Abstract
Because of their extremely low nucleotide mutation rates, plant mitochondrial genes are generally not expected to show variation within species. Remarkably, we found nine distinct cytochrome b sequence haplotypes in the gynodioecious alpine plant Silene acaulis, with two or more haplotypes coexisting locally in each of three sampled regions. Moreover, there is evidence for intragenic recombination in the history of the haplotype sample, implying at least transient heteroplasmy of mitochondrial DNA (mtDNA). Heteroplasmy might be achieved by one of two potential mechanisms, either continuous coexistence of subgenomic fragments in low stoichiometry, or occasional paternal leakage of mtDNA. On the basis of levels of synonymous nucleotide substitutions, the average divergence time between haplotypes is estimated to be at least 15 million years. Ancient coalescence of extant haplotypes is further indicated by the paucity of fixed differences in haplotypes obtained from related species, a pattern expected under trans-specific evolution. Our data are consistent with models of frequency-dependent selection on linked cytoplasmic male-sterility factors, the putative molecular basis of females in gynodioecious populations. However, associations between marker loci and the inferred male-sterility genes can be maintained only with very low rates of recombination. Heteroplasmy and recombination between divergent haplotypes imply unexplored consequences for the evolutionary dynamics of gynodioecy, a widespread plant breeding system.
Plant mitochondrial genomes are characterized by very low nucleotide mutation rates, in contrast to their fast structural evolution attributable to intramolecular rearrangements (1–5). Given the slow pace of sequence evolution, coexistence of distinct mitochondrial sequence haplotypes within species would imply their ancient coalescence, presumably because of particular selective or demographic reasons. In the absence of negative frequency-dependent selection (or other types of balancing selection), population genetic theory predicts little or no within-species DNA sequence polymorphism for plant mitochondrial genes. As far as we are aware, no study has sequenced plant mtDNA from multiple individuals and populations to date, presumably because of the prevailing assumption that little mitochondrial polymorphism will exist.
The unusual mode of sex determination in gynodioecious plants, together with patterns of selection at the phenotypic level, might help to maintain within-species DNA polymorphism. Populations of our study species Silene acaulis (moss campion, Caryophyllaceae) typically contain hermaphroditic and female plants, and thus exhibit a gynodioecious breeding system (6–8). In several agricultural model systems, so-called cytoplasmic male sterility (CMS) factors have been shown to be chimeric mitochondrial genes, whose protein products are believed to disrupt proper pollen development, thus converting otherwise hermaphroditic plants to functional females (9, 10). Under the cytonuclear system of sex determination believed to be prevalent in gynodioecious populations, individuals bearing a CMS gene are females (“male-steriles”) unless they carry appropriate nucleus-encoded “restorer” alleles (11–13). Progeny sex-ratio data from crosses within populations of other gynodioecious species suggest the frequent coexistence of two or more functionally different CMS cytotypes and their corresponding nuclear restorer loci (13–15), but the actual CMS genes have not been identified in any of these natural systems.
Mathematical models designed to capture the evolutionary dynamics of cytonuclear sex determination, in particular the required joint polymorphism for nuclear restorer genes and cytoplasmic male-sterility factors, have suggested the possibility of negative frequency-dependent selection on functionally different CMS genes (16–18). Intuitively, this prediction can be understood in terms of an interplay between time-lagged arrival of appropriate restorer alleles (through mutation or migration) in response to CMS factors, and the diminishing transmission advantage of particular CMS cytotypes once these become locally common. The latter (frequency-dependent) effect might be mediated by pollen limitation at high female frequencies and/or the typically lower seed fitness of hermaphrodites (6–8, 11, 19). Under such a scenario, different CMS cytotypes might be maintained for very long periods of time, particularly under metapopulation structure (20, 21). Provided there is long-term cotransmission by means of uniparental inheritance (22, 23), one would expect nucleotide variants at other mitochondrial genes to accumulate between the CMS cytotypes that are under selection.
Here, we present evidence for abundant mitochondrial sequence diversity in natural populations, which tends to be partitioned into few, distinct haplotypes. Jointly, these results are broadly consistent with the operation of negative frequency-dependent selection on linked mitochondrial genes that are involved in sex determination.
Materials and Methods
Southern Hybridization.
Using conventional Southern hybridization techniques, we discovered two mitochondrial restriction fragment length polymorphism (RFLP) haplotypes in a Colorado population of the circumpolar, long-lived alpine plant Silene acaulis (unpublished data). Briefly, total genomic DNA was extracted from 1–3 g of fresh leaf tissue from individual plants grown at the Indiana University greenhouse, by using a modified hexadecyltrimethylammonium bromide (CTAB) procedure and CsCl-gradient ultracentrifugation (24). The genomic DNA was digested with one of several restriction enzymes (HindIII, BamHI, PvuII, EcoRI, PstI), electrophoresed through 0.8% agarose gels, and transferred to Nytran membranes by standard capillary blotting procedures. Membranes were prehybridized and hybridized at 65°C, and washed in 2× SSC at 65°C after DNA hybridization. Hybridization probes (S. acaulis PCR products from cob, coxI, coxII, atp6, and atp9) were made by 32P-labeling using random oligonucleotide primers. We obtained strong hybridization with all gene probes, indicating that these genes in fact reside in the mitochondrial genome (25).
