Abstract
The genes that determine the development of the male or female sex are known in Caenorhabditis elegans, Drosophila, and most mammals. In many other organisms the existence of sex-determining factors has been shown by genetic evidence but the genes are unknown. We have found that in the fish medaka the Y chromosome-specific region spans only about 280 kb. It contains a duplicated copy of the autosomal DMRT1 gene, named DMRT1Y. This is the only functional gene in this chromosome segment and maps precisely to the male sex-determining locus. The gene is expressed during male embryonic and larval development and in the Sertoli cells of the adult testes. These features make DMRT1Y a candidate for the medaka male sex-determining gene.
The overwhelming majority of multicellular animal species occur as both sexes, and in many cases the decision of whether an organism becomes a male or a female is determined by the genome. In most mammals, several flies, and the worm Caenorhabditis elegans the cascades of sex-determining genes are reasonably well understood. However, the master regulator encoded by the sex-determining locus (SD) on the Y chromosome of most mammalian species, SRY, is not functioning like that in some mammals (1). In nonmammalian species, which also have a XX/XY sex-determination system, SRY is not present at all. Neither the sxl gene of Drosophila nor xol of C. elegans, the genes at the top of the sex-determination cascade in these organisms, are functioning in the same way in more distantly related species (for review see ref. 2). This finding indicates that on the molecular level another dimension of diversity is added to the complex situation of multiple genetic systems for sex determination. In between worms and flies on the one side and mammals on the other side, there is a large gap in our knowledge about sex-determination genes.
Fishes are an attractive group of organisms for studying the evolution of sex determination because members of this class exemplify a broad range of various types of sex determination from hermaphroditism to gonochorism and from environmental to genetic sex determination (for review see refs. 3 and 4). The structure and expression of genes involved in sex determination and differentiation can be compared in species, which exhibit either similar or divergent sex-determination systems. Unfortunately, in both main fish models, the pufferfish (Takifugu rubripes) and the zebrafish (Danio rerio), no information exists on the mode of sex determination, the potential presence of sex chromosomes, and the process of sex differentiation.
The situation for a molecular analysis of sex determination is much more favorable in another fish model, the medaka (Oryzias latipes) (for review see ref. 5). Medaka has a XX/XY sex determination system like mammals. Male and female medakas are easily distinguished by a number of secondary sex characters (see ref. 6). A linkage map of the sex chromosomes with several molecular and phenotypic markers is available (7, 8). Especially useful is the quart strain, where the sex chromosomes express different alleles of the lf pigment marker. The presence of leucophores in the male and their absence in the female allow differentiation of both sexes as early as at 2–3 days of embryonic development (9).
From the genes known to be involved in sex determination/differentiation the DMRT genes are of special interest because of their widespread distribution. They form a family of genes, which share a highly conserved DNA-binding domain (10), the DM domain. DMRT1 is a candidate downstream sex-determination gene in mammals and appears to be involved in a certain type of XY sex reversal in humans (11–13). It is conserved in a wide range of animals with diverse sex-determining mechanisms, including C. elegans, Drosophila, fish, reptiles, birds, and mammals (14–19).
We have investigated a potential role for DMRT genes in sex determination of medaka and found that a duplicated copy of DMRT1 is present at the male SD and shows all features of a sex-determining gene.
Materials and Methods
Medaka Fish.
All experimental animals were from inbred lines of Northern (Kaga, HNI) and Southern medakas (i-3, quart, HB32C, SOK) except for the Carbio strain, which is an outbred strain derived from the Southern population of medaka.
Cloning and Sequence Analysis.
To obtain the Y-chromosomal DMRT gene the male-specific band from Southern blots of genomic DNA was excised and cloned. Briefly, the 3.3-kb male-specific EcoRI fragment from medaka (strain Carbio) was cloned by excising the 3- to 4-kb EcoRI fragments from a preparative 0.8% agarose gel. The DNA was ligated into λZapII EcoRI arms (Stratagene) and packaged in vitro. The resulting subgenomic library was screened with the 4-kb EcoRI fragment from medaka cosmid 73K2481, which contains exons 1–3 of the autosomal DMRT1 gene.
