clones. For each Per gene quantitation we used one sense 5′ oligo and two antisense 3′ oligos (3′[1] and 3′[2]), giving rise to two distinct products (450–600 bp). One set of oligos (5′ and 3′[1]) was used for the harderian gland and the other (oligos 5′ and 3′[2]) for the liver, in the same PCR tube. A second PCR was carried out by changing the 3′ oligo between the harderian and liver tissues. In each PCR, a 300-bp fragment of actin, as an internal control, was also synthesized, using specific actin oligos. Each cDNA was first tested for different PCR amplification cycles with the different sets of oligos. The final experimental PCRs were performed at the logarithmic phase of the reaction for each specific cDNA of interest (18–22 cycles). Each experiment was carried out on two RNA samples taken from two different individuals, and each PCR was repeated three times. The PCR products were then run on ethidium bromide/1.3% agarose gels. The gels were subjected to quantitation of the specific bands by the Eagle Eye II system (Stratagene). The system integrates the density of the ethidium bromide of a rectangle limiting a specific band in pixel values. The values received for the specific Per bands were normalized according to the values received for the actin bands (which were statistically equal in the different tissues and in the different reactions).
Results
Cloning and Structural Analysis of the Three Spalax Per (sPer) Genes.
Analysis of the ORFs of Spalax Per cDNAs revealed transcripts of 1,062 aa residues for sPER1 (GenBank accession no. AJ345059) and 1,248 aa for sPER2 (GenBank accession no. AJ345060). For sPER3 we isolated two truncated deduced proteins, one with 489 aa (isoform a, GenBank accession no. AJ345061) and the other with 583 aa (isoform b, GenBank accession no. AJ345062). Identified functional domains like the PAS domain and the basic helix–loop–helix motif are highly similar in sPER1 and sPER2, but the homology in sPER3 is low. The recently identified casein kinase 1ɛ (CK1ɛ) binding site of human PER2 and the five putative phosphorylation sites (AA 668, 671, 674, 677, and 680) (38) are conserved in PER proteins of Spalax, mice, and humans with the exception of sPER3. Hence sPER3 is probably not a substrate for the Spalax CK1ɛ ortholog.
Evolutionary Analysis of the Spalax Per Genes.
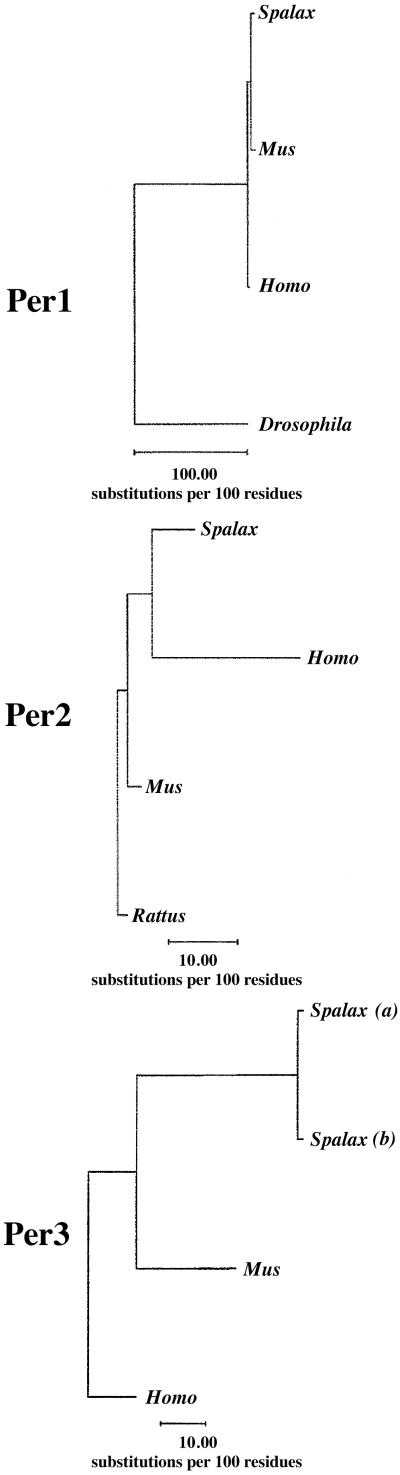
Protein trees of PER1, PER2, and PER3 in Spalax, mice, rats, and humans appear in Fig. 1. The Drosophila Per (GenBank accession no. X03636) was also compared, but it was very different from the three mammalian Per proteins. The computer program used (GCG10) estimated the distance of the dPer from its mammalian counterparts as maximal and beyond the accuracy of the method.
