Abstract
Programmed cell death involves the activation of caspase proteases that can mediate the cleavage of vital cytoskeletal proteins. We have recently reported that, in failing cardiac myocytes, caspase-3 activation is associated with a reduction in contractile performance. In this study we used a modified yeast two-hybrid system to screen for caspase-3 interacting proteins of the cardiac cytoskeleton. We identified ventricular essential myosin light chain (vMLC1) as a target for caspase-3. By sequencing and site-directed mutagenesis, a noncanonical cleavage site for caspase-3 was mapped to the C-terminal DFVE135G motif. We demonstrated that vMLC1 cleavage in failing myocardium in vivo is associated with a morphological disruption of the organized vMLC1 staining of sarcomeres, and with a reduction in myocyte contractile performance. Adenoviral gene transfer of the caspase inhibitor p35 in vivo prevented caspase-3 activation and vMLC1 cleavage, with positive impact on contractility. These data suggest that direct cleavage of vMLC1 by activated caspase-3 may contribute to depression of myocyte function by altering cross-bridge interaction between myosin and actin molecules. Therefore, activation of apoptotic pathways in the heart may lead to contractile dysfunction before cell death.
Heart failure is a leading cause of mortality that ensues following the chronic activation of biomechanical stress pathways, resulting from various forms of myocardial injury (1). Histological evidence of apoptosis has been identified in several cardiovascular disorders leading to congestive heart failure (CHF) (2, 3). Myocardial apoptosis represents a highly complex cell death program, whose execution is regulated by the caspase family of cysteine proteases. Caspase-3 is a key effector enzyme and cleaves downstream critical cellular targets involved in chromatin condensation, DNA fragmentation, and cytoskeletal destruction, thereby expressing the dramatic morphological changes of apoptosis (4). Caspase-3 activation has been documented in the myocardium of end-stage heart failure patients (5), and caspase-3 expression is increased in patients with right ventricular dysplasia, a disease associated with progressive cell loss and sudden death (6). Recently, myocyte apoptosis, assessed by different biochemical hallmarks, including caspase-3 activity, has been described in pacing-induced heart failure models in animals, and correlates with the time-dependent deterioration of cardiac function (7, 8). Moreover, we showed that caspase-3 activation directly influences contractile performance of failing ventricular myocytes, and can be corrected via adenovirus-mediated gene delivery of the potent caspase inhibitor p35 with a positive impact on contractility (8).
The molecular mechanism by which activated caspase-3 causes a deterioration of cardiac function has not yet been established. In an attempt to answer this question, we performed a screening for caspase-3-interacting proteins expressed in the heart, using a modified yeast two-hybrid system. We identified vMLC1 (ventricular essential myosin light chain) as a target for caspase-3, and investigated whether a correlation between caspase-3 activation, vMLC1 cleavage, and contractile performance exists in failing myocytes.
Materials and Methods
Yeast Two-Hybrid Screening.
Yeast two-hybrid screening using pBTM-casp3-p12p17m as bait vector was performed with a human heart cDNA library fused to the Gal4 activation domain in the pACT2 plasmid (CLONTECH), following the Hybrid Hunter two-hybrid system protocol (Invitrogen) in L40 yeast cells (MATa trp1 lue2 his3 ade2 LYS2∷4lexAop-HIS3 URA3∷8lexAop-lacZ). A total of 30 × 106 independent clones were screened by selective growth on Trp−/Leu−/His−/Ura−/Lys−/Ade+ synthetic dropout medium plates and expression of β-galactosidase activity.
In Vitro Cleavage of Positive Clone Products by Recombinant Caspase-3.
To construct expression plasmids for positive clones obtained from the two-hybrid screening, EcoRI–XhoI fragments of positive clones were inserted into the EcoRI–XhoI cloning sites of pYES2/NT-A plasmid (Invitrogen), in which the sequences were under control of the T7 promoter. Biotinylated lysine-labeled proteins were prepared from expression plasmids by using a TNT T7 Quick Coupled Transcription/Translation System (Promega), according to the manufacturer's instructions. Five microliters of biotinylated lysine-labeled protein were incubated for 1 hr at 37°C with 15 ng/μl recombinant active caspase-3 (BD PharMingen) and optionally with 25 μM caspase-3 inhibitor DEVD-fmk (fluoromethyl ketone) in a Tris⋅Cl reaction buffer, pH 7.5 (6 mM Tris⋅Cl, pH 7.5/1.2 mM CaCl2/5 mM DTT/1.5 mM MgCl2/1 mM KCl). Samples were size fractionated by SDS/15% PAGE and blotted to a nitrocellulose membrane. Colorimetric detection of biotinylated products was performed on blots with Transcend Colorimetric Translation Detection System (Promega).
Site-Directed Mutagenesis and Expression of Mutant Proteins in COS-7 Cells.
