Abstract
The human prostate gland is an important target organ of androgenic hormones. Testosterone and dihydrotestosterone interact with the androgen receptor to regulate vital aspects of prostate growth and function including cellular proliferation, differentiation, apoptosis, metabolism, and secretory activity. Our objective in this study was to characterize the temporal program of transcription that reflects the cellular response to androgens and to identify specific androgen-regulated genes (ARGs) or gene networks that participate in these responses. We used cDNA microarrays representing about 20,000 distinct human genes to profile androgen-responsive transcripts in the LNCaP adenocarcinoma cell line and identified 146 genes with transcript alterations more than 3-fold. Of these, 103 encode proteins with described functional roles, and 43 represent transcripts that have yet to be characterized. Temporal gene expression profiles grouped the ARGs into four distinct cohorts. Five uncharacterized ARGs demonstrated exclusive or high expression levels in the prostate relative to other tissues studied. A search of available DNA sequence upstream of 28 ARGs identified 25 with homology to the androgen response-element consensus-binding motif. These results identify previously uncharacterized and unsuspected genes whose expression levels are directly or indirectly regulated by androgens; further, they provide a comprehensive temporal view of the transcriptional program of human androgen-responsive cells.
The androgenic hormones testosterone and dihydrotestosterone exert their cellular effects by means of interactions with the androgen receptor (AR), a member of the family of intracellular steroid hormone receptors that function as ligand-dependent transcription factors (1). Ligand-activated AR, complexed with coactivator proteins and general transcription factors, binds to cis-acting androgen response elements (AREs) located in the promoter regions of specific target genes and serves to activate or to repress transcription (1, 2). During human development, circulating androgens and a functional AR mediate a wide range of reversible and irreversible effects that include the morphogenesis and differentiation of major target tissues such as the prostate, seminal vesicles, and epididimus. The prostate gland has been used extensively as a model system to study androgen effects. In part, this is because of the fact that androgens promote the development and progression of prostate diseases that account for significant morbidity in the population including benign prostatic hypertrophy and prostate adenocarcinoma (2). The recognition that normal and neoplastic prostate epithelial cells depend on circulating androgens for their continued survival and growth led to the development of effective endocrine-based therapy for prostate carcinoma (3). To date, manipulating the androgen pathway by means of surgical or chemical castration remains the primary therapeutic modality for advanced prostate cancer.
In the human prostate, the AR mediates critical processes involved in the normal development, organizational structure, and mature function of the gland. During embryogenesis, the AR is expressed in mesenchymal cells of the urogenital sinus with subsequent temporal expression in prostate epithelial cells, leading to a differentiated epithelial phenotype and the production of prostate-specific proteins (4). In the mature gland, androgens promote cell division and the proliferation of prostate epithelial cells. However, androgens also seem to modulate programmed cell death and a “proliferative shut-off” function that leads to a state of cell quiescence (5, 6). Androgens regulate several aspects of prostate cellular metabolism, including lipid biosynthesis (7), and they control the production of specialized secretory proteins with prostate-restricted expression such as prostate-specific antigen (PSA; ref. 1).
The pivotal role of androgens for the regulation of distinct and diverse physiological processes in normal and neoplastic prostate cells has led to investigations designed to identify the molecular mediators of androgen action. Elegant studies have described morphological changes and gross alterations in DNA, RNA, and protein synthesis in the prostate in response to androgen manipulation (8). Our objective in this study was to characterize the temporal program of transcription that reflects the cellular response to androgens and to identify specific androgen-regulated genes (ARGs) or gene networks that participate in these responses.
Materials and Methods
Cell Culture and General Methods.
DNA manipulations including transformation, plasmid preparation, gel electrophoresis, and probe labeling were performed according to standard procedures (9). Restriction and modification enzymes (Life Technologies, Rockville, MD) were used in accordance with the manufacturer's recommendations. Prostate carcinoma cell lines LNCaP, DU145, and PC3 were cultured in phenol red-free RPMI medium 1640 supplemented with 10% (vol/vol) FCS. For androgen-regulation experiments, LNCaP cells were transferred into RPMI medium 1640 with 10% (wt/vol) charcoal-stripped FCS (CS-FCS) (Life Technologies) for 24 h followed by replacement of the media with fresh CS-FCS supplemented with 1 nM of the synthetic androgen R1881 (NEN/Life Sciences Products) or ethanol vehicle control. Cells were harvested for RNA isolation at 0-, 0.6-, 1-, 2-, 4-, 6-, 8-, 12-, 24-, and 48-h time points. Total RNA was purified from experimental and control cells by using Trizol (Life Technologies) according to the manufacturer's protocol. A reference standard RNA was prepared by combining equal quantities of total RNA isolated from LNCaP, DU145, and PC3 cell lines growing at log phase. RNA derived from one single batch of reference standard was used for every microarray hybridization. Northern analysis was performed as described (10). Multitissue Northern blots were obtained from CLONTECH.