Figure 1.
Similarity tree of the three Per-deduced proteins. The unrooted tree depicts the similarity relationships of the three PER proteins (amino acids) in Spalax, mice, rats, humans, and Drosophila.
The estimated divergence time between Spalax and other rodents is much shorter than the divergence between humans and rodents. Therefore, we expected that the genetic distance between Spalax and mice or rats would be considerably smaller than between humans and these two rodents. We also expected that the distance between humans and Spalax and between humans and mice or rats would be similar. Any divergence from these expectations can suggest that additional factor(s) influence the rate of evolution of the Per genes of Spalax and, therefore, deviate from the phylogenetic divergence time.
Below we present the evolutionary analysis for each Per gene separately.
Per1.
There is agreement between the above evolutionary expectations and the PER1 proteins (Fig. 1 Top) and Per1 nucleotide sequence (tree not shown). The relative distances in protein and nucleotides between Spalax and mice are 75% (6.85 vs. 9.12) and 80% (10.51 vs. 13.18) of the distances between Spalax and humans, respectively. The distances between rodents (Spalax and mice) and humans are similar: 9.12 and 8.04 in the protein and 13.18 and 13.26 in the nucleotide sequence.
Per2.
Rat PER2 (GenBank accession no. MN031678) was also included and expected to be similar to mouse PER2. As in Per1, PER2 results were in agreement with the phylogenetic expectations (Fig. 1 Middle). The relative distances between Spalax and mice were 42% (11.55 vs. 27.54) or 59% (13.07 vs. 22.09) of the distances between Spalax and humans in protein and nucleotides, respectively. The distances between Spalax and rats were 46% (12.69 vs. 27.54) or 66% (14.6 vs. 22.09) of the distances between Spalax and humans in protein and nucleotides, respectively. The distances between rodents (mice, rats, and Spalax) and humans were also similar: 27.00, 27.70, and 27.54 for protein and 22.56, 23.58, and 22.09 for nucleotides, respectively.
Per3.
As mentioned, we cloned two truncated clones of Spalax Per3 (named a and b). Both clones start at the equivalent of mouse 468 bp (110 bp 3′ to the mouse ATG initiation codon). We could not isolate any further 5′ sequences either through RT-PCR or cDNA library screening. Both clones contain an insertion of 198 bp at position 1211 bp of mice that interrupts the ORF. Furthermore, sPer3a has two deletions, the prominent one is 432 bp in length starting at position 1478 bp of the mouse sequence. At the starting point of this deletion in sPer3a, sPer3b has a cluster of termination codons at any of the three reading frames. The apparent initiation ATG is located immediately after the deletion in the sPer3a or these termination codons in sPer3b. Omitting the changes in the sPer3b clone yielded a Spalax Per3 ORF, which is similar to that of mice and humans. It should be emphasized that similar products have never been obtained in negative control amplifications with templates generated without reverse transcriptase enzyme, eliminating the possibility of genomic DNA contamination. Furthermore, the sPer3 clones that were isolated from the Spalax brain cDNA library contain a shorter 3′ untranslated region than those of mice and humans, and in contrast to them, contain an adenylation site 940 bp 3′ to the termination TAA codon. The published 3′ untranslated regions of mice (1,164 bp) and humans (2,421 bp) do not reach the adenylation site. Southern blot analyses suggest that the Spalax sPer3 is probably a pseudogene (results not shown).
The nucleotide distances between Spalax Per3 and that of mice or humans are similar. The protein distances (Fig. 1 Bottom) between the two Spalax PER3 and mice or humans were 56.21 vs. 57.20 for sPER3a, respectively and 60.41 vs. 57.74 for sPER3b, respectively. Kimura's two-parameter nucleotide distance analysis (33) gave more than two substitutions per bp. Thus, the exact calculated value is meaningless and depends heavily on the assumptions of the correction factors. Nevertheless, the calculated distance of Spalax vs. mice is even larger than the calculated distance of Spalax vs. humans. The same is true for the distances calculated separately for synonymous and nonsynonymous substitutions.