Filters of arrayed medaka genomic cosmid libraries (nos. 73 and 74) were obtained from the Resource Center of the German Human Genome Project (Berlin) and screened with the human DMRT1 cDNA under conditions of low stringency (hybridization: 35% formamide, 42°C; washing: 1× SSC/1% SDS, 63°C). Positive clones were initially characterized and arranged in groups by restriction fragment analysis and Southern blotting. Restriction fragments cross-hybridizing with the human DMRT1 probe were subcloned and sequenced.
For isolation of a medaka DMRT1Y cDNA, total RNA was isolated from 6-day-old embryos (strain Carbio). Reverse transcription was done with oligo(dT)12–18 and Superscript II reverse transcriptase following the supplier's protocol (GIBCO/BRL). For PCR from 3 μl of the reverse transcription reaction, primers based on the genomic sequence of DMRT1Y (DMTYh: TCT GCT GAG CTC CCC GGG; DMTYi: GCC TCG CAG CTT CTC A) were used. The PCR was run for 35 cycles at an annealing temperature of 62°C. The sequence of DMRT1Y is deposited under GenBank accession no. AY129240.
Bacterial Artificial Chromosome (BAC) Isolation and Analysis.
A genomic BAC library of the HNI strain with an average insert size of 160 kb was constructed (20). BAC clones containing the DM domain sequences were screened by colony hybridization under low stringency with the PCR fragment of the DM domain of medaka DMRT4 (20). The clones containing DMRT1Y were selected by PCR screening with a specific primer set (DMTk: CAA CTT TGT CCA AAC TCT GA; DMTl: AAC TAA TTC ATC CCC ATT CC). The contig was extended by using PCR end fragments as probes, and BACs 168M02, 209O12, and 113N21 were shotgun-sequenced.
Sequences were analyzed by application of the nix software tool (www.hgmp.mrc.ac.uk/NIX/). Putative exons and genes were compared at the nucleotide and amino acid levels to known genes by standard GCG programs and by blast and fasta database searches. The relevant part of the contig is deposited under GenBank accession no. AY129241.
Southern Blot Analysis.
DNA from individual fish was obtained from pooled organs as described. Five micrograms of genomic DNA was digested with restriction enzymes, separated on 0.8% agarose gels, and blotted onto nylon membranes (Hybond N+, Amersham Pharmacia). Membranes were hybridized either under conditions of moderate stringency [hybridization in 35% formamide, 0.1% Na-pyrophosphate, 50 mM Tris⋅Cl (pH 7.5), 5 × SSC, 1% SDS, 5 × Denhardt's, 100 μg/ml calf thymus DNA at 42°C, washing in 1 × SSC/1% SDS at 60°C] or high stringency (hybridization in the same buffer except that the formamide concentration was 50% and at the same temperature, washing in 0.1 × SSC/1% SDS at 68°C) with the following probes: OlaDMRT1, 4-kb EcoRI fragment from cosmid 73K2481; OlaDMRT1Y, 1.1-kb NheI/SstI fragment from phage 2.1 of the subgenomic library; and HsaDMRT1, 1.5-kb EcoRI fragment from the human DMRT1 cDNA.
Expression Analysis.
Total RNA was extracted from pooled organs of several adult medaka fish or 10–40 pooled total embryos of defined stages (21) by using the TRIZOL reagent (GIBCO/BRL) according to the supplier's recommendation. After DNase treatment reverse transcription was done with 2 or 4 μg total RNA by using Superscript II reverse transcriptase (GIBCO/BRL) and random primers. cDNA from 10 ng (actin) to 200 ng (adult organs) or 600 ng (whole embryos) of total RNA was used for PCR with gene-specific primers: Ola Actin, MAct1(TTC AAC AGC CCT GCC ATG TA) and MAct 2 (GCA GCT CAT AGC TCT TCT CCA GGG AG) at an annealing temperature of 60°C for 25 cycles; Ola DMRT1, DMT1m (TCC GGC TCC ACA GCG GTC) and DMT1n(CAG ACA GAG GGT TGG GGG G) at an annealing temperature of 64°C for 35 cycles; Ola DMRT1Y, DMTYa (GGCCGGGTCCCCGGGTG) and DMTYc (CTG GTA CTG CTG GTA GTT GTG) at an annealing temperature of 64°C for 35 cycles.
RNA from sex-reversed embryos and adults was obtained after treating embryos from day 1 until hatching with 1 mg/ml 17β estradiol in the rearing medium essentially as described (22).