In our RFLP study on the Colorado population, 42 plants shared one multigene pattern (“majority haplotype”), and 5 plants shared another pattern (“minority haplotype”). These divergent haplotypes exhibit distinct banding patterns when probed with four of the five genes, coxII being the only exception. In this report, we limit ourselves to data from a sequencing study that was motivated by these initial observations. Guided by an apparent polymorphism for a HindIII restriction site within the probe, we chose to sequence the apocytochrome b (cob) gene (see below). However, all sequenced plants proved to be monomorphic for the HindIII restriction site, and none of the uncovered sequence polymorphism should affect the cob RFLP patterns (results not shown). This finding suggests that polymorphisms in the genomic regions flanking the structural genes that were used as probes are responsible for the RFLPs.
Plant Material.
Our Colorado sample is from Pennsylvania Mountain (Park County); sequences were obtained from all five plants of known minority haplotype and six plants of known majority haplotype (see above). The majority haplotype was also found in two small samples from additional Colorado populations, Weston Pass (Park County; n = 2) and Loveland Pass (Clear Creek County; n = 2). Additionally, we included a geographically distant sample from near Kennicott in the Wrangell Mountains, Alaska (n = 20; see ref. 7 for site details). A third, composite sample of northern European plants was obtained from Berlevåg in northeastern Norway (n = 12) and from near Kevo Subarctic Research Institute in northern Finland (n = 8). Plants from Alaska and Europe were subjected to Southern hybridization, yielding many additional RFLP haplotypes not found in the Colorado population (results not shown).
To assess the genealogical affinities of haplotypes found in S. acaulis to those from related taxa, we sequenced four additional caryophyllids. These outgroup sequences were generated from single individuals of the congeneric dioecious Silene latifolia, Dianthus deltoides (Caryophyllaceae), Stellaria sp. (Caryophyllaceae), and Beta vulgaris (Chenopodiaceae). The B. vulgaris sequence matches the sequence previously deposited in GenBank (accession number NC_002511).
PCR Conditions and Sequencing.
PCRs were performed with 20–50 ng of genomic DNA in 25-μl reactions, with 1 mM MgCl2, 1 mM each dNTP, 2 μM each primer, and Taq DNA polymerase in an Idaho Air Thermo-Cycler (Idaho Technology, Salt Lake City). Denaturation was at 92°C for 20 s, annealing at 53°C for 30 s, and extension at 72°C for 1.5 min (40 cycles), followed by a final extension for 5 min. We used two PCR primers (5′-AGCATTTGATAGATTATCCAACC-3′ and 5′-ATTCCTCTTCCAACTCGTCC-3′) and four additional internal sequencing primers (5′-GCTCCTAATGTTTTG-3′, 5′-CAACAGCGTAGAACACATTATGA-3′, 5′-CTCATCTGACCCCAAGG-3′, and 5′-GAATGGGCGTTATGGC-3′). The PCR product spans 88% of the cob coding region, with a sequence length of 1,041 base pairs (bp) between the PCR primers. After gel purification using the Gene-Clean kit (Bio 101), both strands of all PCR products were sequenced directly on an ABI 377 automated sequencer (Applied Biosystems). Where distinct cob sequence haplotypes were found in only one or two plants, we sequenced known maternal siblings of the plants in question. In all five such cases, the original cob sequence was verified.
Statistical Analyses.
All standard population-genetic analyses of the sequence data, including neutral coalescent simulations (26), were performed by using the program package DnaSP, version 3.50 (27). Sequence alignment, including outgroup sequences obtained from GenBank database, was performed with the command xpileup in the GCG package [Wisconsin Package version 10.0, Genetics Computer Group (GCG), Madison, Wisconsin]. Additional methods of inference are described in Results and Discussion.
Results and Discussion
The previously identified Colorado RFLP haplotypes were found to differ at six nucleotide sites of 1,041 bp of the cob coding region. These remarkable findings are strengthened and generalized by the corresponding results from the Alaskan and the two northern European populations. Sequencing the cob gene for 20 randomly selected individuals from each of these two geographically distant regions, we uncovered seven additional cob haplotypes. Overall, the nine haplotypes detected within S. acaulis include 22 segregating sites (Table 1). The RFLP data for haplotype AK-4, however, indicate the presence of two mitochondrial copies of the cob gene. We therefore excluded this haplotype from all quantitative analyses reported in this paper, although none of our conclusions are affected by this treatment (results not shown). Importantly, we found two or more haplotypes coexisting in all sampled populations. Hence, S. acaulis harbors considerable mitochondrial sequence polymorphism within local populations as well as among distant geographic regions.