Synonymous vs. Nonsynonymous Substitutions in the Per Family.
Our calculations show that Per2 has a ratio around 0.2, indicating that it attained optimum function before the divergence of the species. Per1 and Per3 have a ratio of 0.43 to 0.75, a relatively high ratio suggesting adaptive evolution. The Drosophila Per showed a ratio >1.0, indicating positive selection for a functional change.
sPer Genes Oscillate in the Spalax SCN.
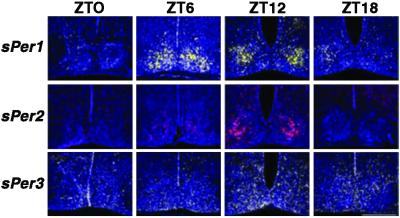
ISH with antisense riboprobes in the brain revealed a rhythmic pattern of expression for sPer1 and sPer2, mainly in the SCN but sPer3 is widely spread in the brain (Fig. 2). Maximal expression for sPer1 was at ZT6 and for sPer2 and sPer3 at ZT12. The amplitude of sPer3 expression was markedly lower (P < 0.05) than that of sPer1 and sPer2. The sense (control) riboprobes of the three sPer had a reproducible background hybridization that did not overlap with the antisense probe. No rhythmic expression with the sense riboprobe hybridization intensity was noted.
Figure 2.
Diurnal expression of sPer1, sPer2, and sPer3 of Spalax in the SCN: coronal sections through the brain. Blue color represents Hoechst-stained nuclei. (Top) The yellow signal shows the expression of Spalax Per1 over 24 h measured at 6-h time intervals (ZT0: lights on; ZT12: lights off). Maximal expression is seen at ZT6. (Middle) The red signal shows the expression of sPer2 in representative sections. Maximal expression is seen at ZT12. (Bottom) The green signal shows the expression of sPer3 in representative sections. Maximal expression is at ZT12. Note that sPer1 and sPer2 expression is mainly in the SCN, whereas sPer3 expression is weaker and spreads in different areas of the brain. (Magnification: ×20.)
sPer Genes Exhibit a Diurnal Oscillation in Spalax Peripheral Tissues.
Significant expression of the three sPer genes was noted, using RT-PCR, in lung, intestine, liver, harderian gland, eye, brain, and skeletal muscle (data not shown).
The Spalax Eye.
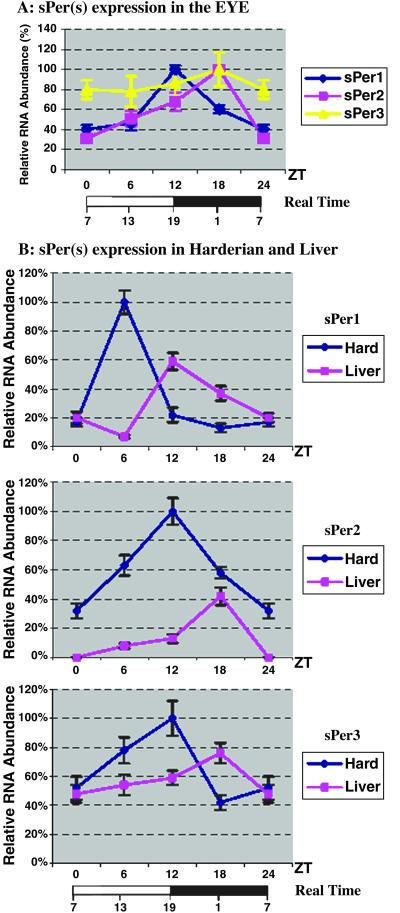
In the Spalax eye (Fig. 3A) ISH revealed a shift of 6 h in the expression maxima of all three Per genes compared with the expression in the SCN. Expression of sPer1 was maximal at ZT12 and of sPer2 and sPer3 at ZT18, but for sPer3, the amplitude of expression was very small. Thus the RNA rhythms for all three Per genes of Spalax are present in the photoperiodic retina, the site of light detection.
Figure 3.
Expression of Spalax Per genes in peripheral tissues. (A) Densitometric analysis of silver grain in situ staining in the eye. The maximal expression in the eye shows that the expression of the three sPer genes is shifted by 6 h as compared with their expression in the SCN (Fig. 2). The oscillation of sPer3 is very weak (P > 0.05). (B) Expression of the three sPER genes in the harderian gland and liver quantitated by RT-PCR. The expression peaks of all three sPer genes in the harderian gland are synchronized with the SCN (see Fig. 2), whereas the peak of expression in the liver is shifted by 6 h. As in the SCN, the difference in sPer3 expression in ZT6 and ZT12 is very small (P > 0.05).