The coding sequence of human wild-type vMLC1 was cloned into the KpnI–XbaI sites of the gfp-expressing pAdTrack vector, between a non-tissue-specific cytomegalovirus (CMV) promoter and a SV40 polyadenylation signal. Site-directed mutagenesis of the Asp at position 132 to Ala (D132A) and of the Glu at position 135 to Ser (E135S) was performed with the QuikChange XL Site-Directed Mutagenesis kit (Stratagene), using the pAdTrack-vMLC1 plasmid and two complementary oligonucleotides containing the desired mutation. Mutated clones were identified by diagnostic StuI and BsrBI digestion (for D132A and E135S, respectively) and verified by sequencing.
For transient expression of wild-type vMLC1 and its mutants vMLC1mD132A and vMLC1mE135S, COS-7 cells were transfected with pAdTrack-vMLC1 or pAdTrack-vMLC1m plasmids by polycationic SuperFect Transfection Reagent (Qiagen, Valencia, CA). Forty-eight hours after transfection, cell protein extracts were prepared and analyzed for caspase-3 cleavage by Western blot.
Antibodies and Western Blot Analysis.
Protein extracts from rabbit ventricle or from COS-7 cells were prepared by homogenization in Tris⋅Cl reaction buffer, pH 7.5. To examine the cleavage by caspase-3, 150 μg of protein were incubated for 1 hr at 37°C with different amounts of recombinant human caspase-3, in the presence or absence of the caspase-3 inhibitor DEVD-fmk (25 μM). After size fractionation by SDS/15% PAGE, proteins were electrophoretically transferred to a nitrocellulose membrane and blots were incubated for 1 hr with mouse monoclonal antibodies against vMLC1 (0.2 μg/ml, clone 2c8, BiosPacific, Emeryville, CA; 1:10 dilution, clones F109.16A12 and F109.17A5, Biocytex, Marseilles, France). Bound antibodies were detected with horseradish peroxidase-conjugated antibody against mouse IgG (1:10,000 dilution, Sigma) and visualized by chemiluminescence. Obtained chemiluminograms were evaluated by densitometric analysis using scion image (Scion, Frederick, MD).
Determination of Caspase-3 Specificity for DFVE.
Ac-DFVE-AMC and Ac-DEVD-AMC were synthesized by Biosyntan (Berlin) with a purity of 94%. Active recombinant human caspase-3 (BD PharMingen) was added at a final concentration of 3 nM to a reaction mixture containing 20 mM Hepes, 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% CHAPS, 10% sucrose (pH 7.2), and both substrates in concentrations ranging from 0 to 400 μM. After a 10-min preincubation at 37°C, the released fluorogenic AMC was monitored every second minute for 20 min at 460 nm with 380 nm excitation. Km and kcat values were determined from plots of activity versus substrate concentration. Absolute kcat values were calculated using a standard curve determined with AMC.
Construction and Purification of Recombinant Adenovirus.
Recombinant (E1- and E3-deficient) adenoviruses (Serotype 5) carrying the green fluorescence protein gfp (Adgfp) or both gfp and the baculoviral apoptotic suppressor p35 (Adp35) were generated as described (8).
Pacing-Induced Heart Failure and Transcoronary Gene Delivery.
Pacemakers were implanted into New Zealand White rabbits, and animals were paced at 340 beats per min for 2 weeks, as described (8). Transcoronary gene delivery of Adgfp or Adp35 (5 × 1010 plaque-forming units) into rabbit myocardium was performed at the time ventricular pacing was initiated, as reported (8). The project was approved by the institutional ethics review board.
Immunoprecipitation of vMLC1.
Left ventricle lysates from control and 15 days paced rabbit myocardium were prepared by homogenization in denaturing lysis buffer (50 mM Tris⋅Cl, pH 7.4/5 mM EDTA/1% SDS/10 mM DTT/1 mM PMSF/2 μg/ml leupeptin/15 units/ml DNase I), heated at 95°C for 5 min, diluted 1:10 with nondenaturing lysis buffer (50 mM Tris⋅Cl, pH 7.4/300 mM NaCl/5 mM EDTA/1% Triton X-100/10 mM iodoacetamide/1 mM PMSF/2 μg/ml leupeptin/0.02% sodium azide), and centrifuged at 15,000 × g for 10 min at 4°C. After dilution to 3.5 mg of protein per ml, supernatants were precleared with excess of protein G-Sepharose beads (Sigma) and incubated for 2 hr at 4°C with protein G-Sepharose beads (30 μl of beads per ml lysate), preconjugated with 100 μg anti-vMLC1 monoclonal antibody (clone 2c8). Beads containing the immunocomplex were subjected to SDS/15% PAGE and immunoblotting for vMLC1.
Preparation and Culture of Adult Rabbit Ventricular Myocytes.
Single myocytes were isolated from the left ventricle of control and 15 days paced failing rabbits, and cultured in modified M199 medium on laminin-precoated glass slides, as described (9). Two hours after plating, cells were subjected to detection of activated caspase-3.
Activated Caspase-3 Detection and Fluorescence Staining.
