Abstract
Coordinate induction of phase 2 proteins and elevation of glutathione protect cells against the toxic and carcinogenic effects of electrophiles and oxidants. All inducers react covalently with thiols at rates that are closely related to their potencies. Inducers disrupt the cytoplasmic complex between the actin-bound protein Keap1 and the transcription factor Nrf2, thereby releasing Nrf2 to migrate to the nucleus where it activates the antioxidant response element (ARE) of phase 2 genes and accelerates their transcription. We cloned, overexpressed, and purified murine Keap1 and demonstrated on native gels the formation of complexes of Keap1 with the Neh2 domain of Nrf2 and their concentration-dependent disruption by inducers such as sulforaphane and bis(2-hydroxybenzylidene)acetone. The kinetics, stoichiometry, and order of reactivities of the most reactive of the 25 cysteine thiol groups of Keap1 have been determined by tritium incorporation from [3H]dexamethasone mesylate (an inducer and irreversible modifier of thiols) and by UV spectroscopy with sulforaphane, 2,2′-dipyridyl disulfide and 4,4′-dipyridyl disulfide (titrants of thiol groups), and two closely related Michael reaction acceptors [bis(2- and 4-hydroxybenzylidene)acetones] that differ 100-fold in inducer potency and the UV spectra of which are bleached by thiol addition. With large excesses of these reagents nearly all thiols of Keap1 react, but sequential reaction with three successive single equivalents (per cysteine residue) of dipyridyl disulfides revealed excellent agreement with pseudo-first order kinetics, rapid successive declines in reaction velocity, and the stoichiometric formation of two equivalents of thiopyridone per reacted cysteine. This finding suggests that reaction of cysteine thiols is followed by rapid formation of protein disulfide linkages. The most reactive residues of Keap1 (C257, C273, C288, and C297) were identified by mapping the dexamethasone-modified cysteines by mass spectrometry of tryptic peptides. These residues are located in the intervening region between BTB and Kelch repeat domains of Keap1 and probably are the direct sensors of inducers of the phase 2 system.
Induction of phase 2 genes is a highly effective strategy for protection against carcinogenesis and other forms of electrophile and oxidant toxicity (1–3). The molecular mechanisms of these inductions have been studied by two complementary approaches: (i) elucidation of the relation between the chemical structures and the potencies of inducers (4–6), and (ii) identification of the molecular elements and cellular events that control the induction (7, 8). There are at least nine structurally dissimilar classes of inducers (4–6). Their single and universal property is reactivity with thiol groups at rates that are correlated closely with their potencies. On this basis we proposed the existence of a cellular “sensor” protein endowed with reactive thiol groups that recognize and interact chemically (by alkylation or oxidation) with inducers, thereby signaling induction of phase 2 proteins (5, 6, 9). Our article addresses the identity of the sensor.
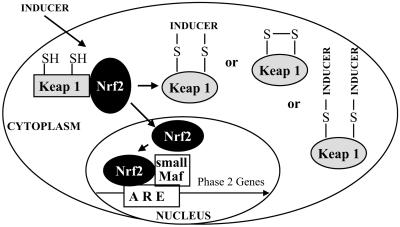
Two proteins participate in the transcriptional activation of phase 2 genes: Nrf2, a member of the NF-E2 family of nuclear basic leucine zipper (bZIP) transcription factors, and Keap1, a cytoplasmic protein homologous to the Drosophila actin-binding protein Kelch (7, 10, 11). Under basal conditions Nrf2 is largely bound in the cytoplasm to Keap1, which is anchored to the actin cytoskeleton. When inducers disrupt the Keap1–Nrf2 complex, Nrf2 migrates to the nucleus where, in heterodimeric combinations with other transcription factors, it binds to the 5′-upstream regulatory antioxidant response element (ARE) regions of phase 2 genes and accelerates their transcription (refs. 11 and 12; Fig. 1). The critical role of ARE is supported both by deletion analysis (13) and the highly quantitative correlation between the potencies of inducers in activating transfected AREs linked to a reporter and in elevating phase 2 enzymes (5, 6).
Figure 1.