Table 1.
Haplotype structure in Silene acaulis and comparison with two caryophyllid outgroup sequences
| Haplotype | Nucleotide at polymorphic position
|
n | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 102 | 241 | 244 | 246 | 499 | 510 | 591 | 592 | 593 | 735 | 858 | 861 | 894 | 909 | 966 | 981 | 987 | 997 | 1002 | 1005 | 1033 | 1034 | 1039 | ||
| CO-2 | A | T | T | T | T | C | C | T | T | C | G | A | A | G | C | G | T | A | G | T | A | C | C | C | 15 |
| AK-1 | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 |
| EU-2 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | A | . | C | T | . | . | . | . | . | 6 |
| CO-1 | . | . | . | . | . | . | . | G | G | . | . | . | . | . | A | . | . | C | T | . | T | . | . | . | 17 |
| AK-3 | . | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | . | C | T | . | . | . | . | . | 2 |
| AK-4 | C | C | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | G | . | G | G | A | 1 |
| AK-2 | C | . | . | . | . | . | A | . | . | A | . | . | . | . | . | . | G | C | T | . | . | . | . | . | 2 |
| EU-3 | C | . | . | . | . | . | A | . | . | T | T | T | . | . | . | . | G | C | T | . | . | . | . | . | 1 |
| EU-1 | C | . | C | G | G | . | . | . | . | . | T | T | . | . | . | . | . | C | T | . | . | . | . | . | 13 |
| S. latifolia | . | . | C | G | G | . | . | . | . | . | T | T | G | . | . | . | . | C | T | . | . | . | . | . | 1 |
| Stellaria sp. | . | . | C | G | G | G | . | . | G | . | T | T | . | . | A | . | . | C | T | . | T | . | . | . | 1 |
| Substit. type | r | s | r* | r | s | r | s | r | r | r,r | s | r | s | s | s | s | s | s | r | s | r | r | r | r | |
The first nine haplotypes represent the diversity found in S. acaulis, while the last two represent single individuals of the congeneric S. latifolia and Stellaria sp., respectively. S. acaulis haplotypes are named after the three geographic regions (CO, Colorado; AK, Alaska; EU, northern Europe). Dots indicate identity with the reference sequence CO-2, which was found both in Colorado (Colorado minority haplotype) and Alaska (n = 13 of 20 plants); all other haplotypes are exclusive to one of the three regions. To achieve equal representation of all geographic regions for statistical tests, the Colorado sample was considered to consist of 17 individuals with haplotype CO-1 and two individuals with haplotype CO-2; these proportions reflect the larger RFLP sample (see text). On the last line, nucleotide substitutions are categorized as either synonymous (s) or replacement (r). Only the polymorphism at position 241 is likely to be affected by RNA editing (28, 29), thus converting a replacement into a synonymous substitution (asterisk). Note that there is but a single fixed difference to each of the two outgroup sequences [positions 499 (Stellaria) and 861 (S. latifolia)]; the remaining 22 variable sites are polymorphic within S. acaulis.
Evidence for Recombination.
Intragenic sequence variation is needed for testing for recombination. It seems likely that genetic exchanges among mitochondrial genomes can occur as nonreciprocal gene conversion, rather than exclusively as homologous crossing-over (30–32), but for simplicity we refer to all such processes as “recombination.”
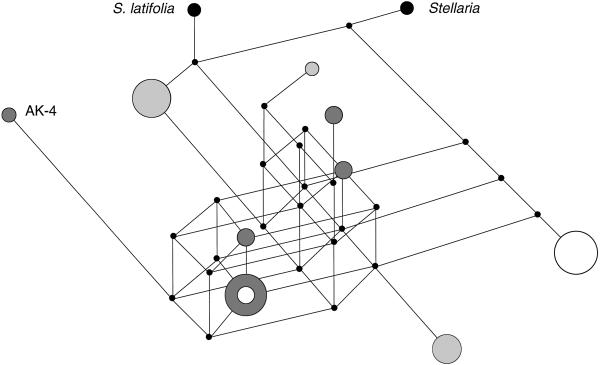
In population-genetic analyses, recombination within a segment flanked by a pair of polymorphic nucleotide sites can be inferred when all four possible gametic (haploid) combinations are observed (“four-gamete test”; ref. 33). Table 2 details the evidence for intragenic recombination based on eight S. acaulis haplotypes (excluding AK-4; see above). There are eight pairs of polymorphic sites satisfying the four-gamete criterion, and their respective locations and overlap along the gene translate into a minimum of three recombination events in the history of the sample (33). The considerable level of homoplasy entailed in the observed number of four-gamete pairs is evident in a median network depicting all nine haplotypes and the two closest outgroup sequences (Fig. 1).