Microarray Experiments.
A nonredundant set of ≈6,400 prostate-derived cDNA clones was identified from the Prostate Expression DataBase (PEDB), a public sequence repository of expressed sequence tag data derived from human prostate cDNA libraries (11). Microarrays were constructed as described (10). PEDB microarrays were assembled in versions composed of 3,000 or 6,388 cDNAs. A second microarray was constructed in a similar fashion by using a minimally redundant set of 17,630 human cDNA I.M.A.G.E. clones (UG Build 19V5.0, plate 1–48; Research Genetics, Huntsville, AL). Labeled cDNA probes were made from 30 μg of total RNA, as described (10). Probes were hybridized competitively to microarrays under a coverslip for 16 h at 63°C. Fluorescent array images were collected for both Cy3 and Cy5 by using a GenePix 4000A fluorescent scanner (Axon Instruments, Foster City, CA), and image intensity data were extracted and analyzed by using GENEPIX PRO 3.0 microarray analysis software. Each experiment was repeated with a switch in fluorescent labels to account for dye effects.
For each experiment, each cDNA was represented twice on each slide, and the experiments were performed in duplicate to produce four data points per cDNA clone per hybridization probe. Normalization of the Cy3 and Cy5 fluorescent signal in each experiment was determined by assuming equivalent global hybridization of test and reference probes. Data were filtered to remove values from poorly hybridized cDNAs with intensity levels less than 2 SDs above the background local to each spot. Intensity ratios for each cDNA hybridized with probes derived from the experimental time points were calculated as log2 (experimental intensity/reference intensity). Intensity ratios for each cDNA at each time point were compared with the time 0 values, and gene expression differences were considered significant if at least three of the four replicate spots for a given cDNA demonstrated an average log2 ratio of >1 or <−1 (2-fold change). Data from the four replicate cDNAs for each experiment were combined, and the average ratios were used for comparative analyses. To identify genes with similar temporal changes in expression, ratio measurements were imported into the CLUSTER software package (12). The results were visualized by using the TREEVIEW program (12).
Identification of ARE Motifs.
Reference sequences of 28 characterized ARGs with a temporal profile corresponding to that of PSA were obtained from the Ref_Seq database at National Center for Biotechnology Information (13) and used to query the assembled human genome sequence at the University of California, Santa Cruz (http://genome.ucsc.edu/). Approximately 3 kb of genomic sequence upstream of each mRNA sequence was obtained and used to search for similarity to the consensus ARE motif AGAACAnnnTGTTCT (TRANSFAC; http://transfac.gbf.de/TRANSFAC/). Scoring was based on the number of nucleotides in the query sequence that matched the consensus sequence by using the web-based tool PATSEARCH VI.1 (http://transfac.gbf.de/cgi-bin/patSearch/patsearch.pl). High-scoring matches that were homopolymeric in the left or right half of the consensus sequence were excluded. The sequence with the highest score was reported for each gene; only matches with at least 9 identities of the 12 consensus nucleotides were reported (75%). If more than one putative ARE was identified, the motif mapping nearest to the 5′ end of the reference cDNA sequence was reported. All of the putative ARE motifs were aligned by using the online CLUSTALW server at the Baylor College of Medicine (http://searchlauncher.bcm.tmc.edu/). The sequence logos representing the ARE consensus sequences were generated by using the online WEBLOGO sequence generation software (www.bio.cam.ac.uk/seqlogo/). The logo characters represent the sequence as stacked nucleotide residues for each position in the aligned sequences. The height of each letter is proportional to the nucleotide frequency at each position, and the nucleotides are sorted so that the most common one is on top. The height of the entire stack is then adjusted to signify the information content of the sequences at that position. The sequence logo represents the consensus sequence, the relative frequency of bases, and the information content (measured in bits) at every position in a site or sequence (14).
Results and Discussion
Construction of a Human Prostate cDNA Microarray.
cDNA libraries were produced from a variety of normal and neoplastic prostate tissue sources that included the LNCaP prostate cancer cell line. Clones were randomly selected, subjected to single-pass sequencing to generate expressed sequence tags (15), and assembled into distinct clusters based on nucleotide homology (11). Individual cDNAs corresponding to each of the ≈6,300 putative unique transcripts were selected, relocated into 384-well microtiter plates, amplified by PCR, and arrayed onto glass slides in duplicate (PEDB-Array). For some experiments described in this report, arrays consisting of a subset of 3,000 prostate cDNAs were used. For other experiments, the PEDB array was supplemented with a microarray (RG-Array) constructed with 17,630 commercially available cDNAs (Research Genetics). Overall, there are ≈3,000 genes overlapping on both arrays.
Androgen-Mediated Alterations in Gene Expression.
