Abstract
von Willebrand factor (VWF) is synthesized primarily in vascular endothelial cells and secreted into the plasma as unusually large VWF multimers. Normally, these multimers are quickly degraded into smaller forms by a plasma metalloproteinase, VWF-cleaving protease (VWF-CP). Decreases in the activity of this enzyme result in congenital and acquired thrombotic thrombocytopenic purpura (TTP). The human VWF-CP has recently been purified. Cloning of the corresponding cDNA revealed that the 1,427-aa polypeptide is a member of the ADAMTS gene family, termed ADAMTS13. Twelve rare mutations in this gene have been identified in patients with congenital TTP. Here, we report missense and nonsense mutations in two Japanese families with Upshaw–Schulman syndrome, congenital TTP with neonatal onset and frequent relapses. The comparison of individual ADAMTS13 genotypes and plasma VWF-CP activities indicated that the R268P, Q449stop, and C508Y mutations abrogated activity of the enzyme, whereas the P475S mutant retained low but significant activity. The effects of these mutations were further confirmed by expression analysis in HeLa cells. Recombinant VWF-CP containing either the R268P or C508Y mutations was not secreted from cells. In contrast, Q449stop and P475S mutants were normally secreted but demonstrated minimal activity. Genotype analysis of 364 Japanese subjects revealed that P475S is heterozygous in 9.6% of individuals, suggesting that approximately 10% of the Japanese population possesses reduced VWF-CP activity. We report on a single-nucleotide polymorphism associated with alterations in VWF-CP activity; it will be important to assess this single-nucleotide polymorphism as a risk factor for thrombotic disorders.
Thrombotic thrombocytopenic purpura (TTP) is a generalized disorder characterized by microangiopathic hemolytic anemia, thrombocytopenia, neurological dysfunction, renal failure, and fever (1–3). Mortality of patients afflicted with this condition may exceed 90% without plasma exchange, the only effective treatment of TTP currently. Both acquired and congenital types of TTP exist. About half of patients with congenital TTP have their first acute episode in childhood, whereas the other half have their first acute episode in adulthood. Symptoms in adults often develop in association with the stress of infection or pregnancy (4). TTP with neonatal onset and frequent relapses is often diagnosed as Upshaw–Schulman syndrome (USS) (5, 6).
The platelet-adhesive blood coagulation protein, von Willebrand factor (VWF), is synthesized mainly in vascular endothelial cells (7–10). VWF is released into plasma as “unusually large” multimeric forms (UL-VWFM), highly active in interactions with platelets and collagen (11, 12). In normal human plasma, UL-VWFM rapidly depolymerizes to smaller multimeric forms ranging in size from 500 to 20,000 kDa. In patients with TTP, UL-VWFM remains in the circulation, because of the loss of regulated VWF proteolysis. The binding of UL-VWFM to platelets may then promote microvascular thrombosis, platelet consumption, and hemolysis. Enhanced platelet aggregation under high shear stress was also proposed as the mechanism of the clinical symptoms of TTP (13). Proteolytic depolymerization of VWF is catalyzed by VWF-cleaving protease (VWF-CP), partially purified by Furlan et al. (14) and Tsai (15). The cleavage site in VWF is an Y842-M843 peptidyl bond within the central A2 domain (14–17).
Human VWF-CP has recently been purified, allowing the determination of an N-terminal amino acid sequence (18–20). This information identified the cDNA sequence encoding VWF-CP (18, 21). Additional approaches attempting to determine the genetic cause of congenital TTP also discovered the same gene, ADAMTS13 (22). VWF-CP/ADAMTS-13 is a member of the ADAMTS family of metalloproteases, named for the characteristic combination of a disintegrin-like and metalloprotease with thrombospondin type 1 (TSP1) motif (23–26). VWF-CP is predominantly expressed in liver (18, 21, 22, 25), consistent with our previous observation that severely decreased levels of plasma VWF-CP activity in patients with biliary atresia can be restored by living-related liver transplantation.†† VWF-CP is 1,427 aa residues in length, containing a signal peptide, a propeptide, a reprolysin-like metalloprotease domain, a disintegrin-like domain, a TSP1, a cysteine-rich domain, seven additional TSP1 repeats, and two CUB domains identified in many developmentally regulated proteins (27). The C terminus of the VWF-CP propeptide ends with the sequence RQRR, suggesting that furin or a related protease cleaves this region during synthesis to allow the secretion of VWF-CP into the plasma as an active enzyme.