Table 2.
Evidence for intragenic recombination
| Haplotype | Nucleotide at variable position
|
n | ||||||
|---|---|---|---|---|---|---|---|---|
| 10 | 510 | 735 | 858 | 981 | 987 | 997 | ||
| CO-2 | A | C | G | A | T | A | G | 15 |
| AK-1 | . | A | . | . | . | . | . | 2 |
| EU-2 | . | . | . | . | . | C | T | 6 |
| CO-1 | . | . | . | . | . | C | T | 17 |
| AK-3 | . | A | . | T | . | C | T | 2 |
| AK-2 | C | A | . | . | G | C | T | 2 |
| EU-3 | C | A | T | T | G | C | T | 1 |
| EU-1 | C | . | T | T | . | C | T | 13 |
| Caryophyllids | A | C | T | T | T,G | C,T,G | T,C,G | 4 |
| Other angiosperms | C | C | T | T | T,G | T | T,C | 9 |
| Liverwort | A | T | T | T | T | T | G | 1 |
Only sites constituting the eight pairs with four gametic types are listed. Dots indicate sequence identity with the reference haplotype (CO-2). Formal tests of recombination indicate a minimum of three recombination events between sites (10, 510), (510, 735), and (858, 981). None of these five critical sites is potentially affected by RNA editing (see Table 1 and supporting information). For the outgroup sequences considered in the lower third of the table, refer to the total alignment in supporting information, which is published on the PNAS web site. Within caryophyllids and other angiosperms, all encountered nucleotide variants are listed.
Figure 1.
Median network depicting all nine cob haplotypes from S. acaulis and the two closest outgroup sequences. White, Colorado; dark gray, Alaska; light gray, Europe. The median network was generated with NETWORK3.0, using the RM algorithm and setting the reduction threshold r > 2 (ref. 34; http://www.fluxus-engineering.com). The extensive homoplasy is visualized by the network reticulations (“cycles”) in the lower part of the diagram. Small black circles represent median vectors. The reference haplotype (CO-2) was found in both Alaska and Colorado (small white disk within larger dark-gray disk). Haplotype AK-4 was not included for tests of recombination and other analyses (see text).
The four-gamete test rests on the applicability of the infinite-sites model, and an alternative explanation for the observed level of homoplasy would be recurrent mutation at the nucleotide sites in question. However, our analysis of available outgroup sequences from angiosperms and the liverwort Marchantia polymorpha gives no indication that these sites are hypervariable (see Fig. 2, which is published as supporting information on the PNAS web site, www.pnas.org). The nucleotide positions critical for the inference of recombination (positions 10, 510, 735, 858, and 981) are well conserved across the assemblage of sequenced angiosperms (Table 2). In fact, four of the five nucleotide bases found in the liverwort are shared with S. acaulis.
Moreover, tests of recombination that do not rely on the infinite-sites model also indicate that recombination has occurred in the history of the haplotype sample. The homoplasy test compares the observed number of homoplasies in a maximum-parsimony tree of the sequences with the number expected under clonal evolution, based on an estimate of effective site number (35). It has been shown to be a powerful test under conditions of low sequence divergence, but heterogeneity in the substitution rate among sites would cause the expected number of homoplasies to be underestimated (36). Successive analyses using all sites, using only third-codon positions, and using only synonymous sites all reject the null hypothesis of no recombination (Table 3). Hence, evidence for recombination is unlikely to have been falsely generated by selection for certain replacement polymorphisms. Similarly, a recently published method for detecting recombination on the basis of the permutation of segregating sites by location yields significant support for recombination in our data (likelihood permutation test, ref. 32; Table 3). This method appears to be more powerful than other permutation-based tests, and unlike the homoplasy test, is robust to misspecification of the underlying model of sequence evolution (32).
Table 3.
Formal tests for recombination
| Parameter | Homoplasy test (ref. 35) | LPT (ref. 32) |
|---|---|---|
| True homoplasies | 5 | — |
| Expected homoplasies | 0.42 | — |
| Polymorphic sites | 17 | 17 |
| Se | 330 | — |
| Probability, P | <0.001 | ∼0.001–0.004 |
Se, effective number of sites, estimated by using the liverwort cob sequence as an outgroup (35). LPT, Likelihood permutation test (32). These tests were conducted with all 1,041 sites—i.e., including both silent and replacement polymorphisms. In addition, the homoplasy test yields significant results when applied to only third-codon positions (P = 0.001), and synonymous sites (P < 0.01). —, Not applicable.
Given the collective weight of this evidence, it appears likely that intragenic recombination and/or gene conversion, rather than recurrent mutation, generated the high homoplasy in our data. Recent claims of recombination in hominid mtDNA (37, 38) have motivated a flurry of subsequent analyses, highlighting the uncertainties regarding the expected relationship between measures of linkage disequilibrium and physical distance between segregating sites (31, 32, 39). Although our approach is, by necessity, still indirect and statistical, we stress our emphasis on homoplasies at particular segregating sites for which multiple outgroup sequences are available for comparison (Table 2). Moreover, in direct contrast to animal mtDNA, plant mtDNA is characterized by very low mutation rates for structural genes (1–5).