To assess the transcriptional response of prostate epithelial cells to androgen, hormone-responsive LNCaP prostate cancer cells were exposed to the synthetic androgen R1881 for specific time periods. The LNCaP cell line was chosen because it is one of the most widely used models for the study of prostate carcinoma and of the direct effects of androgens on human cells (16). Overall, 4,439 of 6,388 genes on the PEDB-Array (69%) and 5,642 of 17,630 genes on the RG-Array (32%) exhibited detectable transcripts in the LNCaP cells for a total assessable LNCaP transcriptome representing ≈8,000 genes (the two clone sets had ≈2,000 detectable cDNAs in common). A comparison of the expression profiles at specific time points after androgen stimulation demonstrated that the vast majority of transcripts (>96%) did not change by more than 2-fold compared with untreated cells. In contrast, 3.7% of the expressed transcripts were reproducibly altered more than 2-fold at one or more time points. After 24 h of androgen stimulation, the expression of 262 genes changed by >2-fold; 183 genes increased >2-fold, and 79 genes decreased >2-fold. After either 24 or 48 h of androgen exposure, the expression of 146 genes changed by ≥3-fold; 119 transcripts increased 3-fold, and 27 transcripts decreased 3-fold. Of these, 102 are genes with described functional roles (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), and 46 represent previously uncharacterized transcripts or putative proteins. These findings support the results from a recent report describing the use of Serial Analysis of Gene Expression to identify ARGs in prostate cells at one time point 24 h after stimulation with 10 nM R1881 (17). Of approximately 15,000 expressed genes assayed, 2.3% (351 distinct transcripts) were found to be either induced or repressed by androgen.
To determine whether androgen exposure induced distinct temporal patterns of gene expression, we used microarrays composed of 3,000 prostate-derived cDNAs to identify transcripts with a ≥2-fold change and grouped the resulting expression profiles of characterized genes by using hierarchical clustering methods (Fig. 1A; ref. 12). Four distinct clusters emerged from this analysis, with the largest group representing a cohort with members whose expression levels gradually increased from 4 h through 48 h (cluster C). This cohort includes the vast majority of genes previously shown to be androgen-regulated in prostate epithelium, such as KLK3/PSA, KLK2, NKX3A, TMPRSS2, TMEPA1, and SPAK (Fig. 1B; refs. 17–19). Other distinct clusters were composed of genes with a gradual decrease in transcript levels (cluster A), genes with a rapid decrease in transcript levels (cluster B), and genes with a rapid increase in transcript levels (cluster D). Genes comprising these clusters are listed on the PEDB web site (www.pedb.org/AR/microarray).
Figure 1.
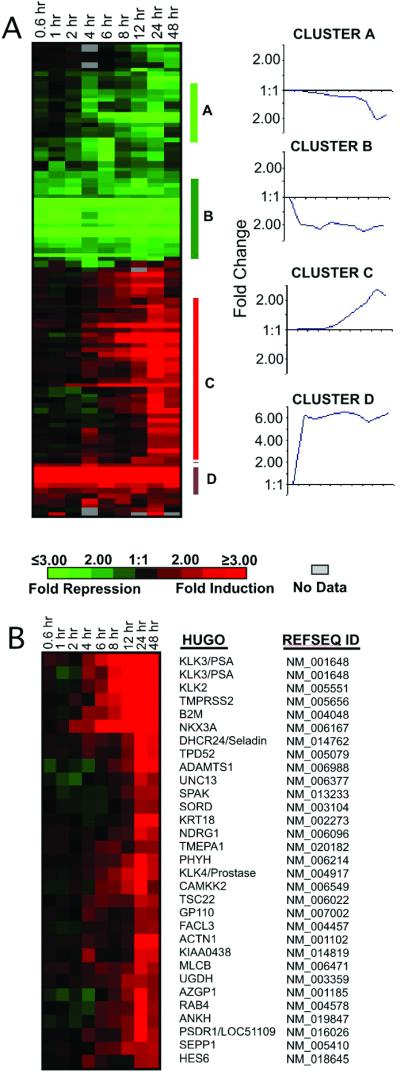
Temporal expression profiles of ARGs. (A) Microarrays composed of 3,000 prostate-derived cDNAs were used to acquire serial measurements of androgen-induced transcripts in the LNCaP cell line. RNAs showing at least a 2-fold change in expression after androgen exposure were clustered. Groups of genes with similar patterns of expression are indicated by vertical bars (A–D). Tick marks on the x axis of clusters A–D temporal profiles indicate the same time intervals as depicted at Left. (B) The expanded cohort of characterized genes with a temporal profile of expression corresponding to PSA are shown and named according to the HUGO gene nomenclature (www.gene.ucl.ac.uk/nomenclature/).
Androgen-Regulated Expression of Characterized Genes.