The human ADAMTS13 gene contains 29 exons, spanning ≈37 kb on chromosome 9q34 (18, 21, 22). Levy et al. (22) identified 12 mutations of ADAMTS13 in seven families of patients affected by congenital TTP (22). Two frameshift mutations were identified in exons 19 and 27, and one splice mutation was found in intron 13. The remaining nine mutations all result in nonconservative amino acid substitutions (H96D, R102C, T196I, R398H, R528G, R692C, C951G, C1024G, and C1213Y). These 12 mutations were excluded as common sequence polymorphisms. They also identified seven single-nucleotide polymorphisms (SNPs) associated with amino acid substitutions (R7W, Q448E, P618A, R625H, A732V, A900V, and A1033T) in 92 unrelated normal control DNA samples, although their effects on VWF-CP activity have not been assessed (22).
Here, we report three missense and one nonsense novel mutations in Japanese families with congenital TTP. Altered activities of the expressed recombinant VWF-CP mutants accounted for the patients' symptoms. Notably, one of the missense mutations is a common SNP with approximately 10% heterozygosity.
Materials and Methods
Sequencing of ADAMTS13 Gene.
All experiments using human materials were performed with the permission of the ethics committees at the National Cardiovascular Center, Yodogawa Christian Hospital, and Sapporo Kosei General Hospital. Human genomic DNA was isolated from whole blood by using an automated nucleic acid isolation system, NA-3000 (Kurabo, Japan). All exons of the ADAMTS13 gene, including the intron–exon boundaries, were PCR-amplified with corresponding intron primers (Table 1) by using either FastStart TaqDNA polymerase (Roche Diagnostics), the GC-RICH PCR system (Roche Diagnostics), or HotStarTaq DNA polymerase (Qiagen, Chatsworth, CA). Products were sequenced in both directions by using a BigDye Terminator Kit (Applied Biosystems) and a 3700 DNA Analyzer (Applied Biosystems).
Table 1.
Primers used for the amplification of ADAMTS13 exons and intron–exon boundaries
| Exon (size, bp) | Forward primer | Reverse primer | Product, bp |
|---|---|---|---|
| 1 (549) | GATTGCCAGGCCGTTTGTGAT | GCAAACCCCAAAGCTGATGTA | 768 |
| 2 (67) | CCTCGGTCTCCCCAAGTGTTA | GAACCCTGGCCTGGCTGGAAC | 348 |
| 3 (158) | GGTGGGGGTGACACGCAATGT | CCAGGGGAGGGAGGGAGAAGA | 437 |
| 4 (84) | TGTTTTCCTTGCGTTAGTTGG | GAGGATGGAGATGCGATGACT | 382 |
| 5–6 (125, 147) | AACAAACCGACCGCAGTCAGC | GGTTCCCCTGTCCTCACACCT | 638 |
| 7 (138) | GCTGGCGCTGCGGCACTAGGG | GTTGGACGGAGGGGTGGGTTG | 399 |
| 8 (163) | ACTCCTCCGTCCCGCCTCCTC | GGCCCAGTCAAACAAAAATGT | 446 |
| 9 (105) | GTGCAGAGTGTTGGCTGTGTC | CTCTGCCCCATACTGGTCCTG | 334 |
| 10–11 (152, 64) | TGAGGATGTTGGGGGACTCTC | CAAATGTGTCCTGGTGTGAAC | 512 |
| 12 (127) | TGAGGCCACACCCACATCTTG | ATGCCAGAGCCTGAACCACTT | 366 |
| 13 (149) | ATAGAAACCCTTGCCCCAGAT | ATCCTTTTCCCCAGCACCACT | 390 |
| 14 (121) | CAGGGCTGCAGAGTCATTGAG | GAAGGGTGGCGAAGTGGAAGA | 358 |
| 15 (81) | CTCCCTTTGTCTGTGGTGTGG | ACTATCAAGCCTGAGGGTGGT | 279 |
| 16 (182) | GGGACCCCGGGAAGGAGAGTC | GTAAGTGACCGCTGAATGAAT | 393 |
| 17–18 (136, 130) | GGCCAGGCTGGAGTGCTATGT | CAGAATGGGGGCACTCACAGA | 770 |
| 19 (186) | ACCAGCCTGTGATTCGGTTGT | AGGAACTCTGACAGCAGCACT | 548 |
| 20 (190) | CTCTTTGGGCTCCTGGATGTT | CAATGGGTGCTCCTCGTTCTC | 386 |
| 21 (121) | AAGGATACCCGCTGCGAGACC | AGCCAATCAACACCCACATTT | 489 |
| 22 (130) | CCATGCGGGCCTTATGTGCTA | TCTGGGTTGCAGTCCTCAAAG | 439 |
| 23 (183) | GGGGGCCTCCAGAAAGAGAAC | GTGTTGCCCAGGTTGGACTTG | 476 |
| 24 (205) | GGCTCAGTGGCTGCACTTTCC | TCCAGCGTCCCCAAACCTAAG | 576 |
| 25 (319) | GACAGGGACCCAGACTTGAAT | AAGTTACTTCCCCTTGATAGT | 729 |
| 26 (147) | CTGCATGTGCCCCCTCTTGCT | TGGGCACATCACTTAATCTCT | 574 |
| 27 (177) | GTGCATTCCCACCTGTAGTTT | TCCCTGGCACGTGCAGACTGA | 583 |
| 28 (185) | CCAGAGCCCAGAACATTTAGC | GCCACTATTTCACTCTTGTAG | 581 |
| 29 (412) | GTGTCCTTGGGGAAGTGATGT | GATTGGATTTTCTTCCTGGAT | 757 |
Transient Expression of VWF-CP.