Departures from Neutral Expectation.
In an attempt to formally test for selective neutrality and demographic equilibrium, we performed neutral coalescent simulations (26, 27). These simulations essentially embody the “haplotype number test” (K test), which is conditional on sample size and the number of segregating sites under an infinite-sites model (40). For two of three geographic regions, the observed number of haplotypes is significantly lower than expected under the K test, whereas it is low without reaching significance in Alaska (Table 4).
Table 4.
Expected number of haplotypes (K test)
| Parameter | Colorado | Alaska (AK-4 excluded) | Norway–Finland |
|---|---|---|---|
| Sample size, n | 19 | 19 | 20 |
| Segregating sites, S | 6 | 7 | 11 |
| Observed haplotypes, Ko | 2 | 4 | 3 |
| Probability, P (K ≤ Ko) | 0.010 | 0.195 | 0.011 |
| Expected Ke | 5.15 | 5.59 | 7.12 |
| 95% C.I. for Ke | 3–7 | 3–8 | 4–10 |
Expectations (means) of parameters, P values, and 95% confidence intervals (C.I.) are based on 10,000 coalescent simulations assuming no recombination, as implemented in DnaSP (27). P (K ≤ Ko) refers to the proportion of simulations yielding a value as low as or lower than the observed Ko. When treated separately, the K test remains significant for the European samples (Norway: Ko = 3, S = 11; Finland: Ko = 2, S = 8), as is true for the Colorado sample when only the 11 sequenced plants are considered.
These coalescent-based expectations (Ke; Table 4) are likely to be conservative, because they are based on the assumption that recombination has not occurred in the history of the sample. Allowing for reciprocal recombination would increase the expected number of haplotypes, although the situation is less straightforward under gene-conversion models (31). Despite these uncertainties, our data clearly suggest that coexisting haplotypes tend to be more divergent than expected under neutrality and large, stable population size, and that polymorphic sites are partitioned into few, distinct haplotypes (Fig. 1).
Ancient Divergence of Extant Haplotypes.
A crude estimate of phylogenetic antiquity of the assemblage of haplotypes we have uncovered in S. acaulis can be based on absolute rate estimates for plant mitochondrial genes. On the basis of weighted means of seven mitochondrial genes from rice and maize (and assuming a divergence time of 50 million years), Gaut (4) estimated an absolute rate of 0.33 × 10−9 synonymous substitutions per site per year. Our independent analysis of 10 mitochondrial genes from rice and maize yields a weighted mean of 0.40 × 10−9 synonymous substitutions per site per year (Table 5). This estimate is derived from the weighted mean number of synonymous substitutions per synonymous site, Ks = 0.040, in pairwise sequence comparisons. Our rice–maize divergence estimate for cob is Ks = 0.0241, in correspondence with cob exhibiting lower-than-average divergence in angiosperm–liverwort multigene comparisons (3).
Table 5.
Evolutionary rates in plant mitochondrial genes (rice–maize comparison)
| Gene (rice–maize) | bp used | Ks | Ka |
|---|---|---|---|
| atpA* | 1,521 | 0.0598 | 0.0146 |
| atp6* | 687 | 0.0860 | 0.0307 |
| atp9* | 216 | 0.0748 | 0.0126 |
| coxI | 1,572 | 0.0321 | 0.0025 |
| coxII* | 777 | 0.0221 | 0.0000 |
| coxIII* | 780 | 0.0103 | 0.0018 |
| nad3* | 342 | 0.0122 | 0.0039 |
| orf25 | 357 | 0.0262 | 0.0713 |
| rps12 | 375 | 0.0676 | 0.0036 |
| cob* | 1,155 | 0.0241 | 0.0028 |
| Sum/weighted means | 7,782 | 0.0400 | 0.0106 |
| cob–S. acaulis | 1,038 | 0.0118 | 0.0038 |
Ks, number of synonymous substitutions per synonymous site; Ka, number of nonsynonymous substitutions per nonsynonymous site. Both Ks and Ka were estimated according to ref. 41, as implemented in DnaSP (27). Mean values were obtained by weighting the individual gene estimates by the number of bp used. Estimates for cob in S. acaulis (last line) refer to the average of 28 pairwise comparisons among eight intraspecific haplotypes. Sequences for the 10 listed genes were obtained from the GenBank database (Oryza sativa, Zea mays) and aligned in the GCG package. All codons with potential RNA editing sites were removed from the alignments prior to analysis.
Genes previously used by Gaut (4).