The Spalax Harderian Gland.
Expression maxima in the harderian gland could be observed by quantitative RT-PCR (Fig. 3B) and ISH (data not shown) at the following ZTs: sPer1 at ZT6 and sPer2 and sPer3 at ZT12. Quantitative RT-PCR analysis in the liver (Fig. 3B) revealed rhythmic expression of sPer genes with maxima of sPer1 at ZT12 and of sPer2 and sPer3 at ZT18. The oscillation in the Spalax liver, as in its eye, shows a 6-h delay compared with the Spalax SCN. However, the circadian rhythm in the Spalax harderian gland is synchronous with the expression pattern in the Spalax SCN.
The Circadian Oscillation of sPer Genes Is Maintained in Constant Darkness.
sPer gene RNA levels in the SCN, eye, and harderian gland were studied at four time points over a 24-h period, on the second day in constant darkness (not shown). ISH revealed that RNA levels of all three sPer genes were rhythmic and the oscillation pattern under constant darkness was similar to that under 12-h light/12-h dark conditions. Highest levels were observed during the subjective day in the SCN at circadian time 6 for sPer1 and circadian time 12 for sPer2 and sPer3. The peak levels of mRNA in the eye were 6 h later than in the SCN, but were synchronized with the SCN in the harderian gland. The amplitude of sPer3 rhythmicity was markedly lower than that of sPer1 and sPer2 in all three tissues studied and only nearly significant (P > 0.05).
Differential Light Regulation of Spalax Per Genes.
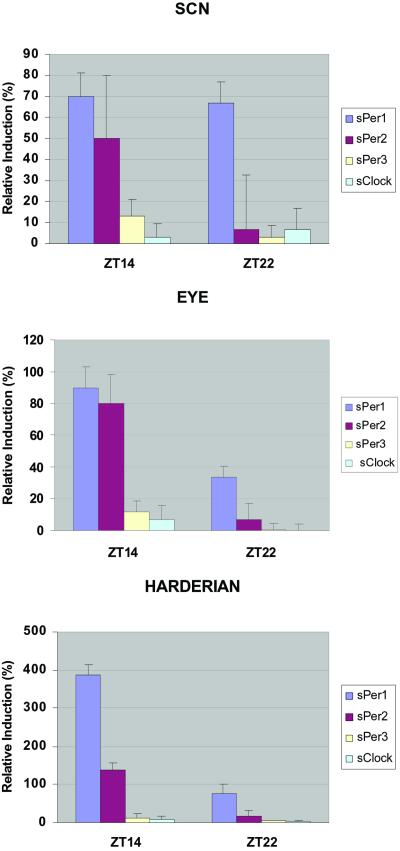
Previous studies have shown that mPer1 and mPer2 expression in the SCN is induced by exposure to light at night (39, 40), whereas mPer3 is unaffected (41). We examined inducibility of the sPer genes in the SCN, eye, and harderian gland by nocturnal light pulses at ZT14 and ZT22 (Fig. 4). These time points were chosen for study as light pulses at these times produce phase delays and advances in locomotor activity.
Figure 4.
Light inducibility of the three Spalax Per genes' expression. Animals were given a 15-min light pulse at ZT14 and ZT22 and killed 1 h later. Coronal sections through the brain and ISH were performed on treated (ZT14 pulse and ZT22 pulse) and control animals that were killed at the same time. Relative mRNA induction was analyzed by densitometric analysis of in situ silver grain staining. sClock inducibility is given as negative control. (Top) In the SCN, sPer1 is equally inducible at ZT14 and ZT22. sPer2 is inducible only at ZT14, whereas sPer3 gene shows no measurable changes in expression levels at the two time points. (Middle) Inducibility of sPer1, sPer2, and sPer3 in the eye is similar to that in the SCN. (Lower) In the harderian gland, sPer1 is highly inducible at ZT14 and less at ZT22 whereas sPer2 expression is light sensitive only at ZT14. sPer3 shows no light inducibility at either time point. Note the significantly (P < 0.01) higher induction of sPer1 in the harderian gland compared with the other tissues after light pulse at ZT14. Note the different y-axis scales in SCN, eye, and harderian.