Mechanism of phase 2 response regulation. Nrf2 is anchored in the cytoplasm by binding to Keap1, which is attached to the actin cytoskeleton. Inducers disrupt the Keap1–Nrf2 complex, and Nrf2 migrates to the nucleus where it forms heterodimers with other transcription factors such as small Maf that bind to the ARE regions of phase 2 genes and accelerate their transcription. Several types of modifications of Keap1 by inducers are shown.
Keap1 represses Nrf2 function, and the Keap1/Nrf2 system is essential for regulation of phase 2 proteins. Mice in which the nrf2 gene was deleted (11, 12) have low basal and mostly uninducible levels of phase 2 proteins and are much more susceptible to hepatic toxicities of acetaminophen (14, 15), pulmonary toxicity of hyperoxia (16), and forestomach carcinogenesis by benzo[a]pyrene (17, 18). Unlike wild-type controls these animals are not protected against tumor formation by phase 2 gene inducers such as oltipraz and sulforaphane (17, 18).
The 624 aa of murine Keap1 comprise five domains. (i) The N-terminal region (NTR, amino acids 1–60). (ii) BTB (amino acids 61–179), an evolutionarily conserved protein–protein interaction motif found in actin-binding proteins and zinc finger transcription factors, often involved in complexes with other BTB domains. (iii) Intervening region (IVR, amino acids 180–314), an especially cysteine-rich region (eight cysteines in 102 aa). (iv) Double glycine or Kelch repeat (DGR), comprising six Kelch motifs (amino acids 315–359, 361–410, 412–457, 459–504, 506–551, and 553–598). Repeated Kelch motifs give rise to a β-propeller structure with multiple protein contact sites (see ref. 19). The DGR of Keap1 combines with Nrf2 through the Neh2 domain of Nrf2, the ≈100 N-terminal aa (10). (v) C-terminal region (CTR, amino acids 599–624).
Murine Keap1 contains 25 cysteines (Fig. 2) that are conserved in human and rat homologues (11). Murine Nrf2 is a 597-aa protein containing seven cysteine residues (10). If inducers regulate the phase 2 response by modifying critical thiol group(s), disruption of the Nrf2–Keap1 complex may result from modification of cysteine thiols of Keap1, Nrf2, or both. Our preliminary experiments using fusion proteins of Keap1 or Nrf2 with maltose-binding protein showed that inducers reacted much more avidly with Keap1, suggesting that Keap1 is the most likely target for inducers (unpublished data). Because inducers disrupted Keap1–Neh2 domain complexes, this separation process does not require the entire Nrf2 structure. Moreover, because the Neh2 domain does not contain cysteine residues (Fig. 2), it is unlikely to be modified by inducers.
Figure 2.
Primary structures of murine Keap1 (Upper) and Neh2 domain of Nrf2 (amino acid residues 1–98) (Lower). Tryptic peptides of Keap1 are numbered, and residues are highlighted: cysteine (yellow), arginine (blue), and lysine (red).
The reactive cysteine residues of Keap1 participating in inducer recognition were identified by kinetic, radiolabeling, and UV spectroscopic studies of Keap1 interaction with selected inducer probes. Here we present evidence that certain highly reactive cysteine thiol groups in the intervening region of Keap1 are probably the primary cellular sensors that recognize and react with phase 2 gene inducers.
Experimental Procedures
Overall Strategy and Selection of Chemical Probes.
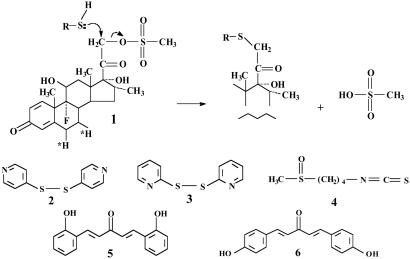
We selected the following chemical probes (Fig. 3) to elucidate the properties of the thiol groups of Keap1. (i) Dexamethasone 21-mesylate (1, Dex-mes), an inducer of phase 2 proteins, in which the mesylate group is displaced by thiols, resulting in their irreversible alkylation by the steroid (20). Reactive cysteines were labeled also with [6,7-3H]Dex-mes. (ii) 4,4′-Dipyridyl-disulfide (2) and 2,2′-dipyridyl-disulfide (3) (weak inducers that react quantitatively and unidirectionally with thiol groups) were used to determine the stoichiometry and reaction rates of the cysteines spectroscopically (21). (iii) The isothiocyanate sulforaphane (4), a very potent inducer isolated from broccoli (22, 23) that reacts rapidly but reversibly with thiols to give rise to a dithiocarbamate, which has high UV absorption near 270 nm. (iv) Two closely related double-Michael reaction acceptors that differ markedly in inducer potencies: bis(2-hydroxybenzylidene)-acetone [2-HBA, 5; CD value (concentration of an inducer that doubles specific activity of NQO1 in Hepa1c1c7 murine hepatoma cells), 0.14 μM] and bis(4-hydroxybenzylidene)acetone (4-HBA, 6; CD value, 14 μM) (9). The UV absorption maxima of these compounds do not overlap significantly with the aromatic region of proteins and are bleached by thiol additions. Thus, they can be used to quantify by direct spectroscopy the kinetics and stoichiometry of reactions of thiols.