Activated caspase-3 was detected in living cells by using CaspaTag Caspase-3 Activity Kit (Intergen, Oxford), according to the manufacturer's instructions. Freshly isolated ventricular myocytes were incubated at 37°C (5% CO2) with FAM-DEVD-fmk or SR-DEVD-fmk, carboxyfluorescein- or sulforhodamine-labeled fluoromethyl ketone tetrapeptide inhibitor of caspase-3. After 1 hr incubation, cells were washed, fixed in 4% paraformaldehyde, permeabilized in 100% methanol (at −20°C), and subjected to Hoechst 33258 staining and either to immunostaining for vMLC1/vMLC2 (ventricular regulatory myosin light chain), or to phalloidin staining. vMLCs were detected with mouse monoclonal antibodies anti-vMLC1 (4 μg/ml, clone 2c8) and anti-vMLC2 (1:2 dilution, clone F109.3E1, Biocytex), followed by incubation with Texas red or Cascade blue goat anti-mouse-IgG conjugate (10 μg/ml, Molecular Probes). Polymerized actin fibers were visualized by Texas red-phalloidin (3 units/ml, Molecular Probes).
Cell Shortening Experiments.
Fractional shortening was measured in rabbit adult cardiomyocytes isolated from left ventricle of control and 15 days paced failing myocardium, after detection of activated caspase-3. Experiments were performed in a temperature-controlled cuvette (37°C), using an electro-optical monitoring system (Scientific Instruments, Heidelberg), as described (9).
Statistical Analysis.
Data represent mean ± SEM and were analyzed by one-way analysis of variance, followed by Scheffé post hoc analysis. Statistical significance was accepted at the level of P < 0.05.
Results
Identification of vMLC1 as Substrate for Caspase-3.
In a rabbit model of CHF obtained by rapid ventricular pacing, we previously demonstrated that caspase-3 activation is associated with a reduction in contractile force of failing myocytes. Using in vivo transcoronary adenovirus-mediated gene delivery of the potent caspase inhibitor p35, we could correct caspase-3 activation in failing myocardium with a positive impact on sarcomeric organization and contractile performance (8). To better understand the mechanism that may cause caspase-mediated sarcomeric disarray, we performed in this study a screening for caspase-3-interacting proteins expressed in the heart. We used a modified yeast two-hybrid system with, as bait vector, the plasmid pBTM-casp3-p12p17m, which has already been successfully used to identify gelsolin as a substrate for caspase-3 (10). Both large (p17) and small (p12) subunits of active caspase-3 were separately expressed in yeast at equimolar ratios under ADH1 promoters. The small subunit was fused to the LexA DNA-binding domain, and a point mutation in the active site of the enzyme (Cys-163 → Ser) prevented proteolytic cleavage of interacting substrates. The bait plasmid was cotransfected into yeast with a human heart cDNA expression library fused to the Gal-4 activation domain. By screening 30 million transformants, we obtained 125 positive clones that were divided into 22 groups on the base of inserted fragment size and restriction enzyme digestion pattern. DNA sequencing analysis showed that six of the positive candidates encoded overlapping C-terminal parts (clones 7, 12, and 20) or the complete sequence (clones 3, 9, and 17) of vMLC1. MLC1 is one of the six polypeptide chains of the myosin molecule, and is proposed to function as an actin/myosin tether regulating cross-bridge cycling events (11). In this study we further analyzed the vMLC1 clones, and the others will be described elsewhere.
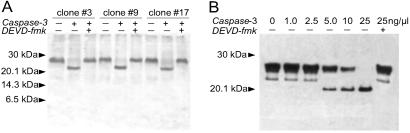
To examine cleavage of vMLC1 by caspase-3 in vitro, proteins encoded by the cDNAs were produced by in vitro transcription/translation-reaction. As shown in Fig. 1A, clones 3, 9, and 17, which contained the complete sequence of human vMLC1, were cleaved by human recombinant active caspase-3, and this cleavage was blocked in the presence of its tetrapeptide inhibitor DEVD-fmk, suggesting that vMLC1 is a possible substrate for caspase-3. Immunoblot analysis of protein extracts from left ventricle, incubated with active caspase-3, confirmed this result (Fig. 1B). An ≈20-kDa cleavage product for vMLC1 was already evident with 5 ng/μl active caspase-3. Indeed, other structurally related sarcomeric proteins, ventricular vMLC2 or β-myosin heavy chain, were not cleaved, demonstrating that cleavage of vMLC1 was not due to a generalized degradation of proteins (data not shown).
Figure 1.
In vitro cleavage of vMLC1 by recombinant active caspase-3. (A) SDS/15% PAGE of biotinylated lysine-labeled proteins from three positive clones encoding human vMLC1 after 1 hr incubation with 15 ng/μl human recombinant active caspase-3 in presence or absence of 25 μM DEVD-fmk. (B) Immunoblot analysis of native vMLC1 cleavage in protein extracts from rabbit left ventricle, incubated with indicated concentration of recombinant active caspase-3 for 1 hr. DEVD-fmk was applied in a concentration of 25 μM.
Mapping of the Caspase-3 Cleavage Site in vMLC1.
