Abstract
In the last few years, several genes involved in X-specific mental retardation (MR) have been identified by using genetic analysis. Although it is likely that additional genes responsible for idiopathic MR are also localized on the autosomes, cloning and characterization of such genes have been elusive so far. Here, we report the isolation of a previously uncharacterized gene, MEGAP, which is disrupted and functionally inactivated by a translocation breakpoint in a patient who shares some characteristic clinical features, such as hypotonia and severe MR, with the 3p− syndrome. By fluorescence in situ hybridization and loss of heterozygosity analysis, we demonstrated that this gene resides on chromosome 3p25 and is deleted in 3p− patients that present MR. MEGAP/srGAP3 mRNA is predominantly and highly expressed in fetal and adult brain, specifically in the neurons of the hippocampus and cortex, structures known to play a pivotal role in higher cognitive function, learning, and memory. We describe several MEGAP/srGAP3 transcript isoforms and show that MEGAP/srGAP3a and -b represent functional GTPase-activating proteins (GAP) by an in vitro GAP assay. MEGAP/srGAP3 has recently been shown to be part of the Slit-Robo pathway regulating neuronal migration and axonal branching, highlighting the important role of MEGAP/srGAP3 in mental development. We propose that haploinsufficiency of MEGAP/srGAP3 leads to the abnormal development of neuronal structures that are important for normal cognitive function.
Remarkable progress in understanding the genetic traits of idiopathic mental retardation (MR) has occurred in recent years. Several genes could be identified on the human X chromosome by using positional cloning strategies, and their contribution to the mental handicap was revealed by mutation detection (1, 2). These analyses showed for most of these genes an apparent common molecular mechanism in contributing to the same intracellular pathways involving the Rho-class of small GTP-coupled proteins. This family of ras-homologous proteins is composed of several members, with RhoA, Rac1, and Cdc42 the best studied examples. Their functions are diverse but have been ascribed roles in outgrowth of axons and dendrites in vivo and the activation of the MAP kinase cascades and cytoskeleton organization (3–5). The correct temporal and spatial activity is accomplished by the tight regulation of the active GTP-bound and the inactive GDP state. This situation is achieved by the mutual function of GEF (guanine exchange factors) and GAP (GTPase-activating proteins) proteins. It has become evident that MR seems to be a consequence of the improper regulation of the small GTPases or their downstream targets. For example, ARHGEF6 represents a GEF for Rac1 and Cdc42 and was identified by the molecular analysis of an X;21 reciprocal translocation (6). In a similar way, oligophrenin1, a GAP for RhoA, was found to be interrupted in a female patient carrying a balanced X;12 translocation associated with MR (7, 8).
In the present study, we analyzed a patient with a balanced de novo translocation t(X;3)(p11.2;p25) with one of its breakpoints mapping within the 3p− syndrome deleted region (9–12). This young woman shows hypotonia and severe MR, features characteristic for 3p− patients, but not microcephaly, growth failure, heart and renal defects, and the 3p− typical facial abnormalities (13, 14). The translocation breakpoint on chromosome X is located outside of any coding region. However, the breakpoint on chromosome 3 interrupts a previously unknown gene, which we initially termed MEGAP (MEntal disorder-associated GAP protein). MEGAP shares structural and functional similarities with other members of the RhoGAP protein family and is highly expressed in fetal and adult brain tissue. The C-terminal part of MEGAP corresponds to srGAP3, which recently has been shown to play a critical role in the Slit-Robo signal-transduction pathway (15). We suggest that the phenotype observed in our patient is caused by a misregulation of a neuronal signal-transduction machinery controlling the correct migration of neurons and their axonal connectivity.
Methods
Case Report.
The proband is a 16-year-old female, the second child of healthy nonconsanguineous parents (father, 47 years; mother, 45 years). Her brother of 26 years is not affected, and there are no further affected individuals in this family. Pregnancy was reported to be uneventful. Delivery occurred at 40 weeks of gestation by breech presentation. Birth weight was 3,720 g, length was 55 cm, and head circumference was 35 cm. Her height is 163 cm (25.pc) and head circumference is 55 cm (75.pc). Mental and motor developmental delay was evident in the early stages of life. She could stand freely at the age of 4 years and walk only at the age of 5.5 years. Clinical findings at the age of 16 years included an extremely psychomotoric retardation with an atactic gait, jerky arm movements, very low mental performance, periodical autoaggressive behavior, and absence of speech. She shows a triangular face with a high nasal bridge, micrognathia, short philtrum, high and narrow palate, slender hands, muscle hypotonia, and scoliosis. In the last 3 years, three epileptic seizures occurred. The patient presents severe attention deficits; therefore, intelligence quotient (IQ) testing was not possible. Magnetic resonance imagery scan or further imaging diagnostics have been rejected by the parents.
Case 150500 is a 3-year-old male diagnosed with psychomotor retardation and epicanthus. Cytogenetically, a terminal deletion of chromosome 3p was diagnosed, with the breakpoint residing in band 3p25. Cases P1–P10 have been described (10, 12). Case P11 is an adult without intellectual impairment, who has a cytogenetically visible deletion of chromosome 3p and a normal phenotype.
Genomic Clones and Deletion Mapping.