Whole-mount RNA in situ hybridization on adult testis was performed according to standard protocols (23). Samples were digested with proteinase K for 2 min before hybridization with a 488-nt DMRT1Y antisense riboprobe at 65°C. The riboprobe was generated from a partial cDNA that was obtained by reverse transcription–PCR using the DMTYa and DMTYc primers. Stained tissue samples were paraffin-embedded, sectioned, and counterstained with eosin.
Mapping.
A sex-reversed XY female backcross panel was generated as reported (8). A total of 117 backcross progeny were analyzed for four STS markers (Yc-2, Casp6, SL1, Casp3B) that were previously isolated (7, 8, 24) and two phenotypic markers (lf: leucophore, y: male sex). DMRT1Y was mapped by PCR using primers DMTk and DMTn (TGA TGC AGC ATT TTG ACA CAT TTA). The products were electrophoresed on 6% acrylamide gels. Segregation of the markers was analyzed with Macintosh mapmaker version 2 (25).
Fluorescence in Situ Hybridization.
BAC clones were labeled separately by standard nick translation using biotin-16-dUTP and digoxigenin-11-dUTP. For two-color hybridization equal amounts of labeled probes were mixed with hybridization solution at a final concentration of 10 μg/ml and used at 100 ng per slide. Before hybridizing with denatured medaka mitotic chromosomes the probe mixture was denatured and preannealed in the presence of excess genomic DNA. Hybridization sites for both probes were simultaneously detected by means of rhodamine-conjugated avidin (Vector Laboratories) and antidigoxigenin (monoclonal)-conjugated fluorescein (Sigma). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Digitized images of the FITC and rhodamine signals were captured separately and displayed on DAPI-stained chromosomes by using EASY FISH 1.0 software (Applied Spectral Imaging, Mannheim, Germany).
Results
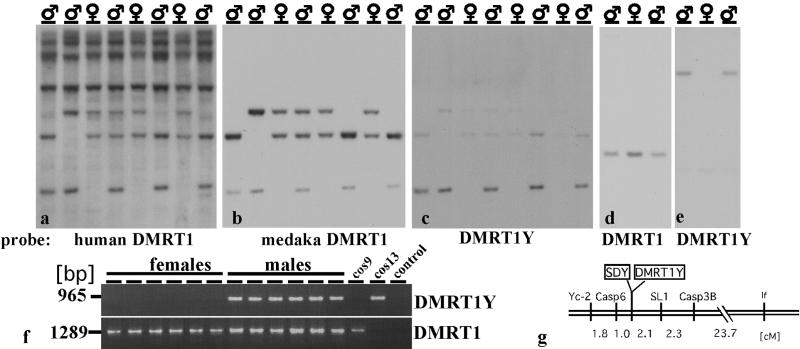
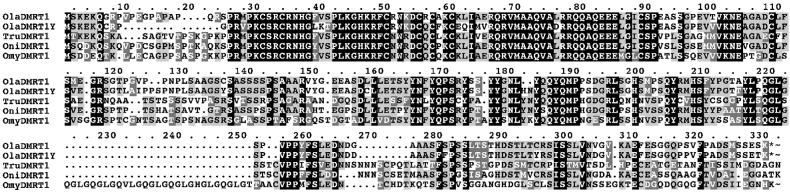
In our studies on sex-determining genes in the medaka we have recently found that the DMRT1 homologue of medaka is located on an autosome in a gene cluster together with its paralogues, DMRT 2 and 3 (26). cDNA probes from human and medaka DMRT1 used on Southern blots of male and female DNA disclosed an additional fragment only in males (Fig. 1 a–e). The male-specific fragment was cloned from a partial genomic library and found to be a duplicated version of DMRT1, showing 93% identity on the nucleotide and 90% similarity on the amino acid level (Fig. 2). It has a nucleotide identity of 96% (92% amino acids) to the DMY gene isolated by Matsuda et al. (27). The sequence differences probably are caused by different strains of medaka that were used for analyses. PCR primers specific to the Y-chromosomal copy (designated DMRT1Y) were used for a linkage analysis (Fig. 1f). DMRT1Y was found in all 81 males but not in 57 females of the i-3 strain and in 226 tested females of seven other strains from the Northern and Southern medaka populations. For mapping we used the sex-reversed (XY female × XY male; ref. 8) backcross mapping panel, which spreads the map distances of Y-chromosomal markers in the vicinity of SD by a factor of 10 compared with the resolution obtained with a male backcross panel. No recombination between SD and DMRT1Y was detected (Fig. 1g), whereas all earlier described markers map with some distance left and right to SD (8, 24). Thus DMRT1Y and the male SD colocalize on the genetic map (linkage <0.24 cM, equivalent to approximately 125 kb according to refs. 7 and 28).