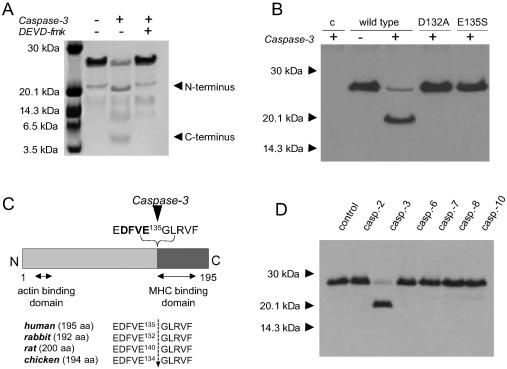
To determine caspase-3 cleavage site of vMLC1, purified human vMLC1 was incubated with recombinant active enzyme (Fig. 2A). Cleavage of purified vMLC1 resulted in two fragments at ≈20 kDa and ≈5 kDa. Edman sequence analysis of the cleavage products revealed that caspase-3 cleaved vMLC1 at E135 of the C-terminal motif DFVE135G, which is highly conserved. This result was confirmed by immunoblot analysis, using a monoclonal antibody for vMLC1 (clone F109.16A12) directed against the sequence V134EGLRV139 at the caspase-3 cleavage site. The antibody detected the intact vMLC1 protein but did not detect either of the two cleavage fragments (data not shown). The mapped cleavage site corresponds to the caspase-3 consensus sequence DXXD (12), with the exception of substituting the last Asp residue for the similar acidic Glu residue at position 135. Caspase-3 is considered to have an almost absolute requirement for an Asp in the P1 and P4 positions (12). To confirm DFVE135 as the caspase-3 cleavage site of vMLC1, we constructed mutant human vMLC1s replacing the Asp in position 132 with Ala (vMLC1mD132A) and the Glu in position 135 with Ser (vMLC1mE135S) by site-directed mutagenesis. As shown in Fig. 2B, wild-type vMLC1 was cleaved by caspase-3 in vitro. In contrast, both mutant proteins exhibited resistance to cleavage. This finding points to the caspase-3 cleavage site of vMLC1 as being C-terminal of E135 (Fig. 2C). Under the same conditions, other executioner caspases, such as caspase-6, -7, and -2, failed to cleave vMLC1 in vitro, suggesting that the protein is a specific substrate for caspase-3 (Fig. 2D).
Figure 2.
Determination of the cleavage site of vMLC1 and its specificity for caspase-3. (A) Cleavage of purified human vMLC1 (10 μg) by recombinant active caspase-3 (20 ng/μl, 1 hr, 37°C), analyzed by SDS/16.5% PAGE and Coomassie blue staining. (B) Immunoblot analysis of wild-type vMLC1, vMLC1mD132A, and vMLC1mE135S proteins expressed in COS-7 cells, after incubation of the cell extracts with recombinant active caspase-3 (20 ng/μl, 1 hr, 37°C). The first line (c) shows extracts from COS-7 cells transfected with control vector plasmid. Mouse monoclonal antibody directed against the residues 1 to 8 (clone F109.17A5) was used for immunodetection. (C) A diagram showing the caspase-3 cleavage site at the C-terminal side of E135 of human, rabbit, rat, and chicken vMLC1. (D) Immunoblot analysis of vMLC1 cleavage in protein extracts from rabbit left ventricle, incubated with different human recombinant active caspases (25 ng/μl) for 1 hr.
Because no other known targets for caspase-3 contain the cleavage site DFVE, we characterized the kinetic constants for the DFVE and the classical DEVD substrates in a fluorimetric in vitro assay. Caspase-3 cleaved efficiently both Ac-DFVE-AMC and Ac-DEVD-AMC with a reproducible Michaelis–Menten kinetic. Calculated Km, kcat, and kcat/Km values, which indicate affinity, turnover, and specificity, respectively, are presented in Table 1. Other caspases did not cleave the Ac-DFVE-AMC substrate (data not shown).
Table 1.
Kinetic constants for Ac-DFVE-AMC and Ac-DEVD-AMC cleavage by caspase-3
| Km, μM | kcat, s−1 | kcat/Km, s−1⋅M−1 | |
|---|---|---|---|
| Ac-DFVE-AMC | 43 | 0.13 | 3,023 |
| Ac-DEVD-AMC | 9 | 0.75 | 83,300 |
Caspase-3 was assayed and kinetic constants were calculated as described in Materials and Methods. kcat values are uncorrected for enzyme purity and fractional activity.
Cleavage of vMLC1 by Activated Caspase-3 in Vivo.