Figure 3.
Irreversible reaction of Dex-mes (1) with a thiol group (Upper) and structures of the chemical probes used: 4,4′-dipyridyl disulfide (2), 2,2′-dipyridyl disulfide (3), sulforaphane (4), 2-HBA (5), and 4-HBA (6).
Purification of Keap1.
Escherichia coli BL21(DE3) was transfected with plasmid pET-21b(+)mKeap1, which encodes a full-length cDNA copy of murine Keap1 inserted between NdeI and BamHI sites. Bacteria were grown with agitation in 4-liter Fernbach flasks in a salt medium (NH4Cl, 1 g; KH2PO4, 1 g; K2HPO4, 3 g; Na2SO4, 0.3 g; MgCl2, 0.05 g; CaCl2, 0.005 g, per liter) and glucose (0.3%) at 37°C to an absorbance of 0.5–0.6 at 600 nm. The flasks then were cooled to 15°C, isopropyl β-d-thiogalactoside was added to a final concentration of 0.5 mM, and the flasks were agitated gently for a further 24 h at 15°C. After raising the pH to 7.5–8.0 with 25 ml of 1.0 M Tris base per liter of medium, the cells were harvested by centrifugation. The pellet was resuspended in 50 mM Tris-Cl/50 mM Bis Tris propane containing 10 mM EDTA and 10 mM DTT, pH 8.4, and the cells were disrupted by sonication at 4°C and centrifuged at 37,000 × g. Keap1 was purified from the crude supernatant fraction by removing DTT by filtration through Sephadex G-25 columns (PD-10, Amersham Pharmacia) equilibrated with 25 mM Tris-Cl/5 mM EDTA, pH 8.0 (no DTT) and then adding ammonium sulfate (adjusted to pH 8.0 with NH4OH) to a final concentration of 300 mM. The precipitate, obtained when these solutions were frozen overnight at −20 or −80°C and then unfrozen, was collected by centrifugation and dissolved in 25 mM Tris-Cl/5 mM EDTA/10 mM DTT, pH 8.0. The precipitation procedure was repeated a second time, and the final product was stored at −80°C.
Binding of [3H]Dex-mes to Keap1 and Competition by Other Inducers.
DTT was removed from purified Keap1 preparations by sequential gel filtration through two small Sephadex G-25 columns (NAP-10, Amersham Pharmacia) equilibrated with 10 mM Tris-Cl/2 mM EDTA/0.01% Tween 20, pH 8.0 and then concentrated in Centricon 30 concentrators (Amicon). Ten microliters of Keap1 (3.5 μg or 50 pmol) was added to 80 μl of 25 mM Tris-Cl/2.0 mM EDTA/0.01% Tween 20, pH 8.0. A range of concentrations of competing inducers (phenylarsine oxide, menadione, 1-nitrocyclohexene, 1-chloro-2,4-dinitrobenzene, 2-cyclohexenone, and α-methylene-γ-butyrolactone) dissolved in acetonitrile/water (1:1) were added in 10-μl volumes, and the reaction mixtures were incubated for 30 min at 25°C. Then 10-μl aliquots of [3H]Dex-mes (27 pmol, ≈600,000 cpm, New England Nuclear) were added, and incubation was continued for 60 min at 25°C. The reaction mixtures (110 μl) were applied to NAP-10 columns followed by 880 μl of buffer, and the protein was eluted with 900 μl of buffer and collected in 7-ml scintillation vials. Scintillation fluid was added, and radioactivity was determined.