YAC clones from Xp11.2 and 3p25 were chosen from the Whitehead radiation hybrid maps WCX.11 and WC3.1 and purchased from the German Resource Center (RZPD). PAC and BAC clones were obtained from the Resource Center, Oakland, (RPCI 1, 3, 11) or from Research Genetics, Huntsville (CIT-B, -C). Cosmids were purchased from the RZPD after screening the LL03NC01 “AC” chromosome 3 and LL0XNC01 “U” chromosome X-specific human cosmid libraries (Lawrence Livermore National Laboratory, Livermore, CA). Overlapping clones were confirmed by PCR and hybridization analysis. Fluorescence in situ hybridization (FISH) was performed as described. Loss of heterozygosity (LOH) analysis was performed as described by Green et al. (12).
Rapid Amplification of cDNA Ends (RACE) Analysis.
Human total RNA of adult brain (Invitrogen) was reverse transcribed by using the gene-specific primer Ex5.1 (5′-GAGCAGGTTCATGCTGAGGT-3′) and Superscript II reverse transcriptase (GIBCO/BRL). After second strand synthesis, the cDNA was ligated with the 2R adapter (GIBCO/BRL) and subsequently amplified with adapter primer 2R (GIBCO/BRL) and nested gene-specific primer Ex5n1 (5′-TCATGCTGAGGTCTCCTGACT-3′). Specific amplification was achieved by using a second round of nested PCR with the nested 2R primer (GIBCO/BRL) and primer Ex4n2 (5′-CTCATTGGTCACCTTCAGGA-3′). The PCR product was cloned into pCR2.1 vector by using the TOPO-TA cloning kit (Invitrogen) and sequenced. Putative exons were aligned with the genomic sequence and confirmed by reverse-transcription (RT)-PCR analysis.
Expression Studies.
Human multiple tissue Northern blots were purchased from CLONTECH. Reverse transcription (RT)-PCR-generated fragments covering exons 1–4 (bp 27–569) or a β-actin cDNA probe were labeled with [α-32P]dCTP (Amersham Pharmacia) by using a random primed labeling protocol. The probes were hybridized to the Northern blots overnight at 65°C, as recommended by the manufacturer, washed with 2× SSC and 0.2× SSC at 65°C, and exposed for 24 hours at −80°C (actin, 2–3 hours at room temperature).
In Situ Hybridization.
Three-month-old male B6 mice were perfused with 4% (wt/vol) paraformaldehyde (PFA) in PBS. After dissection, the brains were postfixed overnight in 4% (wt/vol) PFA and sectioned in 50-μm slices with the vibratome (Leica VT 1000S). Sense and antisense riboprobes for Megap/srGAP3 prepared by in vitro transcription of cDNA encoding for exons 1–19 of the murine homolog were labeled with DIG-11-UTP by T3 or T7 polymerases (Roche Molecular Biochemicals). Floating sections were hybridized overnight at 65°C with the DIG-labeled riboprobes and visualized by alkaline phosphatase conjugated anti-DIG antibody.
Alternative Splicing Analysis.
Total human RNA from different tissues was reverse transcribed with an oligo-dT primer by using Superscript II RT (GIBCO/BRL). PCR amplification of exons 9–14 was carried out in an Eppendorf gradient cycler with primer Ex9for (5′-GCCACCATGCAGACATTACA-3′) and Ex14rev (5′-ACTCTGAAGATCCCCTGCTG-3′) with an initial denaturation step of 94°C for 2 min, 33 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final 3-min extension step at 72°C. PCR products were analyzed on an agarose gel, re-eluted from the gel, and cloned into pCR2.1 vector by using the TOPO-TA cloning kit (Invitrogen). Clones were sequenced and compared with the published genomic sequence.
GAP Assay.
MEGAP/srGAP3b-GST fusion protein was generated by subcloning a SalI fragment of KIAA0411 (residues 1–1468, corresponding to bp 1,377–2,849 of MEGAP/srGAP3b) containing the GAP and SH3 domain into the pGEX4T3 vector (Amersham Pharmacia). Isoform a was generated by replacement of the PsyI/Eco81I fragment (bases 1323–1813). After transfection into Escherichia coli BL21, protein expression was induced with 1 mM isopropyl β-d-thiogalactoside. Recombinant proteins were purified by using glutathione beads in NETN-buffer (20 mM Tris⋅HCl, pH 8.0/100 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/1 mM DTT) and washed with 20 mM Tris⋅HCl/0.1 mM DTT. Vectors containing the full-length coding sequence for the three small GTPases RhoA, Rac1, and Cdc42Hs were obtained from the Guthrie cDNA Resource Center (Sayre, PA) and subcloned into pQE30 vectors (Qiagen). Proteins were prepared under native conditions as recommended, but included 5 mM MgCl2 in each of the buffers (16). Purified proteins were dialyzed overnight against 10 mM Tris⋅HCl, pH 7.5/2 mM MgCl2/0.1 mM DTT. Protein concentrations were determined by SDS/PAGE and bicinchoninic acid assay (Pierce).
GAP assays were performed as described (17). Briefly, 0.3 μM of purified and dialyzed GTPases were incubated for 10 min at 30°C with 10 μCi (1 Ci = 37 GBq) [γ-32P]GTP (Amersham Pharmacia) in 20 μl of 20 mM Tris⋅HCl, pH 7.5/25 mM NaCl/5 mM EDTA/0.1 mM DTT. The reaction was terminated by adding 5 μl MgCl2 (0.1 M) on ice. Three microliters of this reaction were incubated at 30°C with GST-tagged MEGAP/srGAP3 or ARHGAP1-GST protein. Five microliters were removed after 0, 3, 6, 9, 12, and 15 min and mixed with 500 μl of ice-cold stop buffer. The samples were filtered through BA85 nitrocellulose membrane (Schleicher & Schüll) and washed with 10 ml of ice-cold stop buffer (50 mM Tris⋅HCl, pH 7.5/50 mM NaCl/5 mM MgCl2). The amount of radioactivity bound to each of the GTPases was analyzed in a scintillation counter.