Quantitation of the in situ results showed that 1 h after a light pulse at ZT14, sPer1 and sPer2 were significantly induced in all three tissues. Remarkably, the level of sPer1 induction in the harderian gland was significantly (P < 0.05) higher than in the SCN or the eye, reinforcing its great importance for the Spalax clock. One hour after the light pulse at ZT22 only sPer1 was significantly induced in the three tissues examined. Like the sClock gene, sPer3 gene was not light inducible either at ZT14 or ZT22 in the three tissues tested.
Discussion
Adaptive Selection on Per Genes in Spalax to Life Underground in Total Darkness.
Like other mammals, the subterranean blind Spalax has three Per genes.
The distances between sPer1 and sPer2 and those of other rodents or humans are as expected from their divergence time, which is estimated to be 40 million years ago and 80 million years ago, respectively (6). The distances between Spalax or mice and humans are similar, as expected. Generally, the distances calculated for Per2 are larger than the distances for Per1. In our analysis of synonymous vs. nonsynonymous substitutions we relied on Liberles et al. (42), who suggested using this ratio to reveal selection for change in enzymatic function. Data of Makalowski and Boguski (43) show that most rodents and human sequences have a ratio of 0.2. This finding indicates that such proteins, selected over millions of years, attained an optimum function before the divergence of rodents and primates and subsequent evolution was relatively conservative. They also considered ratios between 0.6 and 1.0 as suggesting a relaxation of functional constraint and selection. Our results show that sPer1 and sPer3 have a ratio of 0.4 to 0.6 and sPer2 has a ratio of about 0.2. Hence, presumably, sPer2 has not been changed to function in a visually blind mammal living in a dark environment with negligible light cues. However, the figures obtained for sPer1 and sPer3 may suggest that molecular changes in these genes were necessary to fulfill their expected adaptive function in darkness. If we combine the calculated distances and the high ratio of nonsynonymous substitutions, the result supports the hypothesis of adaptive changes caused by natural selection, possibly in response to life in darkness underground.
sPer3 evinces a different situation. First, this locus in Spalax underwent major changes of deletions and insertions, resulting in two isoforms exhibiting truncated coding regions; somewhat similar results were reported for the hPer4 pseudogene (44). However, sPer3 is very different from hPer4. Its insertion could not be identified with any known sequence, in contrast to the fossil MER-2 mobile element that is within the hPer4 locus (44). When the changes in the sPer3 are omitted, an ORF similar to mPer3 is obtained. When the distances were calculated from the aligned shortened sequences the phylogenetic expectations were not met. The distances between sPer3 and mPer3 are similar or even larger than the distances between sPer3 and hPer3. This finding may support adaptive selective changes in the evolution of this gene and not just the neutral accumulation of substitutions over time. The comparison of sPer3 expression patterns with sPer1 and sPer2, which was described here and is discussed below, raises questions as to the role that sPer3 plays in the Spalax circadian system.
The Functional Circadian Domains of Spalax Per Genes.
sPer1 and sPer2 contain all functionally relevant domains discovered in other mammalian PER proteins so far. The basic helix–loop–helix motif as well as the PAS domain consisting of PAS A, PAS B (39, 45), and PAC CK1ɛ binding and phosphorylation sites (38) are conserved, suggesting their role as a CK1ɛ substrate in the central mechanism of the Spalax circadian clockwork. Remarkably, the whole putative CK1ɛ binding site as well as the N-terminal basic helix–loop–helix motif are missing in sPer3. This finding supports a speculation about its role in the biological clock (45).
Because Spalax is visually blind and lives entirely underground, hence denied outside Zeitgeber information (6), a robust and precise internal clock is necessary for the animal to keep track of time under negligible light cues. Indeed, in situ data revealed that Spalax Per genes' expression is clearly rhythmic and maintained under constant conditions and in constant darkness. sPer1 and sPer2 expression in the Spalax brain is concentrated mainly in the SCN. However sPer3 is widely spread in different areas of the brain, similar to what has been reported for the mouse Per3 (41), and its oscillation levels are less prominent than those of sPer1 and sPer2. These findings raise a question as to the role of Per3 in the circadian system. The central pacemaker of the SCN signals time to the retina and peripheral clocks, as in the liver, where the circadian genes' expression follows its rhythm with a delay of 4–6 h (41, 46). The blind Spalax Per genes' expression is similar, and our results also show a lag of 6 h in the peak expression of the Per genes in the retina and liver. Although the exact role of Spalax Per3 in the maintenance of the circadian rhythm remains unresolved, it may prove substantial for time keeping underground.