ARGs encoding proteins with defined biochemical function(s) were grouped into categories that reflect common functional attributes (Table 1). For clarity, each gene was assigned to only one category, although several of these genes exhibit activities that could be assigned to more than one functional role. The characteristics of the ARGs listed attest to the diverse cellular processes influenced by activation of the AR. In this discussion, we highlight several genes not previously described in the context of the cellular androgen regulation.
Androgen-Responsive Genes Involved in Metabolism.
Androgens induced the expression of transcripts encoding a diverse group of proteins involved in cellular metabolism. It has previously been shown that testosterone regulates genes involved in lipid and fatty acid biosynthesis through a coordinated indirect mechanism that involves the intermediary SREBP transcription factors (20). ARGs mediating fatty acid metabolism include fatty acid synthase and acetyl-CoA-carboxylase (20). ARGs mediating cholesterol metabolism include HMG-CoA-synthase and HMG-CoA-reductase (20). Our microarray studies identified additional ARGs involved in these pathways, including stearoyl-CoA desaturase, an enzyme that functions in the synthesis of unsaturated fatty acids, HELO1, a homolog of yeast long-chain polyunsaturated fatty acid elongation enzyme 2, and long-chain fatty acid CoA ligase 3, an enzyme that converts free long-chain fatty acids into fatty acyl-CoA esters. Of further interest, transcripts encoding 3-β-hydroxysterol-Δ-24 reductase (DHCR24) were increased 4.5- and 6-fold after 24 and 48 h of androgen exposure, respectively (Fig. 2A and Table 1). DHCR24 is a member of the FAD-dependent oxidoreductase family and catalyzes a reduction of the Δ24 double bond of sterol intermediates during cholesterol biosynthesis. DHCR24 is also known as seladin1, a gene shown to be down-regulated in the affected temporal cortex of patients with Alzheimer's disease (21). Expression of the DHCR24 protein protects cells from oxidative stress and amyloid-β peptide-induced apoptosis (21).
Figure 2.
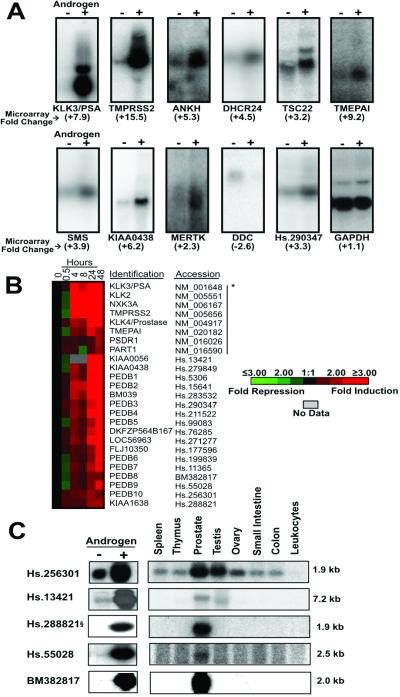
Androgen-regulated expression of characterized and previously uncharacterized genes. (A) Northern analysis confirmation of ARG expression in LNCaP prostate cancer cells treated with the synthetic androgen R1881 (+) or vehicle control (−) for 24 h. For each gene, the corresponding microarray-derived fold change in expression is provided below the gene name. (B) Hierarchical cluster analysis of uncharacterized genes exhibiting a temporal expression profile corresponding to PSA. *, genes to the left of the vertical bar represent characterized ARGs with tissue-expression profiles enhanced or restricted to the prostate. (C) Northern analysis confirmation of uncharacterized ARGs in LNCaP cells treated with androgen (+) or vehicle control (−) for 24 h. Multiple tissue Northern blot of selected uncharacterized ARGs demonstrating prostate-restricted or prostate-enhanced expression relative to other normal human tissues. §, one alternative spliced form of Hs.288821 exhibits prostate-enhanced expression.
The elevated expression of enzymes involved in lipid and cholesterol metabolism may simply reflect the mitogenic or secretory stimulus produced by androgen exposure. Cell division requires the biosynthesis of cell membranes, and the specialized secretory function of prostate epithelial cells requires the synthesis of storage vesicles and secretory components. However, there is emerging evidence that cholesterol and fatty acid metabolizing enzymes and their substrates may play more direct roles in carcinogenesis. Cholesterol seems to be intimately linked with signaling through the Ras pathway (22). Fatty acids also have been identified as signaling molecules which can be recognized by nuclear receptors (23). Numerous studies have reported an association with FAS expression and clinically aggressive cancers; one study correlates high levels of FAS expression with relapse risk in primary prostate carcinoma (24).
Androgen-Responsive Genes Involved in Transport or Trafficking.