Results
Physical Mapping and Characterization of the Translocation Breakpoints.
Cytogenetic analysis of a female patient with severe MR revealed a de novo balanced translocation t(X;3)(p11.2;p25). To characterize the breakpoints on both derivative chromosomes, we performed FISH analysis on the patient's metaphase chromosomes. By using YAC, PAC/BAC, and cosmid clones, we could narrow down the breakpoint region to an interval of approximately 40 kb (Fig. 1a). Subsequent Southern and PCR analyses showed aberrant products in the patients DNA samples but not in controls (data not shown). Sequence comparison of the cloned junction fragments confirmed that the translocation is balanced.
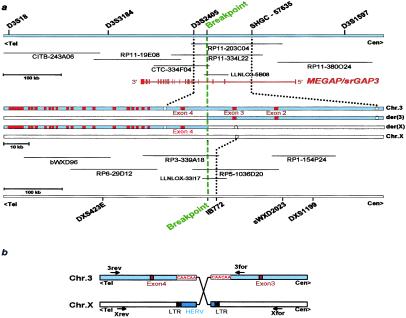
Figure 1.
Genomic organization of the MEGAP/srGAP3 gene on chromosome 3p25 and localization of the translocation breakpoints in the patient. (a) Physical mapping of MEGAP/srGAP3. The precise order of markers for the chromosomal regions 3p25 and Xp11.2 is shown at Top and Bottom, as well as the orientation of telomere and centromere. BAC, PAC, and cosmid clones used for FISH mapping of the respective breakpoints are shown. Both BAC RP11–203C04 and RP5–1036D20 showed signals on the normal and the derivative chromosome, thus containing the translocation breakpoint. The localization of the 22 exons of MEGAP/srGAP3 is indicated as vertical red lines. The entire gene encompasses approximately 250 kb of genomic sequence, with exon 1 separated from exon 2 by about 100 kb. The breakpoint on the derivative chromosome 3 is located between exons 3 and 4 of MEGAP/srGAP3, as shown in the middle part of the scheme. Exons are indicated here as red boxes. Markers D3S2405 and SHGC-57635 (white boxes) are located within intronic sequences of MEGAP/srGAP3. Marker IB772 (white box) resides on the X chromosome. (b) Schematic drawing of the breaking event. The breakpoint on the X chromosome is located in a remnant human endogenous retroviral element (HERV) next to the flanking LTR sequence. The breakpoint region on chromosome 3 shows several CAA trinucleotide repeats on both sides of the translocation. The position of primers used for amplification of the junction fragments are indicated as arrows.
The X chromosomal breakpoint is situated within a truncated HERV element, about 12 kb from the next long terminal repeat (LTR) element (Fig. 1b). We analyzed several hundred kilobases around the breakpoint and excluded this region as causative for the observed phenotype. Analysis of chromosome 3-specific sequences at the breakpoint identified two partially overlapping cDNA clones, KIAA0411 and KIAA1156, derived from adult human brain (Kazusa Institute, Japan). Alignment of these cDNAs to the genomic sequence revealed that both clones reside distal to the breakpoint.
Isolation of a Previously Uncharacterized Gene on Chromosome 3p25.
RT-PCR with primers corresponding to the KIAA clones indicated that both cDNAs, overlapping by 162 bp, are derived from a single transcriptional unit. To identify the 5′ end and the start codon of the transcript, we performed 5′-RACE. The resulting PCR product was sequenced and compared with the genomic sequence. We identified three additional putative exons, all located proximal to the breakpoint (Fig. 1a). Thus, this transcript bridges the breakpoint on chromosome 3 of the patient. Alignment of the cDNA clone KIAA0411 to the genomic sequence established that it contains exons 9 to 22, whereas KIAA1156 contains exons 4–11. Just recently, database searches revealed another unnamed sequence (AX056907) that corresponds to one of the isoforms (see below) that we isolated; however, it lacked the 5′ and 3′ untranslated regions of the gene.
The longest (8,327 bp) mRNA encoded by this gene has a putative start codon in the first exon 103 bp downstream of the consensus cDNA sequence. A stop codon in exon 22 defines an ORF of 3,299 bp which encodes a putative protein of 1,099 amino acids (Fig. 2a). Thus, the mRNA contains a long 3′-untranslated region (UTR) of about 5 kb, encoded by a large part of the last exon.
Figure 2.
Peptide sequence of MEGAP/srGAP3a. FES/CIP4 (FCH), GAP, and SH3 domains are highlighted; inverted carets indicate exon junctions.
Screening of 3p− Patients.