The Harderian Gland: A Circadian Center in Spalax.
Noteworthy, mRNA levels in the Spalax hypertrophic harderian gland oscillate synchronously with the SCN. Similar synchronization in the peak levels of mRNA in the SCN and the harderian gland was seen also for Spalax MOP3 gene (8), the dimerization partner of the CLOCK protein (47) and an essential component of the circadian pacemaker in mammals (48). Furthermore, sPer1 inducibility in the harderian gland after light pulse during the dark phase of the clock cycle is much higher than in the SCN. The harderian gland of Spalax is extraordinarily enlarged and occupies the entire eye socket, presumably as an adaptation to life underground. Previous studies (15) suggested that the harderian gland of Spalax is a possible photoreceptor and photoperiodic organ. Given its exposed position directly behind the atrophied minute eye, it seems likely that the harderian gland has a prominent role in the Spalax clock mechanism. This gland may demonstrate the extreme of evolutionary progression during the adaptive morphological and molecular reorganization for life underground (6).
Differential Regulation of Spalax Per Genes.
Light inducible experiments in Spalax reveal that sPer1 is light inducible both early (ZT14) and late (ZT22) at night, whereas sPer2 is light inducible only at the beginning of the night. This result is in accordance with the findings of Albrecht et al. (39). sPer3 levels are unresponsive to light pulses applied throughout the dark phase of the circadian cycle. This differential regulation among the three sPer genes suggests that each has a different regulatory function in the SCN. The behavioral effects of photic stimuli at these two time points (ZT14 and ZT22) have been characterized in mice (49, 50). Light pulse at ZT14 causes phase delays in behavioral rhythms and light pulse at ZT22 causes phase advances. Our results indicate a role of sPer1 in both phases and of sPer2 in the phase-delay mechanism. Our in situ data provide a molecular confirmation of previous behavioral studies in Spalax (11) and link the activity pattern of these species with the cellular cycling in sighted animals. The poor sPer3 oscillation and the absence of light influence on sPer3 expression levels may suggest a role for sPer3 outside the central pacemaker. This finding is consistent with data from Per3-deficient mice (51, 52) but may prove an adaptation to life underground and deserves further critical studies.
All three Spalax Per genes are expressed in a wide variety of nonneural tissues as previously shown in Drosophila (53, 54) and mice (39, 41). In three of these tissues (liver, eye, and harderian gland) we found that the RNA levels for the three sPer genes oscillate. Oscillation of sPer genes in the eye, the target organ of light absorption, is rational. As we already suggested, the oscillation of the sPer genes in the Spalax harderian gland is in accordance with previous results, suggesting an important role of this tissue in circadian maintenance (15) and merits further intensive study. The oscillation in the liver and the widespread expression in other peripheral tissues that were examined suggest the existence of clocks outside the SCN.
Molecular-Genetic Tinkering of Circadian Genes in a Blind Mammal.
This study substantiates the claim that the blind subterranean Spalax needs a photoperiodic system to perceive daily and seasonal cycles. It has retained a functional retina with effective visual pigment genes signaling to the SCN (14, 17–20, 23, 24) presumably induced by the small amount of photons that penetrate underground to an otherwise dark environment. Circadian genes in the retina, harderian gland, SCN, and other tissues, including Clock (8) and the three Per genes described here, process the light signals and translate them into the unique behavioral repertoire of Spalax, including polyphasic, polymorphic, and seasonal behavioral phenotypes. In this respect we should emphasize that Spalax exhibits naturally occurring predominantly either diurnal or nocturnal individuals. Currently we are studying the expression pattern of the Per genes in naturally occurring nocturnal animals or on diurnal animals after a phase shift of light. The mosaic evolution of the Spalax eye (17, 18), harderian gland (30), and brain (26–28) and its circadian genes provide a striking model of tinkering evolution at both the molecular and organismal levels. From an evolutionary perspective the genetic basis of circadian rhythms in the blind subterranean Spalax may be different from that of strictly diurnal or nocturnal and sighted mammals. Identification of the circadian genes of blind Spalax might advance insights into the structure and evolution of the circadian organization in mammals, including humans, at the molecular level and their ecological causation. Remarkably, the colonization of the subterranean dark ecological zone by Spalax did not obliterate the conservative circadian rhythmicity and its genetic basis of photoreceptiveness and clock genes. All of the circadian machinery was adaptively transformed to perceive light in darkness.