The transcript encoding the FK-506 binding-protein FKBP5 (alias FKBP51) was up-regulated 25-fold in LNCaP cells after 48 h of androgen exposure. FKBP5 is a member of the immunophilin protein family and is involved in protein folding and trafficking. Of interest in the context of hormone-mediated gene expression is a report describing a role for FKBP5 in the earliest known event in glucocorticoid-receptor signaling through participation in the control of receptor subcellular localization and transport (25). To our knowledge, specific FKBP5 interactions with the AR or AR coregulatory proteins have not been described. However, FKBP5 has been shown to be up-regulated in xenograft models of androgen-independent prostate cancer and, thus, also may participate in AR signaling (26).
Among the genes involved in processes of cellular transport, we observed a 6-fold increase in ANKH gene expression (Fig. 2A and Table 1). ANKH encodes a multipass transmembrane protein that regulates the transport of pyrophosphate from the cytoplasm to the extracellular space (27). Mice with mutations in the ANKH gene develop a progressive form of arthritis accompanied by calcium phosphate mineral deposition, tissue calcification, joint destruction, and the formation of bony outgrowths (27). One unique hallmark of metastatic prostate cancer is a strong tropism for bone with the development of osteoblastic lesions characterized by excessive, disorganized deposition of new bone. It is possible that ectopic or altered ANKH expression in metastatic prostate cells or the surrounding bone stromal environment could contribute to the skeletal pathology that predominate in advanced prostate carcinoma.
Androgen-Responsive Genes Involved in Cell Proliferation or Differentiation.
Androgens mediate the disparate functions of prostate epithelial cell proliferation and differentiation (28). Prostate morphogenesis and overall glandular growth depends on circulating androgens during development. Subsequently, androgens maintain the differentiated functional state of the mature gland. Castration leads to the loss of differentiated secretory epithelium in rodent and human prostate tissues. This epithelium can be renewed by the restoration of androgens, but the overall proliferative response is limited to a specified cell mass through mechanisms yet to be characterized. There is evidence from several model systems that androgens also may participate in the negative regulation of cell proliferation (6, 29). A recent study using immortalized nontumorigenic rat prostate cells demonstrated a marked suppression of epithelial cell growth after cellular exposure to androgens that reflected changes in cell morphology consistent with terminal differentiation (30). The signaling mechanism(s) responsible for these distinct cellular responses have not been defined and may, in part, be temporally regulated during development and be dependent upon the cellular context of supporting stromal elements.
In this study, we identified several ARGs that encode proteins involved in cell-cycle regulation or cellular differentiation. Transcripts encoding the Maf oncoprotein were increased 16-fold after 48 h of androgen exposure. The original member of the Maf protein family (v-Maf) was identified as the transduced transforming component of avian musculoaponeurotic fibrosarcoma virus, AS42. Overexpression of Maf has been reported in multiple myeloma (31) and in melanoma cells (32). Functionally, several classes of transcriptional regulators have been shown to interact with Maf and/or Maf family proteins, including the bZip transcription factors Jun, Fos, and Bach1 (33). In addition to a role in oncogenesis, Maf mediates differentiation programs in specific cell types such as monocytes and helper T cells (34). It is hypothesized that Maf and related family members form a network with other classes of transcription factors that allow for the combinatorial fine tuning of regulatory protein interactions that dictate cellular responses that either prohibit or promote specific differentiation or growth programs.
The inhibitor of differentiation 2 (ID2) gene encodes a helix–loop–helix protein that is down-regulated in senescent human fibroblasts and during the differentiation process of lymphocyte development (35). ID2 disrupts the antiproliferative effects of the retinoblastoma protein family and negates the effect of the growth inhibitory protein p16 (36). Androgen induced the expression of ID2 3.5-fold after 48 h of exposure, an event that would be expected to relieve an inhibitory checkpoint for cellular proliferation.
Transcripts encoding several genes with reported roles in cell-cycle regulation were altered by androgen. The expression of cell division cycle 14B (Cdc14B) was up-regulated 3-fold at the 48-h time point. Cdc14 is essential for cell-cycle progression in yeast and encodes a protein tyrosine phosphatase involved in the exit of cell mitosis and initiation of DNA replication. In contrast to ID2 and Cdc14B, androgen decreased the expression of cyclin-dependent kinase 8 (CDK8). CDK8 and its regulatory subunit cyclin C are components of the RNA polymerase II holoenzyme complex which phosphorylates the largest subunit of RNA polymerase II (RNAPII). The cell cycle and transcription by RNAPII are closely related, and yeast orthologs of CDK8 and cyclin C have been implicated in the negative regulation of transcription. The mechanism of CDK8/cyclin C transcriptional regulation involves phosphorylation of the CDK7/cyclin H subunits of the general transcription initiation factor IIH (TFIIH), which results in repression of both kinase activity and the ability to activate transcription (37). Mimicking CDK8 phosphorylation of cyclin H in vivo has a dominant–negative effect on cell growth. Combined, the effect of androgen to down-regulate negative regulators of the cell cycle and induce positive regulators serves to promote a proliferative response. Future work should determine whether these molecular alterations represent specific androgenic cellular responses or rather reflect a more general mitogenic stimulus.