Recent studies (10, 12, 14) indicated that patients carrying terminal deletions of chromosome 3p are associated with MR and further multiple recurring symptoms depending on the size of their deletion. The deletion size is variable but generally encompass several megabases of DNA, as shown by microsatellite analyses. To find out whether MEGAP/srGAP3 is deleted in these patients, we carried out a combined FISH/LOH study in 11 patients diagnosed with terminal deletions of chromosome 3p. Four cases (P1, P2, P5, and P7) were found deleted for D3S1597, a marker approximately 50 kb proximal to MEGAP/srGAP3 (Fig. 1a and Table 1). Cases P4 and P6 were not informative for D3S1597, but showed deletions in the more proximal marker D3S1038. In patients P3 (GM10922), P8 (GM10985), and P150500, we carried out fluorescence in situ hybridizations with BAC clones RP11–203C04 and RP11–334L22, which contain MEGAP/srGAP3. We could show that in all three patients, P3, P8, and 150500, both BACs presented one signal on the normal chromosome 3 but were deleted on the derivative chromosome 3 (Table 1). Most interestingly, we also examined a patient with a terminal deletion of chromosome 3p but without diagnosed intellectual impairment. We could show that marker D3S1597 is still present on both alleles. Together, these data indicate that the loss of one MEGAP/srGAP3 copy is associated with the mental handicap in 3p− patients.
Table 1.
3p−-deletion mapping results
| D3S | cM | 150 500 | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P10 | P11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3691 | 15.6 | − | NI | NI | NI | NI | − | NI | NI | |||
| 1597 | 16.1 | − | − | − | − | NI | − | NI | − | − | NI | + |
| 1038 | − | − | − | − | − | − | − | + | + | |||
| 611 | 17.5 | NI | − | NI | NI | − | − | + | + | |||
| 1263 | 18.4 | − | − | − | − | + | + | + | + | + | ||
| MR | + | + | + | + | + | + | + | + | + | + | NO |
−, deleted; +, retained; NI, uninformative; MR, mental retardation.
MEGAP/srGAP3 Shares Homologies with Other RhoGAP Proteins.
Comparison of the predicted amino acid sequence with the SwissProt database revealed that the protein shows significant sequence similarity to srGAP1 and 2, as well as ARHGAP4 (15, 18). Compared with ARHGAP4, both proteins show 50% amino acid identity and 69% similarity. Strikingly, all sizes of the individual exons 1 to 19 are conserved. ARHGAP4 is an X-linked gene and has been shown to contain three functional domains. All three domains were also identified in the predicted peptide sequence of the gene and of srGAP1 and srGAP2. A RhoGAP motif of 149 amino acids was identified between amino acids 520 and 670, sharing 30–40% identity with other known members of the family of Rho-GTPase-activating proteins (RhoGAP) such as oligophrenin1, BCR1, and N-chimerin (data not shown). Besides the RhoGAP domain, an SH3 and a FES/CIP4 (FCH) domain were identified at the C- and N-terminal ends, respectively (Fig. 3b).
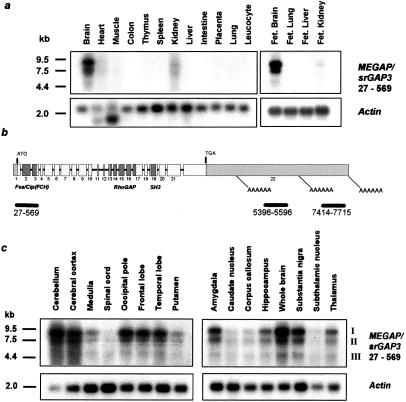
Figure 3.
Expression analysis of MEGAP/srGAP3. (a) Multiple tissue Northern blots from adult and fetal tissues (CLONTECH) containing 1–2 μg polyA+ RNA per lane were hybridized with [α-32P]dCTP-labeled probes containing exons 1 to 4 of MEGAP/srGAP3 (bp 27–569). The filters were hybridized overnight at 65°C, stringently washed at 65°C, and exposed to x-ray films for 24 h at −80°C. A control hybridization with β-actin shows signals at 2.0 kb in all lanes, with additional bands at 1.6–1.8 kb in heart and skeletal muscle. (b and c) Three different isoforms of MEGAP/srGAP3 (I, II, and III). (b) Schematic drawing of exons and motifs of MEGAP/srGAP3. The position of the start (ATG) and stop (TGA) codons are indicated. Introns are depicted as black lines and are not drawn to scale. Light gray boxes, 5′ and 3′-UTR; dark gray boxes, FES/CIP4 (FCH), RhoGAP, and SH3 domains; black bars, localization of probes used for hybridization of the Northern blots; AAAAA, localization of putative polyA tails in the 3′-UTR. (c) Expression of MEGAP/srGAP3 in different brain regions. Hybridization of exons 1–4 results in three signals (I, II, III). The longest band (I) corresponds to the predicted 8.3-kb full-length transcript. Signals at 7 kb (II) and 4.5 kb (III) represent transcripts with differing 3′-UTRs.
Expression Analysis.
Because MEGAP/srGAP3 was identified in a patient with mental and motor deficits, we investigated the MEGAP/srGAP3 mRNA expression in whole brain and in specific brain regions, as well as in a series of other adult tissues. Fig. 3a shows that MEGAP/srGAP3 is predominantly and highly expressed in adult and fetal brain, with low expression detectable in kidney tissue and very low expression in other tissues. In the adult brain, the MEGAP/srGAP3 gene is expressed in all tested regions including cerebellum, cortex, occipital pole, frontal and temporal lobe, amygdala, hippocampus, substantia nigra, and thalamus. Besides the predicted 8.3-kb full-length transcript, two additional isoforms II and III of about 7 and 4.5 kb were observed. The 4.5-kb fragment was not detected by using a probe derived from position 5,396–5,596 of the gene. By using a probe from position 7,414–7,715, the 4.5- and the 7-kb transcripts were not detected (Fig. 3b). These results strongly suggest that the three different transcript sizes, I, II, and III, are due to different polyadenylation sites in the 3′-UTR.