Acknowledgments
We thank Dr. Z. S. Sun for his initial help with the cloning of the Spalax Per1 probe. We are also grateful to Mr. Michael Margulis for his help with the computer graphic work. This work was supported by the Max Planck Society and Deutsche Forschungsgemeinschaft Grant AL549/1-1 (to U.A.) and the Ancell-Teicher Research Foundation for Genetics and Molecular Evolution (to E.N.).
Abbreviations
- SCN
suprachiasmatic nuclei
- ZT
Zeitgeber time
- RT-PCR
reverse transcription–PCR
- ISH
in situ hybridization
- CK1ɛ
casein kinase 1ɛ
Footnotes
References
- 1.Lowrey L P, Takahashi J S. Annu Rev Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- 2.Pittendrigh C S. Annu Rev Physiol. 1993;55:15–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Hastings M, Maywood E S. BioEssays. 2000;22:23–31. doi: 10.1002/(SICI)1521-1878(200001)22:1<23::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Nevo E. Evol Biol. 1991;25:1–125. [Google Scholar]
- 6.Nevo E. Mosaic Evolution of Subterranean Mammals: Regression, Progression, and Global Convergence. Oxford: Oxford Univ. Press; 1999. [Google Scholar]
- 7.Nevo E, Ivanitskaya E, Beiles A. Adaptive Radiation of Blind Subterranean Mole Rats. Leiden: Backhuys; 2001. [Google Scholar]
- 8.Avivi A, Albrecht U, Oster H, Joel A, Beiles A, Nevo E. Proc Natl Acad Sci USA. 2001;98:13751–13756. doi: 10.1073/pnas.181484498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nevo E. In: Principles of Animal Design. Weibel E R, Taylor C R, Bolis C, editors. Cambridge, U.K.: Cambridge Univ. Press; 1998. pp. 288–298. [Google Scholar]
- 10.Nevo E, Guttman R, Haber M, Erez E. J Mamm. 1982;63:453–463. [Google Scholar]
- 11.Tobler I, Herrmann M, Cooper H M, Negroni J, Nevo E, Achermann P. Behav Brain Res. 1998;96:173–183. doi: 10.1016/s0166-4328(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shlomo R, Ritte U, Nevo E. Behav Genet. 1995;25:239–245. doi: 10.1007/BF02197182. [DOI] [PubMed] [Google Scholar]
- 13.Haim A, Heth G, Pratt H, Nevo E. J Exp Biol. 1983;107:59–64. doi: 10.1242/jeb.107.1.59. [DOI] [PubMed] [Google Scholar]
- 14.Sanyal S, Jansen H G, de Grip W G, Nevo E, De Jong W W. Invest Ophthalmol Visual Sci. 1990;31:1398–1404. [PubMed] [Google Scholar]
- 15.Pevet P, Heth G, Haim A, Nevo E. J Exp Zool. 1984;232:41–50. doi: 10.1002/jez.1402320106. [DOI] [PubMed] [Google Scholar]
- 16.Rado R, Gev H, Goldman B D, Terkel J. In: Photobiology. Riklis E, editor. New York: Plenum; 1991. pp. 581–589. [Google Scholar]
- 17.Cooper H M, Herbin M, Nevo E. Nature (London) 1993;361:156–159. doi: 10.1038/361156a0. [DOI] [PubMed] [Google Scholar]
- 18.Cooper H M, Herbin M, Nevo E. J Comp Neurol. 1993;328:313–350. doi: 10.1002/cne.903280302. [DOI] [PubMed] [Google Scholar]
- 19.DeGrip W J, Janssen J J M, Foster R G, Korf H W, Rothschild K J, Nevo E, de Caluwe G L J. In: Signal Transduction in Photoreceptor Cells. Hargrave P A, Hofmann K P, Kaupp U B, editors. Berlin: Springer; 1992. pp. 43–59. [Google Scholar]
- 20.Janssen J W H, Bovee-Geurts P H M, Peeters Z P A, Bowmaker J K, Cooper H M, David-Gray Z K, Nevo E, DeGrip W J. J Biol Chem. 2000;275:38674–38679. doi: 10.1074/jbc.M008254200. [DOI] [PubMed] [Google Scholar]
- 21.Argamaso S M, Froehlich A C, McCall M A, Nevo E, Provencio I, Foster R G. Biophys Chem. 1995;56:3–11. doi: 10.1016/0301-4622(95)00009-m. [DOI] [PubMed] [Google Scholar]