Androgen-Regulated Expression of Uncharacterized Genes.
In addition to genes with defined cellular functions, we identified 46 ARGs with homology only to hypothetical proteins or uncharacterized expressed sequence tags. Our criteria for prioritizing ARGs for verification and biochemical characterization centered on those with a tissue-distribution profile enhanced or specific to prostate tissue. Such genes may provide biological insights into unique facets of prostate physiology, or they may provide diagnostic or therapeutic targets for the treatment of prostate carcinoma. As a first step toward identifying genes with enhanced expression in the prostate, we selected ARGs with a temporal pattern of expression similar to that of PSA, a gene shown to be directly regulated by androgens and used extensively in clinical applications as a diagnostic and prognostic marker of prostate carcinoma. Seventeen uncharacterized ARGs grouped with KLK3/PSA and other well described prostate ARGs, such as KLK2 and NKX3A (Fig. 2B). We further investigated the tissue distribution of these ARGs by Northern analysis by using RNAs representing eight different human tissues (Fig. 2C). Five genes exhibited exclusive or high expression levels in the prostate relative to all other tissues studied, and these were confirmed to be induced by androgens in the LNCaP cell line (Fig. 2C). KIAA0056, originally identified through large-scale sequencing of cDNA clones from the immature myeloid cell line KG-1, encodes a putative protein of 1,498 residues (38). The Unigene sequence represented by Hs.256301 encodes the putative protein MGC13170 cloned from a retinoblastoma cDNA library. Interestingly, this sequence maps to chromosome 19q13.3, a region harboring several androgen-regulated prostate proteases including PSA/KLK3, KLK2, and KLK4/prostase. The protein predicted to be encoded by Hs.288821 does not exhibit overall similarity with any characterized human protein but does have WD-domains and has significant homology to proteins predicted in the Drosophila melanogaster and Caenorhabditis elegans genomes. By Northern analysis, several isoforms of Hs.288821 were shown to be present in human tissues with one isoform showing prostate specificity (Fig. 2C). The cDNA corresponding to Unigene sequence Hs.55028 maps to chromosome 16 and encodes a predicted polypeptide of 33 amino acids with similarity to a protein candidate for X-linked retinopathies (GenBank accession no. A46010). The cDNA for PEDB8 was derived from a library constructed from normal prostate tissue and does not exhibit significant homology with sequences in the public nucleotide databases.
Regulation of Androgen-Responsive Gene Expression.
Androgenic hormones exert their biological effects through the regulated expression of specific effector proteins and the initiation of signaling cascades. One mechanism of androgen-mediated gene expression involves the direct interaction of hormone with the AR protein, resulting in nuclear translocation and interactions with specific DNA sequences located near or within androgen target genes (1). Binding to these promoter and enhancer sequences, known as AREs and androgen regulatory regions, facilitates interactions with the general transcriptional machinery leading to gene transcription. Androgens also can affect gene expression through posttranscriptional (39) and genome-independent mechanisms (40). Alternatively, androgen target genes may be regulated indirectly: as a secondary or tertiary event through the initial direct up-regulation or liberation of a transcription factor(s) that in turn regulates the expression of other target genes (20). Such a network allows for layers of regulatory control that may be advantageous for the temporal direction of protein synthesis, the amplification of androgen signaling, and the coordinated expression of genes involved in common metabolic processes.
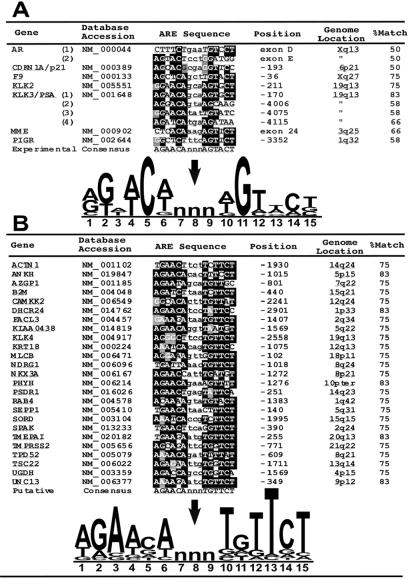
To gain an understanding of potential regulatory mechanisms operative in the ARGs identified in this study, we sought to identify sequences with similarity to known AREs that could support a mechanism of direct, rather than indirect, transcriptional control. Most AREs conform to a consensus sequence composed of two 6-base asymmetrical elements separated by three spacer nucleotides; 5′-AGAACAnnnTGTTCT-3′ (http://transfac.gbf.de/TRANSFAC/). To date, AREs in seven human genes have been characterized experimentally by others using reporter gene and gel-shift experiments (Fig. 3A). The AR and PSA genes have been shown to contain multiple functional AREs (41, 42). Importantly, operational human AREs that do not conform to the consensus mammalian ARE sequence have been described (42). A CLUSTALW alignment of the experimentally confirmed human AREs diverges from the consensus ARE with particular variability in positions 3 and 13 of the 15-nucleotide motif (Fig. 3A).