To further investigate the temporal and spatial expression pattern of MEGAP/srGAP3, we isolated its mouse ortholog. We used RT-PCR with mouse brain cDNA to clone and sequence the entire ORF of the murine homolog. The mouse Megap/srGAP3 ORF shows 92% nucleotide homology with the human cDNA; the predicted proteins share 97% identity (data not shown). These data were confirmed by a partial database entry (AF338472), which corresponds to the human isoform a but lacks the 5′-part of the sequence. We then performed a series of RNA in situ hybridizations on sections of the adult mouse brain. Megap/srGAP3 shows a distinct expression pattern with the highest level of expression seen throughout the neurons of the hippocampus and specific parts of the cortex (see Fig. 4) and olfactory bulb. The high expression in these brain structures strongly suggests that MEGAP/srGAP3 may play a role in processes underlying synaptic plasticity, higher cognitive function, learning, and memory.
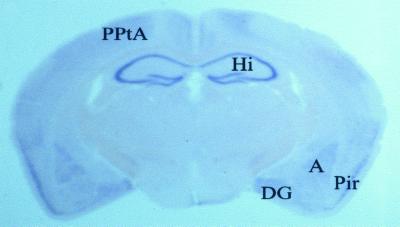
Figure 4.
Distribution of Megap/srGAP3 mRNA in adult mouse brain. In situ hybridization with digoxigenin-labeled Megap/srGAP3 antisense riboprobe on coronary section showing strong expression in the amygdala, piriform cortex, posterior parietal associative area, the dentate gyrus, and particularly strong expression in the hippocampal formation. Hybridization with Megap/srGAP3 sense DIG-RNA gave no signal. A, amygdala; DG, dentate gyrus; Hi, hippocampus; Pir, piriform cortex; PPtA, posterior parietal associative area.
Multiple MEGAP/srGAP3 Isoforms Are Generated by Alternative Splicing.
Besides the different transcript sizes detected by Northern analysis, RT-PCR analysis indicates that MEGAP/srGAP3 mRNA contains three alternatively spliced forms (Fig. 5a). These isoforms differ in the use of exon 12. The largest and most abundant transcript MEGAP/srGAP3a comprises exon 12a (Fig. 2). The smaller exon 12b is present in both KIAA cDNA clones and in AX056907. In isoform MEGAP/srGAP3c, exon 12 is skipped completely, leading to a frame shift and a premature stop nine amino acids after the splice acceptor site, just before the RhoGAP domain (Fig. 5b). Interestingly, this truncated form can be detected in all analyzed tissues by RT-PCR (e.g., adult kidney), but is absent in whole brain and in all analyzed brain subregions (data not shown). Among the investigated tissues, the MEGAP/srGAP3a isoform seems to be the most prominent transcript, as determined by RT-PCR (data not shown). Taking into account the different isoforms I, II, and III, and a, b, and c, nine different transcripts and three protein isoforms of MEGAP/srGAP3 may exist.
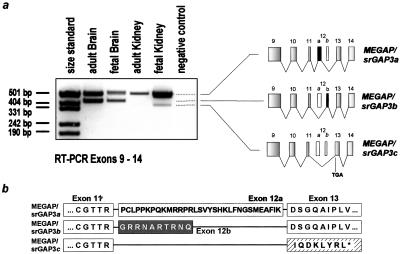
Figure 5.
Alternative splicing of exon 12. (a) Alternative splicing forms in brain and kidney of fetal and adult origin. RT-PCR (33 cycles) was performed with oligo-dT-transcribed cDNAs by using primers derived from exons 9 and 14. PCR products at 482, 412, and 383 bp correspond to isoforms a, b, and c, respectively. Isoforms a and b of MEGAP/srGAP3 differ in the use of an alternative exon 12. The presence of exon 12b leads to a transcript 72-bp shorter than isoform MEGAP/srGAP3a, leading to an ORF of 3,227 bp and coding for a predicted protein of 1,075 amino acids. Isoform MEGAP/srGAP3c lacks exon 12, leading to a frame shift and a premature stop as indicated. (b) Predicted peptide sequences of exons 11–13 of isoforms MEGAP/srGAP3a, -b, and -c. *, stop codon due to the frame shift in exon 13.
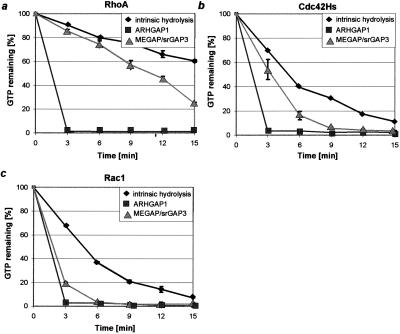
MEGAP/srGAP3 Enhances the GTPase-Activity of Rac1 and Cdc42Hs but Not of RhoA Substrates.