Preparation of Neh2 Domain of Nrf2.
A PCR fragment of the N-terminal residues (1–98) of Nrf2 was modified at the 5′ end by addition of 6-His tags and a thrombin cleavage site and at the 3′ end by addition of three stop codons. This construct was inserted between the NdeI and XhoI sites of pET-15b (Novagen) to construct the plasmid pET-15b-Neh2. E. coli (BL21/DE3) was transformed, and expression was induced with 0.5 mM isopropyl β-d-thiogalactoside. The cells were disrupted sonically and treated with DNase I, and the supernatant fraction underwent nickel-resin chromatography. After dialysis against 20 mM sodium phosphate buffer (pH 7.2) containing 10% glycerol and 10 mM β-mercaptoethanol, the His tag was removed by the addition of thrombin and a serine protease inhibitor. The resulting purified protein contains 98 N-terminal amino acid residues of Nrf2 preceded by Gly-Ser-His (from the thrombin cleavage site). Neh2 protein was concentrated by membrane centrifugation and purified by ion-exchange chromatography and gel filtration.
Keap1–Neh2 Binding Studies.
DTT was removed from Keap1 as described above, and 10 μl (100 pmol) of Keap1 and 10 μl (50–60 pmol) of the Neh2 domain of Nrf2, both in 10 mM Tris-Cl/1 mM EDTA/0.005% Tween 20, pH 8.0, were placed in 0.6-ml Eppendorf tubes with or without DTT, flushed with N2, and incubated for 16 h at 4°C. Inducers (sulforaphane, 2-HBA, or 4-HBA) were added in 10 μl of the same buffer, or water, and the mixtures were incubated for 60 min at 25°C. Glycerol and tracking dye were added, and the mixtures were applied to native gels.
Gel Electrophoresis.
Electrophoresis was carried out in a Protean II apparatus (Bio-Rad). For native gels, the separating gel (12% acrylamide/0.08% bis-acrylamide in 100 mM Tris-Cl, pH 8.7) was polymerized with ammonium persulfate and prerun for 4 h at 200 V in the same buffer. The stacking gel (7% acrylamide/0.047% bis-acrylamide in 100 mM Tris-Cl, pH 7.5) was polymerized with riboflavin. The reservoir buffer was 189 mM glycine/25 mM Tris base, pH 8.5. All gel solutions and electrophoresis buffers were deaerated and gassed with N2 just before use. Native gels were run for 6–7 h at 100 V with water cooling. SDS gels (10% acrylamide/0.067% bis-acrylamide) were run at 200 V for 4 h. All gels were stained with Coomassie blue R-250 and then destained.
Radioactive gels were sliced into 2-mm sections, solubilized in 7-ml scintillation vials with 0.75 ml of 30% H2O2 for 2 h at 90°C, and counted.
Protein Measurements.
Protein concentrations were measured according to Bradford (24) with crystalline BSA as standard. We also determined the absorption coefficient of native Keap1 from its tyrosine, tryptophan, and cysteine composition and its absorbance change in guanidinium-HCl (25). The Bradford value was 13.8% higher.
Tryptic Digestion of Keap1.
Unmodified or Dex-mes-labeled Keap1 (50 μl, 300 pmol) in 10 mM Tris-Cl, pH 8.0, was incubated for 30 min at 37°C with 50 μl of 10 mM N-ethylmaleimide/9 M urea in the same buffer. Then 100 μl of 50 mM Tris-Cl/50 mM Bis Tris propane, pH 8.4, and 1 mg of trypsin (Worthington, 3× crystallized) were added. After incubation at 37°C for 4 h, a second aliquot of trypsin was added, and digestion was continued for 14 h. The reaction was stopped with 2 μl of trifluoroacetic acid (TFA). The tryptic peptides were separated on a reversed-phase (C8) HPLC column with a gradient of 0–80% acetonitrile/0.1% TFA at a flow rate 0.2 ml/min. Fractions (0.2 ml) were analyzed by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry (Voyager DE STR, Applied Biosystems).