Figure 3.
Identification of ARE motifs. (A) Functional human AREs verified through experimentation are shown with positions relative to the transcriptional start site and the approximate genome location. A CLUSTALW alignment identifies highly conserved residues in black. A consensus sequence generated by WEBLOGO displays the frequency of each base in the consensus proportional to the character height with the height of the entire stack adjusted to signify the information content of the sequences at that position. (B) Putative human AREs identified by searching the 5′ regulatory regions of androgen target genes for a motif corresponding to the Transfac ARE consensus sequence. A CLUSTALW alignment identifies highly conserved residues in black. A consensus sequence indicates the relative frequency and importance of nucleotides in the motif.
To obtain DNA sequences containing putative gene regulatory elements, we searched the assembled human genome with mRNA reference sequences encoded by the 28 characterized ARGs with temporal expression profiles corresponding to that of PSA (Fig. 1B). Approximately 3 kb of sequence upstream of the putative transcriptional start sites were examined for homology to the consensus ARE obtained from the TRANSFAC database of eukaryotic cis-acting regulatory DNA elements. We identified 25 genes containing a motif comprising at least 9 of 12 nucleotides corresponding to the consensus ARE (Fig. 3B). A CLUSTALW alignment of these putative AREs suggests a high conservation of the “right-half” sequence, TGTTCT. Direct repeats of this motif have been shown to confer high-affinity AR binding that may contribute to androgen-selective responses in vivo (43). The biochemical characterization of these putative AREs should be useful for the delineation of critical nucleotides mediating DNA–AR interactions and allow for further studies of the contextual arrangement of AREs and other regulatory motifs in relation to transcriptional control.
Supplementary Material
Acknowledgments
We thank Jeff Delrow, Cassie Neal, Ryan Bosum, and Jan Kim for assistance with microarray experiments, and Mike Eisen for the public availability of microarray analysis software. This work was supported by the CaPCURE Foundation, National Cancer Institute Grants CA75173 and CA85286, and Department of Defense Grants DAMD17-98-1-8499 and DAMD17-00-1-0050 (to P.S.N.). P.S.N. is supported by a scholar award from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation.
Abbreviations
- AR
androgen receptor
- ARE
androgen response element
- PSA
prostate-specific antigen
- ARG
androgen-regulated gene
- PEDB
Prostate Expression DataBase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. BM382817).
References
- 1.Prins G S. Mayo Clin Proc. 2000;75:S32–S35. [PubMed] [Google Scholar]
- 2.Eder I E, Culig Z, Putz T, Nessler-Menardi C, Bartsch G, Klocker H. Eur Urol. 2001;40:241–251. doi: 10.1159/000049782. [DOI] [PubMed] [Google Scholar]
- 3.Huggins C, Hodges C V. Cancer Res. 1941;1:293–297. [Google Scholar]
- 4.Bonkhoff H, Remberger K. Prostate. 1996;28:98–106. doi: 10.1002/(SICI)1097-0045(199602)28:2<98::AID-PROS4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs J T, Lundmo P I, Berges R, Martikainen P, Kyprianou N, English H F. J Androl. 1992;13:457–464. [PubMed] [Google Scholar]
- 6.Geck P, Szelei J, Jimenez J, Lin T M, Sonnenschein C, Soto A M. J Steroid Biochem Mol Biol. 1997;63:211–218. doi: 10.1016/s0960-0760(97)00122-2. [DOI] [PubMed] [Google Scholar]
- 7.Swinnen J V, Verhoeven G. J Steroid Biochem Mol Biol. 1998;65:191–198. doi: 10.1016/s0960-0760(97)00187-8. [DOI] [PubMed] [Google Scholar]
- 8.Bruchovsky N, Lesser B, Van Doorn E, Craven S. Vitam Horm (San Francisco) 1975;33:61–102. doi: 10.1016/s0083-6729(08)60951-6. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Lin B, Ferguson C, White J T, Wang S, Vessella R, True L D, Hood L, Nelson P S. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 11.Hawkins V, Doll D, Bumgarner R, Smith T, Abajian C, Hood L, Nelson P S. Nucleic Acids Res. 1999;27:204–208. doi: 10.1093/nar/27.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt K D, Maglott D R. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider T D, Stephens R M. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson P S, Ng W L, Schummer M, True L D, Liu A Y, Bumgarner R E, Ferguson C, Dimak A, Hood L. Genomics. 1998;47:12–25. doi: 10.1006/geno.1997.5035. [DOI] [PubMed] [Google Scholar]