To produce recombinant VWF-CP, the ORF of human ADAMTS13 was PCR-amplified from cDNA (18) with or without a C-terminal FLAG (DYKDDDDK) tag sequence. This fragment was inserted into a mammalian expression vector, pCAGG (28). We used PCR-based single-nucleotide mutagenesis to create the R268P, Q448E, Q449stop, P475S, and C508Y mutants. The direction and sequence of all inserts were confirmed by sequencing.
HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) on 60-mm dishes in humidified air with 5% CO2 at 37°C. Two micrograms of each expression plasmid was transfected into subconfluent cells by using FuGENE6 (Roche Diagnostics). After a 4-h incubation, culture media were replaced with 2.5 ml of serum-free OPTI-MEM I (Invitrogen) and incubated for 44 h. The media were then collected and centrifuged to remove any cellular residue. The cells were washed with PBS and lysed with 300 μl of SDS-sample buffer (10 mM Tris⋅HCl/2% SDS/50 mM DTT/mM EDTA/0.02% bromophenol blue/6% glycerol, pH 6.8).
Western Blot Analysis.
The collected culture media and cell lysates were subjected to SDS/PAGE (2–15% gradient) and transferred to a poly(vinylidene difluoride) membrane (Bio-Rad). After blocking with 3% skim milk, the membrane was incubated with 1 μg/ml anti-FLAG M2 mAb (Sigma) and then with 0.1 μg/ml peroxidase-labeled goat anti-mouse IgG (Kirkegaard & Perry Laboratories). Chemiluminescence was developed by using the Western Lightning Chemiluminescence Reagent Plus (Perkin–Elmer), and detected on an image analyzer LAS-1000 PLUS (Fujifilm).
Deglycosylation.
The culture media and cell lysates were treated with peptide:N-glycosidase F (PNGase F, NEB, Beverly, MA). Ten microliters of the samples was boiled for 10 min in 1× denaturing buffer, followed by a 1-h incubation with or without 1,000 units of PNGase F in 1× G7 buffer and 1% Nonidet P-40. These samples were then subjected to Western blot analysis.
Purification of VWF.
Human VWF was purified by two-column chromatographic steps. Fresh frozen plasma was thawed overnight at 4°C to obtain cryoprecipitate by centrifugation. The cryoprecipitate was dissolved in 25 mM Tris buffer (pH 7.3) containing 0.5 mM EDTA, 0.15 M NaCl, and 1 mM PMSF, and applied to a Gelatin Sepharose 4B (Amersham Pharmacia) column to remove fibronectin. The flowthrough fraction was precipitated by 40% saturated ammonium sulfate, and the precipitate was dissolved in a buffer (20 mM imidazole/20 mM ɛ-aminocaproic acid/1 M NaCl/10 mM sodium citrate, pH 6.5). The sample was applied to a Sepharose 4B (Amersham Pharmacia) gel filtration column to remove factor VIII and a small amount of VWF-CP. VWF was eluted in the void volume.
Analysis of VWF-Multimeric State.
The culture media of HeLa cells expressing recombinant VWF-CP were concentrated to one-tenth the original volume by using Centricon YM-30 (Millipore). VWF-CP activity was assayed as described by Furlan et al. (29) with slight modification (5). In brief, 8 μl of concentrated culture medium was added to 92 μl of purified VWF (1 μg) dissolved in reaction buffer (1.5 M urea/5 mM Tris⋅HCl/10 mM BaCl2/1 mM PMSF/0.05% NaN3, pH 8.0), and incubated at 37°C for 24 h. Next, 10 μl of 100 mM EDTA (pH 8.0) quenched the reaction. Under these conditions, the Y842-M843 peptidyl bond of VWF is cleaved specifically (14). A portion of each reaction mixture was analyzed by SDS-agarose gel electrophoresis; the multimeric state of VWF was visualized by chemiluminescent Western blot analysis by using anti-VWF primary antibodies.
Results
Mutations in ADAMTS13 Gene of Two Japanese Families.