Figure 1.
Southern blot of EcoRI-digested male and female DNA with a human DMRT1 cDNA (a), and rehybridized with medaka genomic DMRT1 (b) and DMRT1Y (c) probes. DMRT1 and DMRT1Y show some cross-hybridization to each other even under conditions of highest stringency because of the high sequence similarity. The hybridization conditions in a were moderate stringency and high stringency in b and c. (d and e) Hybridization under even further increased stringency conditions of PstI-digested DNAs. (f) PCR from DNA of female and male medakas (strain i-3) with DMRT1Y- and DMRT1-specific primers. Cosmid 9 contains the autosomal DMRT1 gene (26) and cosmid 13 contains DMRT1Y. (g) Genetic linkage map of the region flanking the male SD, based on meiosis in sex-reversed XY females. Numbers indicate genetic distances in centimorgans (cM). Markers DMRT1Y and SDY (locus determining the male sex phenotype) showed no recombination.
Figure 2.
Sequence comparison of the medaka DMRT1 and DMRT1Y genes with the DMRT1s from other teleosts. Ola, Oryzias latipes (Medaka); Tru, Takifugu rubripes; Oni, Oreochromis niloticus (Tilapia); Omy, Oncorhynchus mykiss (rainbow trout).
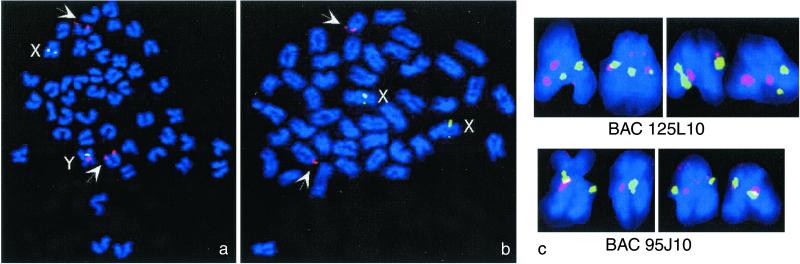
Sequence analysis of BACs from a genomic library of the HNI strain, cosmids from two independent genomic libraries of Northern (Kaga strain) and Southern (Carbio strain) medaka, and a full-length testis cDNA further confirmed that DMRT1Y has all of the features of a functional gene and is not corrupted by mutation. BAC 15H17, which contains DMRT1Y, and adjacent BACs were used for fluorescence in situ hybridization analysis (Fig. 3) on male and female metaphases. BACs containing the marker sequences flanking the male-determining locus [SL1, Casp3B (data not shown), Casp6 (data not shown)] gave signals on both the X and Y chromosomes. Neighboring BACs that overlap with l5H17 hybridized to the Y and X chromosome as well. However, BAC 15H17 gave a strong hybridization signal only on the Y, but not the X. Weak signals were obtained with BAC 15H17 at the subtelomeric region of a chromosome pair that is equivalent to linkage group 9 of medaka. This is the location of the autosomal DMRT cluster.
Figure 3.
Identification of the medaka Y chromosome: metaphases from male (a) and female (b) showing the hybridization signals of two BAC probes (15H17: DMRT1Y; 98C17: SL1). Note the presence of three hybridization spots for the BAC 15H17 in males as compared with the two spots in female (red signal). The additional fluorescence in situ hybridization signal in male is on the Y chromosome. The two relatively weak signals (arrows) in both male and female metaphase spreads represent the autosomal DMRT1 locus (linkage group 9). The SL1 marker containing BAC 98C17 detects both sex chromosomes (green signal). (c) Two highly enlarged XY chromosome pairs from two metaphases of male medaka showing hybridization to both sex chromosomes of BACs, which contain sequences flanking the Y-specific region on either side (95J10 and 125L10) (red signals). The SL1 marker containing BAC98C17 (green signals) was used to identify the sex chromosomes.