Prior analysis of proteins containing a RhoGAP domain has revealed that some are substrate-specific whereas others offer a broad spectrum of reactivity. To test the ability of MEGAP/srGAP3 to enhance the GTPase activity of RhoA, Rac1, and Cdc42Hs in vitro, we performed a GAP assay by using recombinant proteins expressed in E. coli. GST-bound MEGAP/srGAP3 isoform a (amino acid residues 426–939) and isoform b (amino acid residues 426–915) were incubated with [γ-32P]GTP preloaded GTPases at 30°C, and the amount of GTP hydrolysis was measured in a filter binding assay. As control, we used GST-tagged ARHGAP1 (residues 198–439 amino acids), which has been shown to enhance the hydrolysis rate of Cdc42Hs, RhoA, and Rac1 (19). As shown in Fig. 6, MEGAP/srGAP3 strongly activates Rac1 GTPase function and, to a lower extent, Cdc42Hs. In contrast, MEGAP/srGAP3 shows no activity toward RhoA (Fig. 6c), whereas ARHGAP1 is able to enhance its intrinsic GTPase activity efficiently . No difference in activity was detectable between the two isoforms a and b, suggesting that the alternative use of exon 12a or -b does not influence its activity in the in vitro assay.
Figure 6.
Filter binding assay showing GAP activity of MEGAP/srGAP3 toward Rac1 and Cdc42Hs. As described, ARHGAP1 (■) shows strong activity toward RhoA, Rac1, and Cdc42Hs. However, MEGAP/srGAP3 (▴) shows almost no activity toward RhoA, a strong activity toward Rac1, and a lower but still significant activity toward Cdc42Hs. [γ-32P]GTP-labeled p21 (0.3 μM) was incubated at 30°C in the absence or presence of equimolar concentrations of the GAP domains of MEGAP/srGAP3a (amino acid residues 426–939) or ARHGAP1 (amino acid residues 198–438). Aliquots of the reaction were taken every 3 min and immediately passed through a 0.45-μm nitrocellulose membrane. The amount of radioactivity bound to the filter was measured in a scintillation counter. Activity at t = 0 min was set to 100%.
Discussion
Some 2–3% of all humans have an IQ of less than 70 (moderate MR), and 0.3% have an IQ of less than 50 (severe MR; ref. 2). Given the ease of determining X-linked disorders in hemizygous males, the X chromosome has become a focus for mapping and isolating genes involved in MR in recent years (1, 2, 20–23). No autosomal nonspecific MR genes have been identified as yet. It has been speculated that the identification of autosomal genes for cognitive function will build on the knowledge accumulating in the field of X-linked MR, with X-linked and autosomal loci likely to be coded for proteins in the same or similar molecular pathways (24). This prediction holds true very much for the MEGAP/srGAP3 gene. The identification of a chromosomal breakpoint within the 3p− syndrome region in a patient with severe MR led us to the positional cloning of a previously unknown gene. The MEGAP/srGAP3 gene is commonly deleted in patients with the 3p− syndrome, a syndrome characterized by multiple congenital abnormalities and MR. Previous studies have shown that all analyzed 3p− patients had a minimal terminal deletion ranging from the telomere to marker D3S1597 (10, 11). The deleted region in 3p− patients spans approximately 12 megabases of DNA and encompasses many genes, some of which may contribute to the phenotype (25–27). As the most proximal marker D3S1597 that is consistently deleted in all 3p− deletion patients resides only a few kilobases proximal to MEGAP/srGAP3, we speculated that the loss of MEGAP/srGAP3 is crucial for the clinical description of the cognitive defect in the 3p− syndrome. To prove this idea, we screened a subset of 11 patients with terminal deletions of chromosome 3p by a combined FISH/LOH analysis. Indeed, all of the examined patients with severe mental handicap showed a deletion of MEGAP/srGAP3. The only 3p− patient who did not show a loss of MEGAP/srGAP3 was the one with normal cognitive function (P11), supporting our hypothesis of a critical role of MEGAP/srGAP3 in cognitive impairment.
As it has been shown that MEGAP/srGAP3 is deleted in 3p− patients and inactivated by a balanced translocation in a patient with severe MR, we propose that haploinsufficiency is the genetic mechanism underlying the disease. Nevertheless, we cannot completely rule out possible dominant-negative or -positive effects caused by expression of a truncated form of the protein. The situation at the translocation site presents two possibilities. First, a transcript containing the first three exons plus additional, unknown material could be expressed by the MEGAP/srGAP3 promoter. Such a construct remaining stable in the cell is questionable, because it lacks a suitable polyA tail. The second possibility is the expression of a truncated MEGAP/srGAP3 mRNA containing exons 4 to 22 driven by a possible promoter element in the flanking HERV (Fig. 1b). This situation is unlikely, as the putative promoter element resides approximately 12 kb apart from exon 4. We could not detect any MEGAP/srGAP3 transcript on Northern blots of the lymphoblastoid cell line derived from the patient's blood material. For obvious reasons, other material is not available. Nonetheless, the truncated transcript would miss the entire N-terminal FCH domain. This part of the protein is likely to confer some specific function, because it is conserved also in the homologous proteins srGAP1, srGAP2, and ARHGAP4.
We have found that MEGAP/srGAP3 is specifically and predominantly expressed in different portions of the brain and have identified three transcripts differing in the length of their 3′-UTRs. The functional relevance of these mRNA isoforms remains unclear, but it could include a posttranscriptional regulation of the MEGAP/srGAP3 expression or stability on the RNA level or differential targeting of MEGAP/srGAP3 mRNA isoforms in the neurons (28). In addition, RT-PCR experiments have identified three further MEGAP/srGAP3 mRNA isoforms which differ in the use of exon 12. This finding suggests that at least three protein isoforms, MEGAP/srGAP3a, -b and -c are encoded by the MEGAP/srGAP3 gene. MEGAP/srGAP3a likely represents the most abundant product, as it is present in all tissues tested, whereas MEGAP/srGAP3b is predominantly expressed in brain and only to a lower extent in other tissues. Both MEGAP/srGAP3a and -b represent fully functional GAP proteins, as shown by the in vitro GAP assay.