To amplify the 29 exons of the ADAMTS13 gene, we designed and synthesized 26 pairs of oligonucleotide primers on the basis of the two public genomic (GenBank accession nos. AL158826 and AC002325) and cDNA sequences (GenBank accession nos. AB069698, AF414401, and AY055376) (Table 1). PCR using these primers successfully amplified all exons, despite the failure of the previous report to obtain exon 7 (22). The PCR products (279–770 bp in size) amplified from the genomic DNA of seven individuals in two unrelated USS families (Fig. 1) were fully sequenced. Single-nucleotide mutations at 10 sites were identified (Table 2).
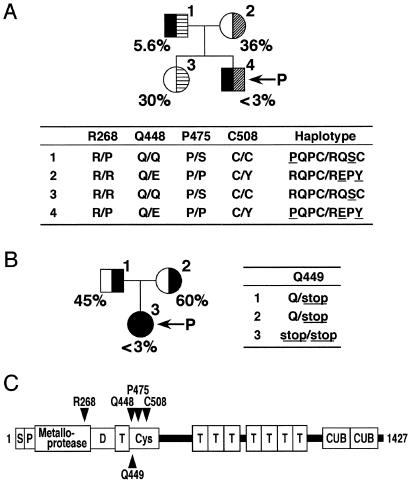
Figure 1.
Pedigrees and haplotypes of patient families. (A) Family A. (B) Family B. Squares and circles indicate male and female, respectively, and arrows indicate the probands. The VWF-CP activities determined by multimer analysis, as reported (5), are shown as a percentage of the normal control. Missense and nonsense mutations found in the ADAMTS13 gene are shown in one-letter amino acid residues numbering from the initiation methionine. (C) Schematic domain structure of VWF-CP. S, signal peptide; P, propeptide; D, disintegrin-like; T, TSP1; Cys, Cys-rich.
Table 2.
ADAMTS13 mutations in two TTP families
| Exon | Family | Nucleotide | Amino acid | Frequency | Heterozygosity |
|---|---|---|---|---|---|
| 5 | A | 420 T>C* | Silent | ND | – |
| 6 | A | 546 C>T | Silent | ND | – |
| 7 | A | 803 G>C | R268P | GG:364, CG:0, CC:0 | <0.3% |
| 12 | A | 1342 C>G* | Q448E | CC:321, CG:125, GG:8 | 31.2% |
| 12 | B | 1345 C>T | Q449stop | CC:364, CT:0, TT:0 | <0.3% |
| 12 | A | 1423 C>T | P475S | CC:328, CT:35, TT:1 | 9.6% |
| 13 | A | 1523 G>A | C508Y | GG:364, AG:0, AA:0 | <0.3% |
| 15 | A | 1716 G>A* | Silent | ND | – |
| 19 | A | 2280 C>T* | Silent | ND | – |
| 29 | A, B | 4221 C>A* | Silent | ND | – |
ND, not determined.
Reported by Levy et al. (22).
Five silent mutations, 420T>C, 546C>T, 1716G>A, 2280C>T, and 4221C>A, were discovered in the two families examined. The nucleotides are numbered from the A of the initiation Met codon. These silent mutations will not be associated with the structure and function of VWF-CP itself, although it may affect the expression level through gene transcription or RNA processing. Four of them, excluding 546C>T, have been reported as SNPs (22). We also identified five additional mutations: four missense mutations in family A and one nonsense mutation in family B. Among these, the 1342C>G mutation (Q448E) has been reported as an SNP (22). The effect of this mutation on VWF-CP activity, however, has not been described. Then, we assessed the effects of the 1342C>G (Q448E) mutation and four additional mutations, 803G>C (R268P), 1345C>T (Q449stop), 1423C>T (P475S), and 1523G>A (C508Y), on VWF-CP activity, comparing the plasma VWF-CP activities of seven individuals in the two USS families with their genotypes.
Assessment of the Mutations in Family A.
The plasma VWF-CP activities of family A were reported (5) (Fig. 1A). The proband with USS possessed an activity below 3% of normal, a detection limit of the assay. The VWF-CP activities in proband's mother and sister were moderately decreased to 36% and 30% of normal levels, respectively. Because the activity in normal subjects ranged from approximately 60% to 150% of pooled normal plasma (30), we currently consider the individuals with 30–70% activity to be heterozygotes if they have no inhibitors. Subclinical father had extremely low activity (5.6%).
We discovered four missense mutations in this family: R268P, Q448E, P475S, and C508Y. All of these mutations were detected as heterozygous; no other mutations in the gene were identified. Haplotype was determined by linkage analysis. The proband possessed the PQPC/REPY (underlines show the mutated positions) diplotype configuration, implying that the R268P mutation completely abrogated VWF-CP activity as well as either Q448E or C508Y (or their combination). Because Q448E and C508Y were located on a single allele, the effect of each of these two mutations was indistinguishable. The diplotype of proband's mother, RQPC/REPY, was consistent with the intermediate effect of the Q448E/C508Y mutation. The diplotype of his sister, RQPC/RQSC, suggested that the P475S mutation also results in decreased VWF-CP activity.