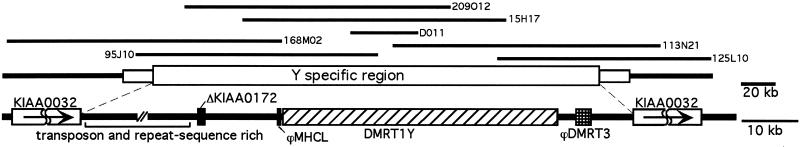
Analysis of the contiguous sequence of three overlapping BACs (covering 380 kb, roughly equivalent to 0.74 cM; refs. 7 and 28) revealed the size and the borders of the duplicated region on the Y (Fig. 4). Downstream of the Y-specific region a gene is encoded that is the medaka orthologue of human KIAA0032 (GenBank accession no. BAA04945). This gene (as well as the next one, KIAA0914, GenBank accession no. XP_003489) is also present on the X, thus defining the downstream limit of the Y-specific segment. At the upstream border of the Y-specific fragment another copy of KIAA0032 is present. Thus, the size of the Y-specific fragment is about 260 kb. In the Y-specific region DMRT1Y is the only gene that is not corrupted by mutations. There are remnants of three other genes that are also found adjacent to DMRT1 on the autosome and a fourth one from elsewhere in the genome, but they are all nonfunctional. From the BAC sequences no other functional gene is predicted by using 11 different gene or exon/intron prediction programs. However, a strikingly high number of transposons and other repetitive sequences were noticed, consistent with the expected genetic degeneration and recombinational isolation of the chromosomal region surrounding SD on the male-determining chromosome (29).
Figure 4.
Schematic representation of the DMRT1Y-containing region. Lines above show the analyzed BAC and cosmid clones. Genes and sequences with predicted homology to known genes are shown as boxes: striped, DMRT1Y; hatched, ϕDMRT3; light gray, KIAA0032; black, ΔKIAA0172 and ϕMHCL. The region upstream of ΔKIAA0172 contains only repetitive DNA and sequences with similarity to transposable elements of various organisms. Two genes upstream of DMRT1, a myosin heavy chain like gene (MHCL) and an ankyrin repeat containing gene (orthologous to human KIAA0172), are part of the duplicated fragment from linkage group 9 on the Y. The duplicated Y-chromosomal copy of MHCL is, however, destroyed by insertion of a poseidon element, a non-long terminal repeat retroposon (40), in Southern medaka and additionally a TX-1-related transposon in Northern medaka. The Y-chromosomal version of KIAA0172 is corrupted by a deletion that takes out two exons. In addition, the 5′ part of the gene is missing, indicating the border of the duplicated fragment. In intron 4 of DMRT1Y an insertion has occurred. This insertion contains a duplicated copy of the putative medaka homologue of the human brain and testes antigen gene MAP1 that is located on medaka linkage group 19 (M.K., H. Mitani, A.S., and M.S., unpublished work). The Y-chromosomal copy of MAP1, however, has a frameshift mutation that leads to a prematurely terminated protein. Downstream of the Y-chromosomal DMRT1 a copy of DMRT3 is found. But its coding sequence is lacking the ATG start codon and it has several frameshifts. DMRT2, which is the next gene following DMRT3 on the autosomal cluster, is not found in the Y-specific region.
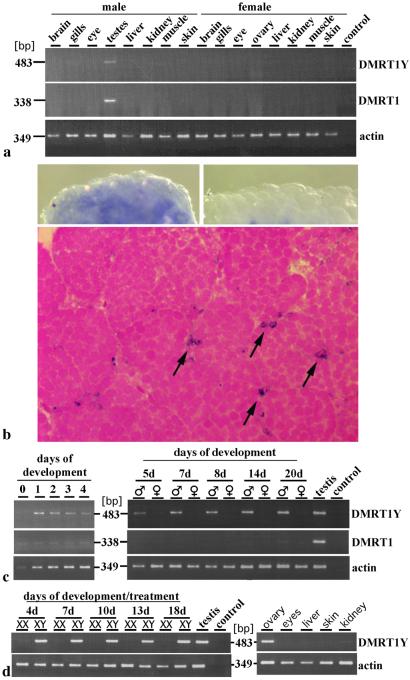
In adult fish DMRT1Y is expressed only in testes like the autosomal DMRT1 (Fig. 5a). The transcript is localized in the Sertoli cells (Fig. 5b). During development DMRT1Y expression starts at the neurula stage and persists during embryogenesis and larval stages to adulthood (Fig. 5c). It is expressed only in male embryos. The expression of the autosomal DMRT1 starts much later, around day 20.