The mean pairwise estimate among eight S. acaulis haplotypes is Ks = 0.0118 (Table 5). Hence, using the conservatively high mitochondrial average of Ks = 0.040 and the rice–maize calibration, the proportion of synonymous substitutions among the eight haplotypes would imply an average divergence time on the order of 15 million years, and even older coalescence for the more distinct pairs of cob haplotypes. For several reasons, these seem likely to be underestimates (42), but the implications of coexisting, highly divergent mitochondrial haplotypes are not diminished by this uncertainty. Such divergence times are remarkable, given the generally smaller effective population size of mitochondrial genes compared with nuclear genes (43, 44).
Further evidence of ancient coalescence is provided by the paucity of fixed nucleotide differences relative to the two closest outgroup sequences (Table 1, Fig. 1). Incidentally, this lack of differentiation prevents formal tests of the ratio of synonymous to replacement substitutions that are polymorphic within S. acaulis to the equivalent ratio for sites fixed between S. acaulis and related species (45). Our results are reminiscent of the well-characterized instances of trans-specific evolution observed at vertebrate major histocompatibility and plant self-incompatibility loci, which are explained by the origin of allelic lineages predating the species in which they are currently found (46–48). Because the expected persistence time for neutral variation is short relative to the age of most speciation events, trans-specific evolution is considered a molecular signature of selectively maintained genetic polymorphisms.
Our evidence for old mitochondrial haplotypes has implications for theoretical models of gynodioecy that assume cytonuclear determination of gender. Although some stochastic models predict fairly rapid turnover of CMS factors and thus limited within-population variability at any given time (17), other models predict single-point equilibria (16, 18) or permanent cycling of CMS factors caused by negative frequency-dependent selection (18). The long retention times suggested by our data imply that the nonstochastic models are a better approximation of what occurs in natural populations of S. acaulis.
Ks/Ka Ratio.
Excluding one codon likely to be affected by RNA editing (28, 29), there are eight synonymous and nine nonsynonymous substitutions across the eight S. acaulis haplotypes (Table 1). However, the mean number of nonsynonymous sites (n = 794) is much larger than the number of synonymous sites (n = 244), and the mean between-haplotype number of nonsynonymous substitutions per nonsynonymous site, Ka, = 0.0038, whereas the Ks estimate is 0.0118 (Table 5). Hence, the ratio Ks/Ka is ≈3.1, compared with ≈7.1 for the mean of angiosperm–liverwort comparisons (3) and ≈8.6 for the rice–maize pair (Table 5), suggesting that purifying selection may have been somewhat relaxed throughout the history of the extant cob haplotype assemblage. This relaxation might have been mediated by small effective population size, leading to more rapid accumulation of mildly deleterious mutations (49).
Population Structure and Open Questions.
Although levels of population subdivision appear to be high (results not shown), there is little congruence between the geographic origin of haplotypes and their position in the median network (Fig. 1). In view of the inferred ancient coalescence of extant haplotypes, the assumption that they diverged in situ does not seem plausible. Rather, it seems more likely that the distribution of sampled cob haplotypes was generated by interactions between mutational/recombinational proliferation, stochastic loss, selection, and metapopulation dynamics, including large-scale changes of the species range over evolutionary time.
Collectively, our results seem qualitatively consistent with expectations under the hypothesis of negative frequency-dependent selection on linked CMS genes (see Introduction; refs. 16–23). However, some aspects of the data defy easy interpretation, primarily because of lack of relevant information. Linkage to a locus under balancing or frequency-dependent selection is expected to increase neutral diversity, but this effect is rapidly diminished with increasing recombination distance from the site of selection (50–52). Given our evidence for intragenic recombination in cob and our ignorance regarding the genomic locations of putative CMS genes in S. acaulis (as well as insufficient information about the nature of the inferred processes of recombination), quantitative assessment is not yet feasible.
Foremost, we need to ascertain whether the mitochondrial polymorphism we have uncovered is truly exceptional or actually more widespread than generally assumed. Gynodioecy is likely to be the ancestral breeding system in the genus Silene (which contains several hundred species), and it is also found in related genera (53, 54). Hence, the putative CMS factors, their corresponding nuclear restorer genes, and the predicted microevolutionary processes might have shaped the genetic structure of cytoplasmic genomes in Silene for long periods of time. Future research should contrast gynodioecious species with those exhibiting other breeding systems, including hermaphroditic or dioecious taxa in groups with no gynodioecious relatives.
Evolutionary Implications of Recombination.
Uncovering population-genetic evidence for intragenic recombination was entirely contingent on finding multiple (≥4) differentiated haplotypes. Although intramolecular recombination between repeat regions, leading to structural rearrangements, has long been known to occur in plant mtDNA (2, 55), such processes fail to explain our data. Population-genetic signatures of recombination, such as pairs of sites with all four gametic types, require recombination among appropriately diverged haplotypes, mediated by heteroplasmy of distinct sequences.