Assessment of the Mutation in Family B.
The plasma VWF-CP activities of family B were also reported (5) (Fig. 1B). The proband with USS possessed VWF-CP activity less than 3% of normal. The activities of proband's father and mother were moderately decreased to 45% and 60% of control levels, respectively. They did not contain inhibitors of VWF-CP activity.
In this family, we identified only one nonsense mutation, Q449stop. The proband was homozygous for the mutation, whereas each of the parents was heterozygous. Therefore, the Q449stop seriously affects VWF-CP activity.
Expression of Recombinant VWF-CP and Its Mutants.
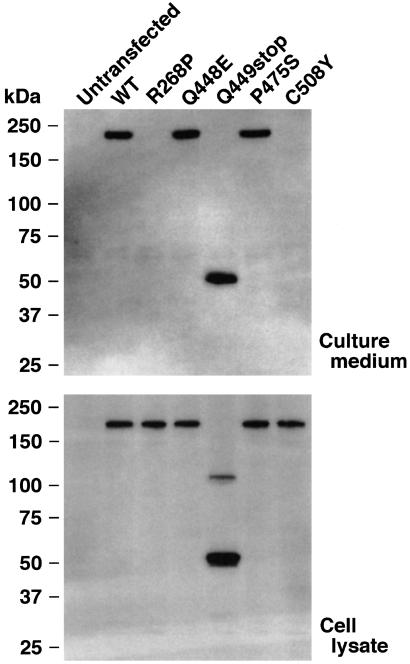
To confirm the functional effects of the identified mutations, wild-type (WT) and mutant human VWF-CP forms were transiently expressed in HeLa cells. The FLAG-tag sequence was added to the C terminus to aid in immunochemical detection. Transient expression of WT VWF-CP produced a single immunoreactive band with a molecular mass of approximately 230 kDa in culture medium (Fig. 2 Upper). Because the band was not detected in untransfected cells, this protein is considered to be the expected product of the transfected plasmid. Upon expression of the Q448E and P475S mutants, a single band of the same size appeared in culture medium. The Q449stop mutant exhibited a 54-kDa band. This finding suggested that each of these mutants was secreted from cells. No bands, however, were detected in the culture medium of cells expressing either the R268P or C508Y mutants. As these mutants were synthesized within the cells (Fig. 2 Lower), both the R268P and C508Y mutations affected some aspect of the secretion pathway possibly including changes in protein folding and stability. This observation explains the absence of activity in the plasma of family A members with these alleles.
Figure 2.
Expression of recombinant VWF-CP. The human WT VWF-CP and mutants with C-terminal FLAG-tag were transiently expressed in HeLa cells. The culture media (Upper) and cell lysates (Lower) were analyzed by Western blot with an anti-FLAG antibody. The representative of three repetitive experiments is shown. The sizes of the protein standards are indicated at the left.
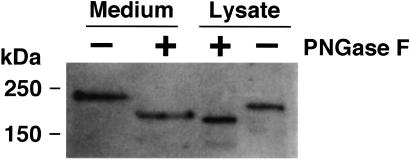
Side-by-side analysis of WT bands in both culture media and cell lysates clearly demonstrated significant differences in size; the protein possessed a molecular mass of 230 kDa in media vs. 200 kDa in lysates (Fig. 3). To examine the possibility that this difference was caused by differences in glycosylation structure, both were treated with PNGase F that removes N-linked carbohydrate residues from polypeptides. This treatment lowered the apparent molecular masses of the bands in the medium and lysate to 180 and 170 kDa, respectively, indicating both were N-glycosylated. Even after PNGase treatment, however, a 10-kDa difference between them remained. This difference may result from a posttranslational modification such as O-linked glycosylation.
Figure 3.
N-glycosidase treatment of recombinant VWF-CP. FLAG-tagged WT VWF-CP derived from culture media and cell lysates were treated with PNGase F and analyzed by Western blot with an anti-FLAG antibody. The sizes of the protein standards are indicated at the left.
Because Q448E, Q449stop, and P475S mutants were efficiently secreted (Fig. 2), the deleterious effect of these mutations on plasma VWF-CP activity cannot be explained by abnormal secretion. Therefore, we examined the VWF-CP activity of these recombinant proteins.
Cleavage of VWF by Recombinant VWF-CPs.