MEGAP/srGAP3 shares homology with oligophrenin1, another RhoGAP-containing protein previously identified in a patient with X-specific MR (7, 8, 29). It has been suggested that Oligophrenin1, like many of the known genes with a role in cognitive impairment, participates in intracellular signaling. These genes regulate shared signaling pathways to modulate the actin cytoskeleton involving the Rho-family of GTPases (6, 30, 31). This common functional characteristic has strongly implied that the primary cellular defect in MR is a result of the impaired ability of the cytoskeleton to support neurite outgrowth and structural changes associated with neuronal and synaptic plasticity (4, 32). The SH3 domain of MEGAP/srGAP3 has been identified recently as a binding partner for the Robo transmembrane receptor (15). Robo is a member of a conserved family of repulsive axon guidance receptors that respond to secreted Slit proteins (33). The secreted Slit proteins repel neuronal precursors migrating from the anterior subventricular zone in the telencephalon to the olfactory bulb (34). Loss of MEGAP/srGAP3, a direct intracellular part of this signal-transduction pathway, may, therefore, inhibit the correct migration of neuronal cell from their birthplace to their final positions in the nervous system. MEGAP/srGAP3 is homologous to srGAP1, another Robo-interacting GAP protein, but the biochemical data show that both proteins act in different pathways. Whereas MEGAP/srGAP3 is mainly a repressor of Rac1-function, srGAP1 has been shown to act primarily on Cdc42. This finding renders it unlikely that srGAP1 compensates for the loss of one MEGAP/srGAP3 copy in the affected patients.
Despite the convincing structural and functional evidence for the role of MEGAP/srGAP3 in neuronal development, mutations of the gene in additional patients with idiopathic MR or families mapping in the respective genetic regions have not been demonstrated so far. Because of an enormous genetic and phenotypical heterogeneity as already demonstrated for the X-linked forms of MR, mutation analysis of MEGAP/srGAP3 is not a suitable approach (23). We propose that exploiting the extraordinary structural and functional homology between the human and mouse MEGAP/srGAP3 will lead to a much faster and more detailed understanding of the MEGAP/srGAP3 role in neuronal function and cognitive development. We have found that MEGAP/srGAP3 is highly expressed in fetal and adult brain, primarily in areas associated with higher cognitive functions and neuronal plasticity. Similarly, most if not all MRX genes identified so far show prominent expression in the hippocampus and cortex, areas important for higher cognitive functions including learning and memory. In comparison with the expression of the previously studied genes GDI1, IL1RAPL, and TM4SF2 (30, 31, 35), MEGAP/srGAP3 mRNA is particularly highly expressed in the hippocampus, cortex, and amygdala. By generating targeted gene disruption of MEGAP/srGAP3 mimicking the patient's mutation and by testing the mutants in both electrophysiological and behavioral tests for neuronal functions in the hippocampus, cortex, and amygdala, a mouse model corresponding to the human phenotype may be developed. We hope that such a model will greatly enhance our understanding of the underlying molecular and cellular mechanisms leading to MR.
Acknowledgments
We thank R. Blaschke, N. Muncke, R. Sprengel, and F. Vogel for discussion, D. Hager for patient 150500, and C. R. Bartram for his continuous support. This work was supported by a grant from the Fritz Thyssen Foundation, Bundesministerium für Bildung und Forschung, and the British Heart Foundation.
Abbreviations
- MR
mental retardation
- GAP
GTPase-activating protein
- MEGAP
mental disorder-associated GAP protein
- FISH
fluorescence in situ hybridization
- RT-PCR
reverse transcription–PCR
- UTR
untranslated region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. for: MEGAPa, AF427144; KIAA0411, AB007871; KIAA1156, AL032982; RP3–339A18, Z97054; RP5–1036D20, AL109851; RP11–203C04, AC066583; RP11–334L22, AC037193; RP11–19E08, AC034186; RP11–380O24, AC026191; CTC-334F04, AC055745; srGAP1, XM_051143; and srGAP2, XM_051949).