Two not mutually exclusive processes are candidates for achieving (transient) heteroplasmy. First, the widespread occurrence of subgenomic molecules of angiosperm mtDNA in very low stoichiometry (“sublimons”; ref. 56) might allow for the long-term coexistence of two or more copies of particular genomic regions within individual cell lineages (57). In our case, this hypothesis requires a second copy of cob being maintained continuously for millions of years in low copy number, without being rendered nonfunctional by deleterious mutations. Subsequent amplification of the hypothetical cob-containing sublimons to normal stoichiometry (58) would conceivably introduce “novel” cob sequences into populations. Recombination or gene conversion events among the two copies of cob coexisting within cell lineages would be expected to occur, but are likely to erode sequence divergence between them.
Second, and perhaps more likely, occasional paternal leakage of mitochondria is fully sufficient to generate transient heteroplasmy. Although unequivocal molecular evidence for heteroplasmy has been obtained in technically tractable systems (59), low-level paternal inheritance of angiosperm mitochondria remains a poorly understood phenomenon in terms of mechanisms, taxonomic distribution, and evolutionary significance (60–63).
Regardless of the mechanisms generating heteroplasmy of diverged mitochondrial genomes, our evidence for intragenic recombination in S. acaulis suggests added layers of complexity for the genetic basis of breeding-system evolution in gynodioecious species. While entirely conjectural at present, recombination events might allow for the duplication, elimination, and reciprocal exchange of CMS genes among the interacting mitochondrial genomes. Such processes imply unexplored consequences for the evolutionary dynamics and stability of a widespread plant breeding system.
Supplementary Material
Acknowledgments
We thank W. Morris and D. Doak for providing seeds from the Alaska population, Y. Cho and F. Knapczyk for laboratory assistance, and J. Palmer for generously providing laboratory facilities. We also thank K. Adams, Y. Chen, Y. Kim, J. Palmer, F. Pulido, L. Rieseberg, L. Rose, W. Stephan, C. Vogl, M. Wade, and two anonymous reviewers for valuable discussion and/or comments on a previous version of the manuscript. W. Stephan provided excellent working conditions for T.S. during data analysis and manuscript preparation. This work was supported by National Science Foundation Grants DEB-9319002 and DEB-9629774 to L.F.D.
Abbreviations
- CMS
cytoplasmic male sterility
- RFLP
restriction fragment length polymorphism.
Footnotes
References
- 1.Wolfe K H, Li W-H, Sharp P M. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer J D, Herbon L A. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 3.Laroche J, Li P, Maggia L, Bousquet J. Proc Natl Acad Sci USA. 1997;94:5722–5727. doi: 10.1073/pnas.94.11.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaut B S. Evol Biol. 1998;30:93–120. [Google Scholar]
- 5.Muse S V. Plant Mol Biol. 2000;42:25–43. [PubMed] [Google Scholar]
- 6.Shykoff J A. Am J Bot. 1988;75:844–850. [Google Scholar]
- 7.Morris W F, Doak D F. Am J Bot. 1998;85:784–793. [PubMed] [Google Scholar]
- 8.Delph L F, Carroll S B. Evol Ecol Res. 2001;3:487–505. [Google Scholar]
- 9.Saumitou-Laprade P, Cuguen J, Vernet P. Trends Ecol Evol. 1994;9:431–435. doi: 10.1016/0169-5347(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 10.Schnable P S, Wise R P. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- 11.Couvet D, Atlan A, Belhassen E, Gliddon C, Gouyon P-H, Kjellberg F. Oxford Surv Evol Biol. 1990;7:225–249. [Google Scholar]
- 12.Olson M S, McCauley D E. Evolution (Lawrence, Kans) 2002;56:253–262. doi: 10.1111/j.0014-3820.2002.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth D, Laporte V. Genetics. 1998;150:1267–1282. doi: 10.1093/genetics/150.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koelewijn H P, Van Damme J M M. Genetics. 1995;139:1749–1758. doi: 10.1093/genetics/139.4.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudle D A, Mutikainen P, Delph L F. Heredity. 2001;86:265–276. doi: 10.1046/j.1365-2540.2001.00833.x. [DOI] [PubMed] [Google Scholar]
- 16.Charlesworth D. Heredity. 1981;46:27–39. [Google Scholar]
- 17.Frank S A. Am Nat. 1989;133:345–376. [Google Scholar]
- 18.Gouyon P-H, Vichot F, Van Damme J M M. Am Nat. 1991;137:498–514. [Google Scholar]
- 19.McCauley D E, Brock M T. Evolution (Lawrence, Kans) 1998;52:30–36. doi: 10.1111/j.1558-5646.1998.tb05135.x. [DOI] [PubMed] [Google Scholar]
- 20.McCauley D E, Taylor D R. Am Nat. 1997;150:406–419. doi: 10.1086/286072. [DOI] [PubMed] [Google Scholar]