Figure 5.
Expression of DMRT1Y and DMRT1. (a) Reverse transcription–PCR with DMRT1Y-specific primers of total RNA from organs of adult male and female medaka. Actin expression was determined for calibration. (b) Whole-mount RNA in situ hybridization in adult testes with DMRT1Y antisense probe (Upper Left), DMRT1 sense control (Upper Right), and section of testes showing staining in Sertoli cells (arrows, Lower). (c) Reverse transcription–PCR of medaka embryos and hatchlings with the same primers as in a. (Left) Analysis from Carbio strain samples. (Right) Analysis from Quart strain samples. (d) DMRT1Y expression in 17β estradiol-treated Quart embryos and sex-reversed adult XY females.
To analyze whether DMRT1Y is an upstream sex-determining gene or a more downstream sex-differentiation gene we analyzed the expression of DMRT1Y in XY sex-reversed females. During the estrogen treatment and after hatching the expression of DMRT1Y was not affected. The transcript was even detected in the ovary of the adult XY females at levels comparable to testes (Fig. 5d).
Discussion
In medaka the pseudoautosomal region of the sex chromosomes is very large. In fact, sex chromosomal crossing-over occurs over the entire length of the chromosome, with the possible exception of the region immediately adjacent to SD on the Y chromosome where an extremely high density of markers could not be ordered because of lack of detectable recombination (7, 8, 24). Linkage group 1, which contains SD, is the largest one and is equivalent to one pair of homomorphic chromosomes of the largest group. All markers mapped on linkage group 1 (the sex chromosomes) are shared between the X and Y chromosomes, whereas the markers generated from the Y-specific fragment are present only on the Y chromosome (data not shown). This finding indicates that the Y-specific region may be very small. Together with the fact that DMRT1Y is highly similar to its ancestor DMRT1, which points to a recent rather than to an ancient duplication event, it appears that the sex chromosomes of medaka are at an early stage of evolution.
Gene hierarchy studies in the worm and the fly revealed that the corresponding DMRT1 homologues mab-3 and DSX are placed at the bottom of the sex-determination cascade. From an evolutionary point of view it appears that the genes at the top of the hierarchy, which rule the mechanism of sex determination, have become involved in this process only relatively recently. However, at least some of the downstream genes, like DMRT1, are conserved with respect to sequence and function (2, 30, 31). Based on findings that DMRT1 is Z-linked in chicken it has been suggested that DMRT1 in birds has been recruited as an upstream regulatory sex-determining factor (16, 32). In chicken DMRT1 expression precedes expression of all other potential SD genes, is stronger in male than in female gonads, and is evident before the sex differentiation of the gonad anlage starts (18, 19). However, the exact map position of DMRT1 on chicken Z in relation to the male SD locus remains to be determined. In the temperature-dependent sex determination of alligators and turtles, DMRT1 exhibits the expected expression pattern for a sex-determining gene (19, 33). In rainbow trout it has been shown that DMRT1 is expressed in the developing male gonad before morphological differentiation (15). In the mouse gonad specific expression of DMRT1 is detected at embryonic day 9.5, whereas Sry expression begins around embryonic day 10.5 (18). Also in humans the simultaneous onset of DMRT1 and Sry expression suggested a role of DMRT1 in early events of sex determination (34).
What makes DMRT1Y in medaka a reasonable candidate for a sex-determination gene? The usual experimental tools to confirm a candidate gene cannot be applied here. In medaka, like in many other fish, sex can be experimentally reversed by steroid treatment or interfering with the activity of sex-differentiation genes. This means that transgenic expression of genes that act downstream in the sex-determination cascade or even sex-differentiation genes will lead to full sex reversal as well. Blocking DMRT1Y activity by antisense oligonucleotides cannot be used either, because the gonad is the last organ system to develop in medaka (35), and even morpholinos are not stable enough to be effective (data not shown). Matsuda et al. (27) found that a point mutation in the DMY (DMRT1Y) leads to XY male to female sex reversal. This finding shows convincingly that this gene is necessary for male sexual development. Also in humans the loss of DMRT1 is connected to XY sex reversals (11–13, 36). DMRT1 knockout mice revealed that the gene is essential for testis development (37). Thus, the data are not informative whether the gene in medaka has a different (more upstream) or a similar function (more downstream) like that in mammals. Of course, the linkage of DMRT1Y to the male SD (independently seen in this and another study, ref. 27) is pretty suggestive for its function as the primary sex-determining gene. As genetic mapping has only a certain resolution, a candidate gene may be located very close but not exactly at the locus encoding the phenotype in question. Thus further cumulative evidence should be helpful.