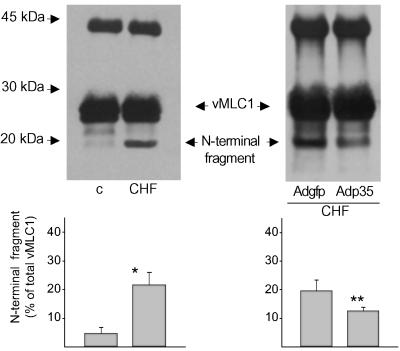
To determine the functional relevance of vMLC1 cleavage by caspase-3 in the heart in vivo, we investigated the evidence of vMLC1 cleavage products in extracts from rabbit failing ventricular myocardium, where we have previously documented a ≈6-fold increase in caspase-3 activity (8). As shown in Fig. 3, the intact vMLC1 protein of ≈27 kDa was relatively stable in healthy control hearts. In contrast, a main ≈20-kDa fragment, corresponding to the N-terminal cleavage product, was present in failing myocardium (% of total vMLC1: 22 ± 5%). Somatic gene delivery of the irreversible caspase inhibitor p35 into failing myocardium reduced vMLC1 cleavage of almost 30% compared with control adenoviral infection, indicating that vMLC1 disruption in failing myocytes is directly mediated by caspase-3 (Fig. 3). The lack of a complete abolition in vMLC1 cleavage by p35 infection can be due to the transcoronary gene transfer protocol used, which affects only a third of the whole left ventricular myocardium (8).
Figure 3.
In vivo cleavage of vMLC1 in failing myocardium and its reduction by p35 overexpression. Immunoblot analysis after immunoprecipitation of native vMLC1 cleavage products in extracts of left ventricle from control healthy (c) and failing (CHF) rabbit myocardium, after adenoviral gene delivery of gfp or p35. Shown are representative data from one of three animals in each group. Bar graph represents mean ± SEM of the percentage of the ≈20-kDa N-terminal fragment. n = 3. *, P < 0.01; **, P < 0.05 (vs. control and CHF+Adgfp, respectively).
Disruption of vMLC1, Sarcomeric Integrity, and Contractile Performance in Failing Myocytes.
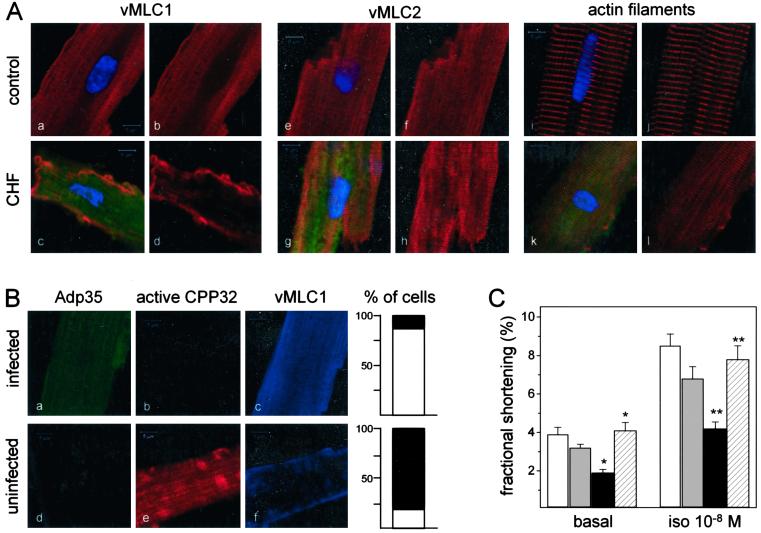
Myosin is the major component of the thick filaments of sarcomeres, and consists of two heavy chains (α and β), each associated with two types of light chains, the essential (MLC1) and the regulatory (MLC2). X-ray crystallographic analyses demonstrated that essential and regulatory myosin light chains are spatially close, and are both associated with the neck region of the myosin heavy chain globular head (13). To examine whether in failing myocytes a morphological disruption of the organized vMLC1 staining of A-bands in sarcomeres occurred, and whether it correlated with caspase-3 activation, single cardiomyocytes from control and CHF hearts were isolated. Fig. 4A shows confocal laser scanning microscopy of isolated ventricular myocytes after staining for activated caspase-3 and immunostaining for vMLC1 or vMLC2. In cardiomyocytes isolated from control hearts, there was no evidence of caspase-3 activation, and both myosin light chains appeared organized in the sarcomeric units (Fig. 4A a–b and e–f). In contrast, failing myocytes with activated caspase-3 presented a loss of the characteristic localization of vMLC1 in sarcomeres, and the A-band vMLC2 staining, which was maintained, showed a reduced sarcomeric organization compared with that of control cells (Fig. 4A c–d and g–h). Sarcomeric disarray in failing cells presenting caspase-3 activation was confirmed by phalloidin staining, which visualizes actin filaments (Fig. 4A k–l).
Figure 4.