- 22.David-Gray Z K, Janssen J W H, Nevo E, Foster R, G. Nat Neurosci. 1998;12:655–656. doi: 10.1038/3656. [DOI] [PubMed] [Google Scholar]
- 23.David-Gray Z K, Cooper H M, Jannsen J W H, Nevo E, Foster R G. FEBS Lett. 1999;461:343–347. doi: 10.1016/s0014-5793(99)01455-6. [DOI] [PubMed] [Google Scholar]
- 24.Avivi A, Joel A, Nevo E. Gene. 2001;264:45–49. doi: 10.1016/s0378-1119(00)00603-x. [DOI] [PubMed] [Google Scholar]
- 25.Vuillez P, Herbin M, Cooper H M, Nevo E, Pevet P. Brain Res. 1994;654:81–84. doi: 10.1016/0006-8993(94)91573-3. [DOI] [PubMed] [Google Scholar]
- 26.Rehkamper G, Necker R, Nevo E. J Comp Neurol. 1994;347:570–584. doi: 10.1002/cne.903470408. [DOI] [PubMed] [Google Scholar]
- 27.Frahm H D, Rehkamper G, Nevo E. J Brain Res. 1997;38:209–222. [PubMed] [Google Scholar]
- 28.Mann M D, Rehkamper G, Reinke H, Fraham H D, Necker R, Nevo E. J Brain Res. 1997;38:47–59. [PubMed] [Google Scholar]
- 29.Ben-Shlomo R, Ritte U, Nevo E. Behav Genet. 1996;26:177–184. doi: 10.1007/BF02359895. [DOI] [PubMed] [Google Scholar]
- 30.Balemans M G M, Pevet P, Legerstee W C, Nevo E. J Neural Transm. 1980;49:247–255. doi: 10.1007/BF01252129. [DOI] [PubMed] [Google Scholar]
- 31.Veres G, Gibbs R A, Scherer S E, Caskey C T. Science. 1987;237:415–417. doi: 10.1126/science.3603027. [DOI] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 33.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 34.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Li W H, Wu C I, Luo C C. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 36.Li W H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 37.Albrecht U, Lu H-C, Revelli J-P, Xu X-C, Lotan R, Eichele G. Human Genome Methods. New York: CRC; 1998. pp. 93–119. [Google Scholar]
- 38.Toh K L, Jones C R, He Y, Eide E J, Hinz W A, Virshup D M, Ptacek L J, Fu Y H. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 39.Albrecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 40.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 41.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 42.Liberles D A, Schreiber D R, Govindarajan S, Chamberlin S G, Benner S A. Genome Biol. 2001;2:1–6. doi: 10.1186/gb-2001-2-4-preprint0003. [DOI] [PubMed] [Google Scholar]
- 43.Makalowski W, Boguski M S. Proc Natl Acad Sci USA. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gotter A L, Reppert M S. Mol Brain Res. 2001;92:19–26. doi: 10.1016/s0169-328x(01)00115-2. [DOI] [PubMed] [Google Scholar]
- 45.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. Nature (London) 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 46.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 47.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunger M K, Wilsbacher L D, Moran S M, Clendenin C, Radcliffe L A, Hogenesch J B, Simon M C, Takahashi J S, Bradfield C A. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz W J, Zimmerman P. J Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albrecht U, Zheng B, Larkin D, Sun Z S, Lee C C. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 51.Shearman L P, Jin X, Lee C, Reppert S M, Weaver D R. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae K, Jin X, Maywood E S, Hastings M H, Reppert S M, Weaver D R. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 53.Hardin P E. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plautz J D, Kaneko M, Hall J C, Kay S A. Science. 1997;279:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]