First, our finding of expression of DMRT1Y in XY sex-reversed females indicates that DMRT1Y is located upstream in the genetic hierarchy and is not one of the male sex-differentiation genes that have to be suppressed by sex-reverting hormone treatment. This experiment, however, does not rule out a difference in activity of DMRT1Y in sex-reversed animals, for instance if DMRT1Y is regulated posttranscriptionally. Second, the expression pattern of DMRT1Y is consistent with a sex-determination function. It is expressed early and exclusively in the male embryo. Most importantly, DMRT1Y in medaka is the only functional gene found in the Y chromosome-specific segment at the sex-determining region. The fact that androgen treatment during the sensitive period produces functional XX sex-reversed males that are fully fertile (6) excludes that spermatogenesis or other sex-differentiation genes are present on the Y, unlike the situation in mammals (38, 39). Hence, DMRT1Y is not involved in these processes but should have a function as a male sex-determination gene.
Experiments on the biochemical function of DMRT1Y are needed to understand how this gene functions in the male sex-determination process. The origin of DMRT1Y from an autosomal gene raises the question of what role gene duplication of sex determination and differentiation genes may play in the evolution of sex-determination systems. Such studies in suitable fish models might also help to contribute to our understanding of the function of DMRTs in mammals.
Acknowledgments
We thank G. Schneider, H. Schwind, and P. Weber for breeding of the fish and G. Scherer for the human DMRT1 cDNA clone. We are especially indebted to the Sequencing Team of Keio University for BAC clone sequencing. This work was supported by grants supplied by the Commission of the European Community (FAIR CT 97-3796) and Fonds der Chemischen Industrie to M. Schartl, a grant from the Deutsche Forschungsgemeinschaft (SCHM 484/18-1) to M. Schmid, grants from the Future Program (JSPS-RFTF96L00401) from the Japanese Society for the Promotion of Science to A.S., and a Grant-in-Aid for Scientific Research on Priority Areas (Area Nr. 813) from the Ministry of Education, Science, Sports, and Culture (to A.S. and S.A.).
Abbreviation
- BAC
bacterial artificial chromosome
Footnotes
References
- 1.Just W, Rau W, Vogel W, Akhverdian M, Fredga K, Graves J A, Lyapunova E. Nat Genet. 1995;11:117–118. doi: 10.1038/ng1095-117. [DOI] [PubMed] [Google Scholar]
- 2.Marin I, Baker B S. Science. 1998;281:1990–1994. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- 3.Baroiller J F, Guigen Y, Fostier A. Cell Mol Life Sci. 1999;55:910–931. [Google Scholar]
- 4.Devlin R H, Nagahama Y. Aquaculture. 2002;208:191–364. [Google Scholar]
- 5.Wittbrodt J, Shima A, Schartl M. Nat Rev Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto T. Medaka (Killifish) Biology and Strains. Tokyo: Keigaku; 1975. [Google Scholar]
- 7.Naruse K, Fukamachi S, Mitani H, Kondo M, Matsuoka T, Kondo S, Hanamura N, Morita Y, Hasegawa K, Nishigaki R, et al. Genetics. 2000;154:1773–1784. doi: 10.1093/genetics/154.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo M, Nagao E, Mitani H, Shima A. Genet Res. 2001;78:23–30. doi: 10.1017/s0016672301005109. [DOI] [PubMed] [Google Scholar]
- 9.Wada H, Shimada A, Fukamachi S, Naruse K, Shima A. Zool Sci. 1998;15:123–126. doi: 10.2108/zsj.15.123. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Wilken J, Phillips N B, Narendra U, Chan G, Stratton S M, Kent S B, Weiss M A. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- 11.Raymond C S, Parker E D, Kettlewell J R, Brown L G, Page D C, Kusz K, Jaruzelska J, Reinberg Y, Flejter W L, Bardwell V J, et al. Hum Mol Genet. 1999;8:989–996. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- 12.Calvari V, Bertini V, De Grandi A, Peverali G, Zuffardi O, Ferguson-Smith M, Knudtzon J, Camerino G, Borsani G, Guioli S. Genomics. 2000;65:203–212. doi: 10.1006/geno.2000.6160. [DOI] [PubMed] [Google Scholar]