In vivo cleavage of vMLC1 in failing cardiomyocytes is prevented by p35 overexpression with positive impact on myocyte contractile performance. (A) Laser scanning fluorescence microscopy of representative ventricular myocytes isolated from the anterolateral wall of control and failing myocardium. Green fluorescence (525 nm) shows activated caspase-3 identified by FAM-DEVD-fmk, blue fluorescence (480 nm) illustrates nuclei by Hoechst 33258, and red fluorescence (620 nm) reflects vMLC1 (a–d) or vMLC2 (e–h), or polymeric actin by phalloidin staining (i–l). (Scale bars = 5 μm.) One hundred cells isolated from three animals were analyzed in each group. (B) Representative ventricular myocytes isolated from failing myocardium after in vivo infection with Adp35 and visualized by laser scanning fluorescence microscopy. Green fluorescence (503 nm) identifies gfp and shows Adp35-infected cells, red fluorescence (580 nm) reflects activated caspase-3 labeled by SR-DEVD-fmk, and blue fluorescence (423 nm) illustrates vMLC1. (Scale bars = 5 μm.) Bar graphs represent the percentage of cells with organized (white pattern) or destroyed (black pattern) vMLC1 staining in the p35-infected and uninfected groups. One hundred fifty myocytes isolated from three animals were analyzed in each group. (C) Contraction amplitude under basal conditions and isoproterenol stimulation (10−8 M) measured in single left ventricle myocytes. White columns, cells from control myocardium; gray columns, cells from failing myocardium negative for activated caspase-3; black columns, cells from failing myocardium positive for activated caspase-3; hatched columns, cells from failing myocardium expressing p35. Data are expressed as mean ± SEM. n = 40 cells from three animals in each group. *, P < 0.005; **, P < 0.001 [in comparison with control or caspase-3-positive failing cells (basal, isoproterenol 10−8 M)].
We have recently demonstrated that adenoviral overexpression of p35 in vivo preserves sarcomeric structure and reconstitutes contractile performance of failing cardiomyocytes (8). Fig. 4B illustrates laser scanning fluorescence images of representative single ventricular myocytes isolated from failing myocardium infected with bicistronic Ad-p35 (5 × 1010 plaque-forming units) and stained for active caspase-3 and vMLC1. The typical vMLC1 staining was markedly destroyed in almost 80% of the uninfected failing cells, which presented activated caspase-3 (Fig. 4B d–f, and adjacent bar graph). On the contrary, in failing myocytes expressing the caspase inhibitor there was no evidence of caspase-3 activation and in 89% of the analyzed cells vMLC1 was localized in the A-bands of the sarcomeric units (Fig. 4B a–c, and adjacent bar graph).
Single-cell shortening experiments in failing cardiomyocytes showed a reduction of basal and isoproterenol-stimulated contraction correlated to the amount of caspase-3 activation in the cytosol of the failing cells (Fig. 4C). Furthermore, blockade of caspase-3 activity by expression of p35 restored contractile performance of failing myocytes (Fig. 4C). Adenoviral infection did not alter caspase-3 activation, vMLC1 staining, or shortening characteristics of the cells.
Discussion
In this study we used a modified yeast two-hybrid system to screen for caspase-3-interacting proteins of the cardiac cytoskeleton. We identified vMLC1 as a target for caspase-3, and demonstrated that its cleavage in failing myocardium in vivo is associated with a morphological disruption of the organized vMLC1 staining of sarcomeres. Furthermore, caspase-3 inhibition by adenoviral overexpression of p35 prevented vMLC1 cleavage with a positive functional effect on contractility.
Interestingly, vMLC1 is cleaved by caspase-3 at a noncanonical cleavage site, DFVE, that has neither been described earlier in other substrates nor identified as a possible caspase-3 cleavage site in combinatorial approaches by using synthetic tetrapeptide substrates (14, 15). It is widely accepted that caspases are “aspases,” enzymes cleaving substrates carboxyl-terminal to an Asp residue, and that caspase-3 consensus motif is DXXD (12). However, recent evidence indicates that caspase-9 and DRONC, a Drosophila caspase, show activity toward sites with Glu and Asp residues at the P1 position (16, 17). Furthermore, cleavage of the transcription factor Max by caspase-5, of tumor necrosis factor receptor-I by caspase-7, and of lens connexin 45.6 by caspase-3 have also been identified carboxy-terminal to a Glu (18–20). These findings suggest that the specificity of at least some caspases might be more complex than implied previously by studies with small synthetic peptides (14). Our data clearly confirm that the proposed requirement of caspase-3 for Asp in the P1 position is not absolute. Kinetic constants Km, kcat, and kcat/Km measured for the synthetic tetrapeptides Ac-DFVE-AMC and Ac-DEVD-AMC in vitro showed that caspase-3 has a cleavage preference for the classical site over the noncanonical (Table 1). However, the differences in enzyme-substrate affinity, turnover, and specificity are in the range of those described for other caspase–substrate interactions (14). Because of the high substrate/enzyme ratio for vMLC1 and caspase-3 in cardiac myocytes, a lower cleavage specificity may serve as a physiological protection against a massive protein disruption by minimal caspase-3 activation. Moreover, other caspases did not cleave either the native vMLC1 (Fig. 2D) or the DFVE peptide, indicating that the atypical vMLC1 cleavage site is strictly caspase-3-specific.