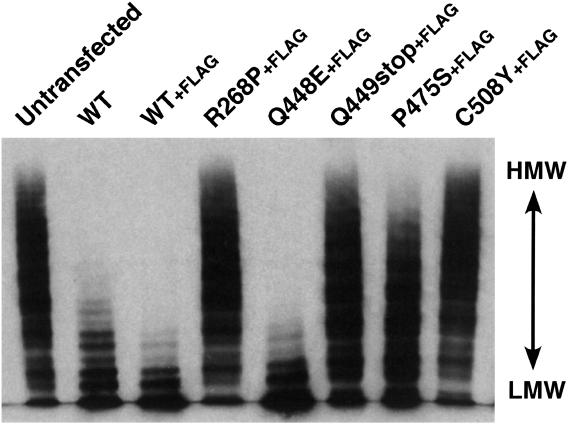
The enzymatic activity of the recombinant VWF-CPs was measured by degradation of VWF multimers. Purified VWF was incubated with the conditioned culture medium, and the multimeric state of VWF was determined by Western blot analysis after SDS-agarose electrophoresis (Fig. 4). After the incubation of VWF with the medium of untransfected cells, protein ladders extended into the high molecular weight area, indicating no VWF-cleaving activity in the medium (Fig. 4, Untransfected). In contrast, the ladders diminished after incubation with WT conditioned medium, demonstrating the VWF-CP activity of the recombinant protein. Because the FLAG-tagged version (WT+FLAG) also possessed cleaving activity, a possible interference effect of the tag was excluded. For R268P and C508Y, no degradation was observed, consistent with the absence of secretion into the medium (Fig. 2).
Figure 4.
Cleavage of VWF multimers. The culture media of HeLa cells either untransfected or transfected with VWF-CP expression plasmids were incubated with purified VWF. A portion of each reaction mixture was then separated by SDS-agarose gel electrophoresis. The multimeric states of VWF were visualized by Western blot analysis with an anti-VWF antibody. The representative of three repetitive experiments is shown. HMW, high molecular weight; LMW, low molecular weight.
The Q448E mutant was fully active in comparison with the WT and WT+FLAG. This mutation was reported as an SNP in the ADAMTS13 gene, although its influence on VWF-CP activity has not been described (22). Our data suggest that the Q448E polymorphism is not involved in alterations of biological activity.
A slight loss of high molecular weight multimers was observed in P475S, suggesting that this mutant had low but significant activity. In the Q449stop, the loss of multimers in high molecular weight was not observed, but the intensities of ladders in the low molecular weight area were increased like the other active mutants such as Q448E and P475S. Neither inactive R268P nor C508Y produced these lower ladders. This finding suggests that the Q449stop mutant possessed quite low VWF-CP activity. Thus, the five mutations we identified can be classified into three categories: (i) Q448E, with no difference from the WT VWF-CP; (ii) R268P and C508Y, with no activity because of secretion problems; and (iii) Q449stop and P475S, with very low activity despite normal secretion.
Frequency of the Mutations in Japanese.
To investigate the frequencies of the five mutations, we sequenced exons 7, 12, and 13 of ADAMTS13 from genomic DNAs isolated from 364 Japanese individuals without TTP (Table 2). The R268P, Q449stop, and C508Y mutations did not appear in the large panel, indicating these critical mutations were rare (<0.3% heterozygosity). We found, however, that the other two mutations, Q448E and P475S, were common. The SNP of Q448E has already been reported, although its frequency is not known (22). Of 364 individuals, we identified 125 heterozygotes and eight homozygotes. The allele frequency and heterozygosity were then calculated to be 19.4% and 31.2%, respectively. The Q448E polymorphism, however, is not associated with reduced VWF-CP activity.
The P475S mutation was also common. Of 364 individuals, 35 were heterozygotes and one was a homozygote. Therefore, the allele frequency and heterozygosity were 5.1% and 9.6%, respectively. Because this mutation results in a nearly complete loss of VWF-CP activity, we calculated that approximately 10% of the Japanese population has significantly reduced VWF-CP activity.
Discussion
In addition to the 12 reported mutations in the ADAMTS13 gene (22), the present study identified three additional mutations responsible for reduced VWF-CP activity. All 15 mutations were excluded as common polymorphisms by the screening of a large panel of unaffected subjects. The mutated sites were not restricted to specific domains but located throughout the molecule. Some mutations such as R268P and C508Y would affect proper folding and secretion of VWF-CP, whereas some likely affect enzymatic activity.
This study and Levy et al. (22) identified eight missense common SNPs, R7W, Q448E, P475S, P618A, R625H, A732V, A900V, and A1033T. We demonstrated that the P475S mutation leads to dramatic decreases in VWF-CP activity, whereas Q448E has no effect. At present, P475S is the only common SNP associated with changes in VWF-CP activity. The functional importance of the remaining SNPs, if any, remains to be uncovered.