- 13.Ottolenghi C, McElreavey K. Mol Genet Metab. 2000;71:397–404. doi: 10.1006/mgme.2000.3060. [DOI] [PubMed] [Google Scholar]
- 14.Guan G, Kobayashi T, Nagahama Y. Biochem Biophys Res Commun. 2000;272:662–666. doi: 10.1006/bbrc.2000.2840. [DOI] [PubMed] [Google Scholar]
- 15.Marchand O, Govoroun M, D'Cotta H, McMeel O, Lareyre J, Bernot A, Laudet V, Guiguen Y. Biochim Biophys Acta. 2000;1493:180–187. doi: 10.1016/s0167-4781(00)00186-x. [DOI] [PubMed] [Google Scholar]
- 16.Nanda I, Shan Z, Schartl M, Burt D W, Koehler M, Nothwang H, Grutzner F, Paton I R, Windsor D, Dunn I, et al. Nat Genet. 1999;21:258–259. doi: 10.1038/6769. [DOI] [PubMed] [Google Scholar]
- 17.Raymond C S, Shamu C E, Shen M M, Seifert K J, Hirsch B, Hodgkin J, Zarkower D. Nature (London) 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 18.Raymond C S, Kettlewell J R, Hirsch B, Bardwell V J, Zarkower D. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- 19.Smith C A, McClive P J, Western P S, Reed K J, Sinclair A H. Nature (London) 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- 20. Kondo, M., Froschauer, A., Kitano, A., Nanda, I., Hornung, U., Volff, J. N., Asakawa, S., Mitani, H., Naruse, K., Tanaka, M., et al. (2002) Gene, in press. [DOI] [PubMed]
- 21.Iwamatsu T. Zool Sci (Tokyo) 1994;11:825–839. [Google Scholar]
- 22.Iwamatsu T. J Exp Zool. 1999;283:210–214. [Google Scholar]
- 23.Hauptmann G, Gerster T. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda M, Sotoyama S, Hamaguchi S, Sakaizumi M. Genet Res. 1999;73:225–231. [Google Scholar]
- 25.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 26.Brunner B, Hornung U, Shan Z, Nanda I, Kondo M, Zend-Ajusch E, Haaf T, Ropers H H, Shima A, Schmid M, et al. Genomics. 2001;77:8–17. doi: 10.1006/geno.2001.6615. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey C E, Shibata N, Asakawa S, Shimizu N, et al. Nature (London) 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsuka M, Makino S, Yoda K, Wada H, Naruse K, Mitani H, Shima A, Ozato K, Kimura M, Inoko H. Genome Res. 1999;9:1277–1287. doi: 10.1101/gr.9.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlesworth B, Charlesworth D. Philos Trans R Soc London B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkins A S. BioEssays. 1995;17:71–77. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- 31.Zarkower D. Nat Rev Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- 32.Graves J A, Shetty S. J Exp Zool. 2001;290:449–462. doi: 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- 33.Kettlewell J R, Raymond C S, Zarkower D. Genesis. 2000;26:174–178. [PubMed] [Google Scholar]
- 34.Moniot B, Berta P, Scherer G, Sudbeck P, Poulat F. Mech Dev. 2000;91:323–325. doi: 10.1016/s0925-4773(99)00267-1. [DOI] [PubMed] [Google Scholar]
- 35.Hamaguchi S. Fish Biol J Medaka. 1992;4:11–17. [Google Scholar]
- 36.Ottolenghi C, Veitia R, Quintana-Murci L, Torchard D, Scapoli L, Souleyreau-Therville N, Beckmann J, Fellous M, McElreavey K. Genomics. 2000;64:170–178. doi: 10.1006/geno.2000.6121. [DOI] [PubMed] [Google Scholar]
- 37.Raymond C S, Murphy M W, O'Sullivan M G, Bardwell V J, Zarkower D. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lahn B T, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 39.Lahn B T, Pearson N M, Jegalian K. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- 40.Volff J N, Hornung U, Schartl M. Mol Genet Genomics. 2001;265:711–720. doi: 10.1007/s004380100468. [DOI] [PubMed] [Google Scholar]