References
- 1.Chelly J. Hum Mol Genet. 1999;8:1833–1838. doi: 10.1093/hmg/8.10.1833. [DOI] [PubMed] [Google Scholar]
- 2.Chelly J. Am J Med Genet. 2000;94:364–366. doi: 10.1002/1096-8628(20001023)94:5<364::aid-ajmg3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Threadgill R, Bobb K, Ghosh A. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 4.Luo L. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 5.Ramakers G J A. Am J Med Genet. 2000;94:367–371. doi: 10.1002/1096-8628(20001023)94:5<367::aid-ajmg4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang H G, Orth U, Boavida M G, David D, Chelly J, Fryns J P, et al. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 7.Bienvenu T, Der-Sarkissian H, Billuart P, Tissot M, Des Portes V, Brüls T, Chabrolle J P, Chauveau P, Cherry M, Kahn A, et al. Eur J Hum Genet. 1997;5:105–109. [PubMed] [Google Scholar]
- 8.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet M C, Zemni R, Crollius H R, Carrie A, Fauchereau F, Cherry M, et al. Nature (London) 1998;392:823–826. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 9.Narahara K, Kikkawa K, Murakami M, Hiramoto K, Namba H, Tsuji K, Yokoyama Y, Kimoto H. Am J Med Genet. 1990;35:269–273. doi: 10.1002/ajmg.1320350225. [DOI] [PubMed] [Google Scholar]
- 10.Phipps M E, Latif F, Prowse A, Payne S J, Dietz-Band J, Leversha M, Affara N A, Moore A T, Tolmie J, Schinzel A, et al. Hum Mol Genet. 1994;3:903–908. doi: 10.1093/hmg/3.6.903. [DOI] [PubMed] [Google Scholar]
- 11.Drumheller T, MacGillivray B C, Behrner D, MacLeod P, McFadden D E, Roberson J, Venditti C, Chorney K, Chorney M, Smith D I. J Med Genet. 1996;33:842–847. doi: 10.1136/jmg.33.10.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green E K, Priestley M D, Waters J, Maliszewska C, Latif F, Maher E R. J Med Genet. 2000;37:581–587. doi: 10.1136/jmg.37.8.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verjaal M, De Nef J. Am J Dis Child. 1978;132:43–45. doi: 10.1001/archpedi.1978.02120260045012. [DOI] [PubMed] [Google Scholar]
- 14.Mowrey P N, Chorney M J, Venditti C P, Latif F, Modi W S, Lerman M I, Zbar B, Robins D B, Rogan P K, Ladda R L. Am J Med Genet. 1993;46:623–629. doi: 10.1002/ajmg.1320460604. [DOI] [PubMed] [Google Scholar]
- 15.Wong K, Ren X R, Huang Y Z, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay S M, Mei L, et al. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 16.Self A J, Hall A. Methods Enzymol. 1995;256:3–10. doi: 10.1016/0076-6879(95)56003-3. [DOI] [PubMed] [Google Scholar]
- 17.Richnau N, Aspenström P. J Biol Chem. 2001;276:35060–35070. doi: 10.1074/jbc.M103540200. [DOI] [PubMed] [Google Scholar]
- 18.Tribioli C, Droetto S, Bione S, Cesareni G, Torrisi M R, Lotti L V, Lanfrancone L, Toniolo D, Pelicci P. Proc Natl Acad Sci USA. 1996;93:695–699. doi: 10.1073/pnas.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamarche-Vane N, Hall A. J Biol Chem. 1998;273:29172–29177. doi: 10.1074/jbc.273.44.29172. [DOI] [PubMed] [Google Scholar]
- 20.Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre A C, Souville I, et al. Hum Mol Genet. 2001;10:941–946. doi: 10.1093/hmg/10.9.941. [DOI] [PubMed] [Google Scholar]
- 21.Merienne K, Jacquot S, Pannetier S, Zeniou M, Bankier A, Gesz J, Mandel J L, Mulley J, Sassone-Corsi P, Hanauer A. Nat Genet. 1999;22:13–14. doi: 10.1038/8719. [DOI] [PubMed] [Google Scholar]
- 22.Gécz J, Barnett S, Liu J, Hollway G, Donnelly A, Eyre H, Eshkevari H S, Baltazar R, Grunn A, Nagaraja R, et al. Genomics. 1999;62:356–368. doi: 10.1006/geno.1999.6032. [DOI] [PubMed] [Google Scholar]
- 23.Chelly J, Mandel J L. Nat Rev Genet. 2001;2:669–680. doi: 10.1038/35088558. [DOI] [PubMed] [Google Scholar]
- 24.Gécz J, Mulley J. Genome Res. 2001;10:157–163. doi: 10.1101/gr.10.2.157. [DOI] [PubMed] [Google Scholar]
- 25.Sotgia F, Minetti C, Lisanti M P. FEBS Lett. 1999;3:177–180. doi: 10.1016/s0014-5793(99)00658-4. [DOI] [PubMed] [Google Scholar]
- 26.Angeloni D, Lindor N M, Pack S, Latif F, Wie M H, Lerman M I. Am J Med Genet. 1999;86:482–485. doi: 10.1002/(sici)1096-8628(19991029)86:5<482::aid-ajmg15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 27.Higgins J J, Rosen D R, Loveless J M, Clyman J C, Grau M J. Neurology. 2000;55:335–340. doi: 10.1212/wnl.55.3.335. [DOI] [PubMed] [Google Scholar]
- 28.Decker C J, Parker R. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 29.Billuart P, Chelly J, Carrie A, Vinet M C, Couvert P, McDonell N, Zemni R, Kahn A, Moraine C, Beldjord C, et al. Ann Genet. 2000;43:5–9. doi: 10.1016/s0003-3995(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 30.D'Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon A K, Oostra B, Wu S K, et al. Nat Genet. 1998;19:134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- 31.Carrié A, Jun L, Bienvenu T, Vinet M C, McDonell N, Couvert P, Zemni R, Cardona A, Van Buggenhout G, Frints S, et al. Nat Genet. 1999;23:25–31. doi: 10.1038/12623. [DOI] [PubMed] [Google Scholar]
- 32.Antonarakis S E, Van Aelst L. Nat Genet. 1998;19:106–108. doi: 10.1038/450. [DOI] [PubMed] [Google Scholar]
- 33.Brose K, Bland K S, Wang K H, Arnott D, Henzel W, Goodman C S, Tessier-Lavigne M, Kidd T. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 34.Wu W, Wong K, Chen J H, Jiang Z H, Dupuis S, Wu J Y, Rao Y. Nature (London) 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zemni R, Bienvenu T, Vinet M C, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, et al. Nat Genet. 2000;24:167–170. doi: 10.1038/72829. [DOI] [PubMed] [Google Scholar]