Although both the USS proband and his father in family A were compound heterozygotes, R268P/C508Y and R268P/P475S, respectively, only the proband was symptomatic (Fig. 1). This finding may result from a differential effect between the C508Y and P475S mutations; the C508Y mutant was not secreted, whereas the P475S mutant was secreted normally and showed low but significant activity (Figs. 2 and 4). In 364 Japanese subjects we analyzed, one individual was homozygous for the P475S mutation, but was asymptomatic. This observation suggests that faint activity of VWF-CP is sufficient for normal hemostasis, consistent with reports (4, 13). Another homozygous mutation, Q449stop, was present in the USS proband of family B. At least in our expression system using HeLa cells, the recombinant Q449stop mutant was secreted from cells but possessed lower enzymatic activity than P475S. The subtle difference in activity may be critical for the onset of neonatal TTP. Alternatively, the absence of the Cys-rich, TSP1, and CUB domains in the Q449stop mutant may mediate the symptomatic difference. These strict correlations between mutation and phenotype may be partly true, especially in neonatal USS. However, much variation exists in the clinical phenotype of TTP, such as the age of onset and severity of symptoms. Therefore, the possibility cannot be excluded that additional genetic or nongenetic factors may also influence the occurrence of congenital TTP.
The P475S allelic frequency of 5.1% was determined from the analysis of 364 individuals. Although none of these patients demonstrated any symptoms of TTP, they were all patients of the Division of Hypertension and Nephrology at the National Cardiovascular Center (Suita, Japan). Therefore, the bias of this population may affect the allelic frequency. Further analysis will be required to determine the relationship between this SNP and hypertension or other related diseases.
A major proportion of TTP cases are not congenital but are acquired in adults possessing underlying conditions, including autoimmunity, pregnancy, and preexisting infection (2, 3, 31). Occasionally, severe TTP is induced as a side effect of drugs such as cyclosporine, ticlopidine, and clopidogrel (32–34). If the occurrence is associated with a particular genetic background, such as P475S polymorphism, genotyping before medication will be useful to prevent the development of drug-associated TTP.
We here demonstrate that some mutants, including Q449stop and P475S, are secreted from cells at levels similar to the WT VWF-CP. This observation suggests that the clinical assay based on the plasma VWF-CP activity is a more accurate prognostic test than the measurement of antigen levels to determine the VWF-CP-dependent hemostatic conditions of patients. Because current methods measuring VWF-CP activity require time and skill, they are not suitable for high-throughput screening. It will be important in the future to develop more rapid, accurate methods to assay VWF-CP activity in contrast to antigen levels.
Acknowledgments
We thank Ms. Yuko Nobe and Akemi Fukumoto (National Cardiovascular Center Research Institute) and Noriko Mimura (The Chemo-Sero-Therapeutic Research Institute) for their technical assistance. This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan; the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for the Promotion of Science; and the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan.
Abbreviations
- TTP
thrombotic thrombocytopenic purpura
- USS
Upshaw–Schulman syndrome
- VWF
von Willebrand factor
- UL-VWFM
unusually large VWF multimer
- VWF-CP
VWF-cleaving protease
- TSP1
thrombospondin-1
- SNP
single-nucleotide polymorphism
- PNGase F
peptide:N-glycosidase F
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 11552.
Matsumoto, M., Chisuwa, H., Nakazawa, Y., Ikegami, T., Hashikura, Y., Kawasaki, S., Yagi, H., Ishizashi, H., Matsui, T., Titani, K. & Fujimura, Y. (2000) Blood 96, 636a (abstr.).