- 16.Horoszewicz J S, Leong S S, Kawinski E, Karr J P, Rosenthal H, Chu T M, Mirand E A, Murphy G P. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 17.Xu L L, Su Y P, Labiche R, Segawa T, Shanmugam N, McLeod D G, Moul J W, Srivastava S. Int J Cancer. 2001;92:322–328. doi: 10.1002/ijc.1196. [DOI] [PubMed] [Google Scholar]
- 18.Nelson P S, Han D, Rochon Y, Corthals G L, Lin B, Monson A, Nguyen V, Franza B R, Plymate S R, Aebersold R, Hood L. Electrophoresis. 2000;21:1823–1831. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1823::AID-ELPS1823>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Qi H, Labrie Y, Grenier J, Fournier A, Fillion C, Labrie C. Mol Cell Endocrinol. 2001;182:181–192. doi: 10.1016/s0303-7207(01)00560-3. [DOI] [PubMed] [Google Scholar]
- 20.Swinnen J V, Ulrix W, Heyns W, Verhoeven G. Proc Natl Acad Sci USA. 1997;94:12975–12980. doi: 10.1073/pnas.94.24.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greeve I, Hermans-Borgmeyer I, Brellinger C, Kasper D, Gomez-Isla T, Behl C, Levkau B, Nitsch R M. J Neurosci. 2000;20:7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox A D, Der C J. Crit Rev Oncog. 1992;3:365–400. [PubMed] [Google Scholar]
- 23.Vanden Heuvel J P. J Nutr. 1999;129:575S–580S. doi: 10.1093/jn/129.2.575S. [DOI] [PubMed] [Google Scholar]
- 24.Shurbaji M S, Kalbfleisch J H, Thurmond T S. Hum Pathol. 1996;27:917–921. doi: 10.1016/s0046-8177(96)90218-x. [DOI] [PubMed] [Google Scholar]
- 25.Davies T H, Ning Y M, Sanchez E R. J Biol Chem. 2001;20:20. [Google Scholar]
- 26.Amler L C, Agus D B, LeDuc C, Sapinoso M L, Fox W D, Kern S, Lee D, Wang V, Leysens M, Higgins B, et al. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- 27.Ho A M, Johnson M D, Kingsley D M. Science. 2000;289:265–270. doi: 10.1126/science.289.5477.265. [DOI] [PubMed] [Google Scholar]
- 28.Davies P, Eaton C L. J Endocrinol. 1991;131:5–17. doi: 10.1677/joe.0.1310005. [DOI] [PubMed] [Google Scholar]
- 29.Sato N, Gleave M E, Bruchovsky N, Rennie P S, Goldenberg S L, Lange P H, Sullivan L D. J Steroid Biochem Mol Biol. 1996;58:139–146. doi: 10.1016/0960-0760(96)00018-0. [DOI] [PubMed] [Google Scholar]
- 30.Whitacre D C, Chauhan S, Davis T, Gordon D, Cress A E, Miesfeld R L. Cell Growth Differ. 2002;13:1–11. [PubMed] [Google Scholar]
- 31.Chesi M, Bergsagel P L, Shonukan O O, Martelli M L, Brents L A, Chen T, Schrock E, Ried T, Kuehl W M. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- 32.Li M, Huang X, Zhu Z, Gorelik E. J Virol. 1999;73:9178–9186. doi: 10.1128/jvi.73.11.9178-9186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kataoka K, Shioda S, Yoshitomo-Nakagawa K, Handa H, Nishizawa M. J Biol Chem. 2001;276:36849–36856. doi: 10.1074/jbc.M102234200. [DOI] [PubMed] [Google Scholar]
- 34.Ho I C, Hodge M R, Rooney J W, Glimcher L H. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 35.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Nature (London) 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 36.Lasorella A, Iavarone A, Israel M A. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akoulitchev S, Chuikov S, Reinberg D. Nature (London) 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 38.Nomura N, Nagase T, Miyajima N, Sazuka T, Tanaka A, Sato S, Seki N, Kawarabayasi Y, Ishikawa K, Tabata S. DNA Res. 1994;1:251–262. doi: 10.1093/dnares/1.5.251. [DOI] [PubMed] [Google Scholar]
- 39.Perry J E, Tindall D J. Cancer Res. 1996;56:1539–1544. [PubMed] [Google Scholar]
- 40.Kousteni S, Bellido T, Plotkin L I, O'Brien C A, Bodenner D L, Han L, Han K, DiGregorio G B, Katzenellenbogen J A, Katzenellenbogen B S, et al. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- 41.Cleutjens K B, van Eekelen C C, van der Korput H A, Brinkmann A O, Trapman J. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 42.Dai J L, Burnstein K L. Mol Endocrinol. 1996;10:1582–1594. doi: 10.1210/mend.10.12.8961268. [DOI] [PubMed] [Google Scholar]
- 43.Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W. J Steroid Biochem Mol Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.