- 21.Couvet D, Ronce O, Gliddon C. Am Nat. 1998;152:59–70. doi: 10.1086/286149. [DOI] [PubMed] [Google Scholar]
- 22.Olson M S, McCauley D E. Proc R Soc London B. 2000;267:1801–1808. doi: 10.1098/rspb.2000.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laporte V, Viard F, Bena G, Valero M, Cuguen J. Genetics. 2001;157:1699–1710. doi: 10.1093/genetics/157.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams K L, Song K, Roessler P G, Nugent J M, Doyle J L, Doyle J J, Palmer J D. Proc Natl Acad Sci USA. 1999;96:13863–13868. doi: 10.1073/pnas.96.24.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams K L, Daley D O, Qiu Y-L, Whelan J, Palmer J D. Nature (London) 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 26.Hudson R R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 27.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 28.Gray M W. Proc Natl Acad Sci USA. 1996;93:8157–8159. doi: 10.1073/pnas.93.16.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowe L M, dePamphilis C W. Mol Biol Evol. 1996;13:1159–1166. doi: 10.1093/oxfordjournals.molbev.a025680. [DOI] [PubMed] [Google Scholar]
- 30.Maynard Smith J. Genetics. 1999;153:1021–1027. [Google Scholar]
- 31.Wiuf C. Genetics. 2001;159:749–756. doi: 10.1093/genetics/159.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McVean G, Awadalla P, Fearnhead P. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson R R, Kaplan N L. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandelt H-J, Forster P, Sykes B C, Richards M B. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maynard Smith J, Smith N H. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 36.Posada D, Crandall K A. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyre-Walker A, Smith N H, Maynard Smith J. Proc R Soc London B. 1999;266:477–483. doi: 10.1098/rspb.1999.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awadalla P, Eyre-Walker A, Maynard Smith J. Science. 1999;286:2524–2525. doi: 10.1126/science.286.5449.2524. [DOI] [PubMed] [Google Scholar]
- 39.McVean G A T. Heredity. 2001;87:613–620. doi: 10.1046/j.1365-2540.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 40.Depaulis F, Veuille M. Mol Biol Evol. 1998;15:1788–1790. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 42.Eyre-Walker A, Gaut B S. Mol Biol Evol. 1997;14:455–460. doi: 10.1093/oxfordjournals.molbev.a025781. [DOI] [PubMed] [Google Scholar]
- 43.Birky C W., Jr . In: Evolution at the Molecular Level. Selander R K, Clark A G, Whittam T S, editors. Sunderland, MA: Sinauer; 1991. pp. 112–134. [Google Scholar]
- 44.Laporte V, Cuguen J, Couvet D. Genetics. 2000;154:447–458. doi: 10.1093/genetics/154.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald J H, Kreitman M. Nature (London) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 46.Takahata N. Proc Natl Acad Sci USA. 1990;87:2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vekemans X, Slatkin M. Genetics. 1994;137:1157–1165. doi: 10.1093/genetics/137.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richman A D, Uyenoyama M K, Kohn J R. Science. 1996;273:1212–1216. doi: 10.1126/science.273.5279.1212. [DOI] [PubMed] [Google Scholar]
- 49.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson R R, Kaplan N L. Genetics. 1988;120:831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charlesworth B, Nordborg M, Charlesworth D. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- 52.Schierup M H, Charlesworth D, Vekemans X. Genet Res. 2000;76:63–73. doi: 10.1017/s0016672300004547. [DOI] [PubMed] [Google Scholar]
- 53.Desfeux C, Maurice S, Henry J-P, Lejeune B, Gouyon P-H. Proc R Soc London B. 1996;263:409–414. doi: 10.1098/rspb.1996.0062. [DOI] [PubMed] [Google Scholar]
- 54.Guttman D S, Charlesworth D. Nature (London) 1998;393:263–266. doi: 10.1038/30492. [DOI] [PubMed] [Google Scholar]
- 55.Palmer J D, Adams K L, Cho Y, Parkinson C L, Qiu Y-L, Song K. Proc Natl Acad Sci USA. 2000;97:6960–6966. doi: 10.1073/pnas.97.13.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small I D, Isaac P G, Leaver C J. EMBO J. 1987;6:865–869. doi: 10.1002/j.1460-2075.1987.tb04832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S. Genetics. 2001;158:851–864. doi: 10.1093/genetics/158.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Small I D, Suffolk R, Leaver C J. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 59.Hattori N, Kitagawa K, Takumi S, Nakamura C. Genetics. 2002;160:1619–1630. doi: 10.1093/genetics/160.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milligan B G. Am J Bot. 1992;79:1325–1328. [Google Scholar]
- 61.Reboud X, Zeyl C. Heredity. 1994;72:132–140. [Google Scholar]
- 62.Birky C W., Jr Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andersson H. J Hered. 1999;90:563–565. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.