References
- 1.Moschcowitz E. Proc N Y Pathol Soc. 1924;24:21–24. [Google Scholar]
- 2.Kakishita E. Int J Hematol. 2000;71:320–327. [PubMed] [Google Scholar]
- 3.Cines D B, Konkle B A, Furlan M. Thromb Haemostasis. 2000;84:528–535. [PubMed] [Google Scholar]
- 4.Furlan M, Lämmle B. Best Pract Res Clin Haematol. 2001;14:437–454. doi: 10.1053/beha.2001.0142. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita S, Yoshioka A, Park Y D, Ishizashi H, Konno M, Funato M, Matsui T, Titani K, Yagi H, Matsumoto M, Fujimura Y. Int J Hematol. 2001;74:101–108. doi: 10.1007/BF02982558. [DOI] [PubMed] [Google Scholar]
- 6.Fujimura Y, Matsumoto M, Yagi H, Yoshioka A, Matsui T, Titani K. Int J Hematol. 2002;75:25–34. doi: 10.1007/BF02981975. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura Y, Titani K. In: Haemostasis and Thrombosis. Bloom A L, Forbes C D, Thomas D P, Tuddenham E G D, editors. New York: Churchill Livingstone; 1994. pp. 379–395. [Google Scholar]
- 8.Furlan M. Ann Hematol. 1996;72:341–348. doi: 10.1007/s002770050184. [DOI] [PubMed] [Google Scholar]
- 9.Ruggeri Z M. J Clin Invest. 1997;99:559–564. doi: 10.1172/JCI119195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadler J E. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 11.Federici A B, Bader R, Pagani S, Colibretti M L, De Marco L, Mannucci P M. Br J Haematol. 1989;73:93–99. doi: 10.1111/j.1365-2141.1989.tb00226.x. [DOI] [PubMed] [Google Scholar]
- 12.Kalafatis M, Takahashi Y, Girma J P, Meyer D. Blood. 1987;70:1577–1583. [PubMed] [Google Scholar]
- 13.Yagi H, Konno M, Kinoshita S, Matsumoto M, Ishizashi H, Matsui T, Titani K, Fujimura Y. Br J Haematol. 2001;115:991–997. doi: 10.1046/j.1365-2141.2001.03222.x. [DOI] [PubMed] [Google Scholar]
- 14.Furlan M, Robles R, Lämmle B. Blood. 1996;87:4223–4234. [PubMed] [Google Scholar]
- 15.Tsai H M. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 16.Dent J A, Berkowitz S D, Ware J, Kasper C K, Ruggeri Z M. Proc Natl Acad Sci USA. 1990;87:6306–6310. doi: 10.1073/pnas.87.16.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai H M, Sussman I I, Nagel R L. Blood. 1994;83:2171–2179. [PubMed] [Google Scholar]
- 18.Soejima K, Mimura N, Hirashima M, Maeda H, Hamamoto T, Nakagaki T, Nozaki C. J Biochem (Tokyo) 2001;130:475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 19.Gerritsen H E, Robles R, Lämmle B, Furlan M. Blood. 2001;98:1654–1661. doi: 10.1182/blood.v98.6.1654. [DOI] [PubMed] [Google Scholar]
- 20.Fujikawa K, Suzuki H, McMullen B, Chung D. Blood. 2001;98:1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 21.Zheng X, Chung D, Takayama T K, Majerus E M, Sadler J E, Fujikawa K. J Biol Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 22.Levy G G, Nichols W C, Lian E C, Foroud T, McClintick J N, McGee B M, Yang A Y, Siemieniak D R, Stark K R, Gruppo R, et al. Nature (London) 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 23.Tang B L, Hong W. FEBS Lett. 1999;445:223–225. doi: 10.1016/s0014-5793(99)00119-2. [DOI] [PubMed] [Google Scholar]
- 24.Hurskainen T L, Hirohata S, Seldin M F, Apte S S. J Biol Chem. 1999;274:25555–25563. doi: 10.1074/jbc.274.36.25555. [DOI] [PubMed] [Google Scholar]
- 25.Cal S, Obaya A J, Llamazares M, Garabaya C, Quesada V, López-Otín C. Gene. 2002;283:49–62. doi: 10.1016/s0378-1119(01)00861-7. [DOI] [PubMed] [Google Scholar]
- 26.Colige A, Vandenberghe I, Thiry M, Lambert C A, Van Beeumen J, Li S W, Prockop D J, Lapière C M, Nusgens B V. J Biol Chem. 2002;277:5756–5766. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 27.Bork P, Beckmann G. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Furlan M, Robles R, Solenthaler M, Wassmer M, Sandoz P, Lämmle B. Blood. 1997;89:3097–3103. [PubMed] [Google Scholar]
- 30.Mori Y, Wada H, Gabazza E C, Minami N, Nobori T, Shiku H, Yagi H, Ishizashi H, Matsumoto M, Fujimura Y. Transfusion. 2002;42:572–580. doi: 10.1046/j.1537-2995.2002.00100.x. [DOI] [PubMed] [Google Scholar]
- 31.Bell W R. Semin Hematol. 1997;34:134–139. [PubMed] [Google Scholar]
- 32.Sugio Y, Okamura T, Shimoda K, Matsumoto M, Yagi H, Ishizashi H, Niho Y, Inaba S, Fujimura Y. Int J Hematol. 2001;74:347–351. doi: 10.1007/BF02982073. [DOI] [PubMed] [Google Scholar]
- 33.Reichenberger F, Wirtz H, Paschke R. Cardiology. 2001;96:51–52. doi: 10.1159/000047386. [DOI] [PubMed] [Google Scholar]
- 34.Medina P J, Sipols J M, George J N. Curr Opin Hematol. 2001;8:286–293. doi: 10.1097/00062752-200109000-00004. [DOI] [PubMed] [Google Scholar]