In a recent study, Communal et al. (28) examined in vitro the cleavage of cardiac myofibrillar proteins by caspase-3. The authors reported that active caspase-3 targets α-actin, α-actinin, and troponin T, with consequences for myocyte function. In our yeast two-hybrid screening, one of the 22 positive candidates encoded the C terminus of troponin T, but neither the protein obtained by in vitro transcription/translation-reaction nor the native protein in ventricular extracts was cleaved by recombinant active caspase-3. Similarly, we did not detect any cleavage product for α-sarcomeric actin in cardiac lysates incubated with the active enzyme (data not shown). These discrepancies could be due to the differing cleavage reaction conditions in vitro or to different antibody sensitivity. In a rabbit model of CHF, we have previously demonstrated that caspase activation contributes to disease progression and influences contractile performance of failing ventricular myocytes by destroying sarcomeric structure (8). It is plausible that numerous combined biochemical pathways are responsible for caspase-mediated dysfunction and sarcomeric disarray of failing cardiac myocytes, and that several components of the myofilaments can directly be disrupted by caspase-3. In this study, we showed that vMLC1 is cleaved by activated caspase-3 in failing myocardium in vivo and its cleavage can be blocked by p35 overexpression with functional consequences on contractility. These findings support the notion that this sarcomeric protein is a target for caspase-3 and plays a central role in the setting of experimental heart failure obtained by rapid ventricular pacing. Generation of a mouse model carrying a cleavage-resistant vMLC1 by cardiac-specific knock-in techniques would specifically allow evaluation of the importance of vMLC1 proteolysis in cardiac apoptosis.
A lot of data suggests that myosin light chains are important for cardiac and skeletal muscle function. Removing MLCs from chicken skeletal muscle myosin reduces the velocity of actin filament movement by 90% in an in vitro motility assay (21). Furthermore, MLC2 removal has little effect on isometric force, whereas MLC1 removal reduces the isometric force by over 50% (22). Mutations in the human essential light chain (Met-149 → Val) or regulatory light chain (Glu-22 → Lys, Pro-94 → Arg) of myosin are associated with rare variants of inherited cardiac hypertrophy, characterized by midventricular cavity obstruction, and correlate with disruption of the stretch activation response of the cardiac papillary muscles (23).
In the human heart, two different essential myosin light chain isoforms exist: (i) an atrial specific isoform (aMLC1, atrial essential myosin light chain), which is expressed in the fetal heart and decreases to undetectable levels during early postnatal development in the ventricle, but persists in the atrium for the whole life, and (ii) a ventricular specific isoform (vMLC1), which is the same isoform present in adult slow skeletal muscle (24). The reexpression of aMLC1 in adult human ventricles has been reported in patients with ischemic or dilative cardiomyopathy (25). Interestingly, in such patients with end-stage heart failure caspase-3 activation has also been documented (5). The isoform shift vMLC1 → aMLC1 correlates with an increase in cross-bridge cycling kinetics, as measured in skinned fibers derived from the diseased muscle (25). Postsurgical return to a normal hemodynamic state decreases aMLC1 expression in these patients (26). The functional significance of this isoform switch is not completely clear, but may be a direct compensatory mechanism to caspase-3-induced vMLC1 cleavage, triggered when the heart attempts to maintain normal cardiac function.
Taken together, these data clearly illustrate that minute changes in vMLC1 structure or composition can have a dramatic impact on myocyte function and heart contractility.
The molecular mechanism for MLC1 to affect the cross-bridge kinetics seems to reside in its Ala- and Pro-rich extended N terminus, which has been shown to interact with the C terminus of actin. The extended MLC1 N terminus may provide a tether between the myosin and actin filaments, serving to position the two filament systems for cross-bridge interaction and to amplify small movements of the myosin globular head (27). The MLC1 C terminus anchors the protein to the myosin globular head (13). One could speculate that destruction of vMLC1 at the C-terminal motif DFVE135G by activated caspase-3 may alter myosin/actin cross-bridge interactions by modifying myosin head stability and thereby lead to reduced force transmission.
In this study, we have demonstrated that vMLC1 is a cellular target for caspase-3. vMLC1 is cleaved, and its localization in sarcomeres is partially lost in failing cardiomyocytes, presenting caspase-3 activation and reduced contractile performance. Adenoviral expression of the caspase inhibitor p35 rescued the myopathic phenotype of the cells. It is plausible that vMLC1 disruption could alter the stiffness of the myosin neck region and therefore reduce the full range of myosin movement during contraction. Our findings suggest that caspase-3-mediated cleavage of vMLC1 may represent a molecular mechanism contributing to the deterioration of cardiac function before myocyte cell death.
Acknowledgments
We thank Dr. S. Kamada (Department of Medical Genetics, Osaka University Medical School) for providing us the plasmid pBTM-casp3-p12p17m and Dr. P. Hutzler (Institut für Biomedizinische Bildanalyse, GSF-Forschungszentrum Neuherberg, Munich) for supporting the laser scanning microscopy analysis. For Edman sequencing of the vMLC1 fragments, we thank Dr. S. Müller (Zentrum für Molekulare Medizin, Köln, Germany). These studies were supported by the Deutsche Forschungsgemeinschaft and Bayerische Forschungsmittel der Technischen Universität München.
Abbreviations
- CHF
congestive heart failure
- fmk
fluoromethyl ketone
- vMLC1
ventricular essential myosin light chain
- vMLC2
ventricular regulatory myosin light chain
Note
During the review process of this article, cleavage of cardiac myofibrillar components by caspase-3 with direct functional effects on skinned fiber contractility was reported by Communal et al. (28).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chien K R. Nature (London) 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- 2.Haunstetter A, Izumo S. Circ Res. 1998;82:1111–1129. doi: 10.1161/01.res.82.11.1111. [DOI] [PubMed] [Google Scholar]
- 3.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara J A, Quaini E, De Loretto C, Beltrami C A, Krajewski S, et al. N Engl J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 4.Hengartner M O. Nature (London) 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 5.Narula J, Pandey P, Arbustini E, Haider N, Narula N, Kolodgie F D, Dal Bello B, Semigran M J, Bielsa-Masdeu A, Dec G W, et al. Proc Natl Acad Sci USA. 1999;96:8144–8149. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallat Z, Tedgui A, Fontaliran F, Frank R, Durigon M, Fontaine G. N Engl J Med. 1996;335:1190–1196. doi: 10.1056/NEJM199610173351604. [DOI] [PubMed] [Google Scholar]
- 7.Cesselli D, Jakoniuk I, Barlucchi L, Beltrami A P, Hintze T H, Nadal-Ginard B, Kajistura J, Leri A, Anversa P. Circ Res. 2001;89:279–286. doi: 10.1161/hh1501.094115. [DOI] [PubMed] [Google Scholar]
- 8.Laugwitz K L, Moretti A, Weig H J, Gillitzer A, Pinkernell K, Ott T, Pragst I, Städele C, Seyfarth M, Schömig A, Ungerer M. Hum Gene Ther. 2001;12:2051–2063. doi: 10.1089/10430340152677403. [DOI] [PubMed] [Google Scholar]
- 9.Laugwitz K L, Ungerer M, Schöneberg T, Weig H J, Kronsbein K, Moretti A, Hoffmann K, Seyfarth M, Schultz G, Schömig A. Circulation. 1999;99:925–933. doi: 10.1161/01.cir.99.7.925. [DOI] [PubMed] [Google Scholar]
- 10.Kamada S, Kusano H, Fujita H, Ohtsu M, Koya R C, Kuzumaki N, Tsujimoto Y. Proc Natl Acad Sci USA. 1998;95:8532–8537. doi: 10.1073/pnas.95.15.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morano I. J Mol Med. 1999;77:544–555. doi: 10.1007/s001099900031. [DOI] [PubMed] [Google Scholar]
- 12.Cohen G M. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayment I, Rypniewski W R, Schmidt-Bäase K, Smith R, Tomchick D R, Benning M M, Winkelmann D A, Wesenberg G, Holden H M. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 14.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 15.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasula S M, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee R, Robbins P D, Fernandes-Alnemri T, Shi Y, Alnemri E S. Nature (London) 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 17.Hawkins C J, Yoo S J, Peterson E P, Wang S L, Vernooy S Y, Hay B A. J Biol Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- 18.Krippner-Heidenreich A, Talanian R V, Sekul R, Kraft R, Thole H, Ottleben H, Lüscher B. Biochem J. 2001;358:705–715. doi: 10.1042/0264-6021:3580705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ethell D W, Bossy-Wetzel E, Bredesen D E. Biochim Biophys Acta. 2001;1541:231–238. doi: 10.1016/s0167-4889(01)00159-8. [DOI] [PubMed] [Google Scholar]
- 20.Yin X, Gu S, Jiang J X. J Biol Chem. 2001;276:34567–34572. doi: 10.1074/jbc.M106073200. [DOI] [PubMed] [Google Scholar]
- 21.Lowey S, Waller G S, Trybus K M. Nature (London) 1993;365:454–456. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]
- 22.VanBuren P, Waller G S, Harris D E, Trybus K M, Warshaw D M, Lowey S. Proc Natl Acad Sci USA. 1994;91:12403–12407. doi: 10.1073/pnas.91.26.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pötter K, Jiang H, Hassanzadeh S, Master S R, Chang A, Dalakas M C, Rayment I, Sellers J R, Fananapazir L, Epstein N D. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 24.Price K M, Littler W A, Cummins P. Biochem J. 1980;191:571–580. doi: 10.1042/bj1910571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morano I, Hädicke K, Haase H, Böhm M, Erdmann E, Schaub M C. J Mol Cell Cardiol. 1997;29:1177–1187. doi: 10.1006/jmcc.1996.0353. [DOI] [PubMed] [Google Scholar]
- 26.Sütsch G, Brunner U T, von Schulthess C, Hirzel H O, Hess O M, Turina M, Krayenbuehl H P, Schaub M C. Circ Res. 1992;70:1035–1043. doi: 10.1161/01.res.70.5.1035. [DOI] [PubMed] [Google Scholar]
- 27.Milligan R A, Whittaker M, Safer D. Nature (London) 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 28.Communal C, Sumandea M, de Tombe P, Narula J, Solaro R J, Hajjar R J. Proc Natl Acad Sci USA. 2002;99:6252–6256. doi: 10.1073/pnas.092022999. [DOI] [PMC free article] [PubMed] [Google Scholar]