Peptide identity was confirmed by liquid chromatography tandem mass spectrometry coupled with electrospray ionization and fragmentation by collision-induced dissociation. Peptides were fractionated by reversed-phase HPLC in a 0–60% methanol gradient containing 0.5% formic acid at 200 nl/min on a 75-μm × 100-mm PepMap column equipped with a FAMOS autoinjector (LC Packings, San Francisco). Eluted peptides were sprayed through a 10-μm Picotip (New Objectives, Woburn, MA) directly into QSTAR (Applied Biosystems/MDS Sciex, Foster City, CA). Fragmentation spectra were obtained for specific doubly or triply charged ions by using 0.5-s survey acquisitions followed by 1.5-s collision-induced dissociation acquisitions in Enhance mode at 25, 30, and 35 V collision energies. Errors on fragmentation ions were less than 0.02 atomic mass unit.
Results
Purification of Keap1.
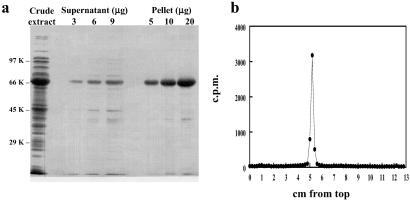
Initial attempts to purify Keap1 were hampered because the overexpressed protein formed insoluble inclusion bodies in LB medium at 37°C. However, growth at 15°C in an inorganic medium with glucose increased the amount of soluble protein substantially. Keap1 was purified easily from crude extracts by precipitation when frozen in the presence of 100–300 mM ammonium sulfate without DTT. The precipitated protein was readily soluble in Tris-Cl buffer, pH 8.0, containing DTT. Below pH 7.0, Keap1 precipitated even in the presence of DTT. Reducing SDS/PAGE of the purified protein showed essentially one band with a molecular weight of ≈64,000 (calculated 69,552) (Fig. 4a). In contrast, in nonreducing SDS/PAGE, Keap1 migrated faster, possibly because of formation of disulfide bridges (26). The N-terminal sequence by Edman degradation was MQPEPKLSGAP-, in perfect agreement with the sequence of Keap1 deduced from its cDNA (11). Electrophoresis of purified Keap1 labeled with [3H]Dex-mes on reducing SDS/PAGE gel gave a single sharp band of radioactivity coinciding with the protein (Fig. 4b).
Figure 4.
(a) SDS/PAGE of purified Keap1. (b) SDS/PAGE of Keap1 after labeling with [3H]Dex-mes: 5 μg of Keap1 was incubated with 50 pmol of [3H]Dex-mes for 60 min at 25°C, and unbound Dex-mes was removed by gel filtration. Labeled Keap1 was separated on a 10% gel and stained with Coomassie blue. The gel lane was cut into 2-mm slices, each digested with 30% H2O2, and radioactivity was counted.
Competition of Phase 2 Inducers with [3H]Dex-mes Binding to Keap1.
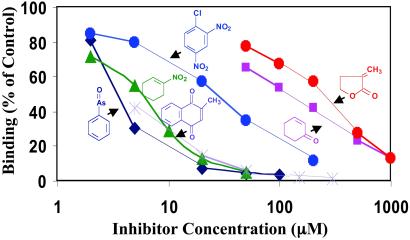
Several inducers of different chemical types and potencies competed with the binding of [3H]Dex-mes to Keap1 (Fig. 5). The approximate concentrations of each competitor required to reduce binding by 50% were: phenylarsine oxide (3.5 μM); 1-nitro-2-cyclohexene (5.8 μM); 1-chloro-2,4-dinitrobenzene (26.5 μM); 2-cyclohexenone (110 μM); and α-methylene-γ-butyrolactone (250 μM). The order of potencies of these compounds as competitors for binding of Dex-mes to Keap1 parallels their potencies for induction of NQO1 in intact cells (4–6). Thus, phenylarsine oxide and 1-nitrocyclohexene are the most potent competitors and inducers, strongly supporting that Dex-mes reacts with Keap1 at sites relevant to the inducer mechanism.
Figure 5.
Inhibition of [3H]Dex-mes binding to Keap1. Each reaction mixture contained 10 μl (3.5 μg, 50 pmol) of Keap1 and 80 μl of 25 mM Tris-Cl/2 mM EDTA/0.01% Tween 20, pH 8.0. A range of concentrations of competitors (10 μl in 50% acetonitrile/50% water) was added, and the mixture was incubated for 30 min at 25°C. Then 10 μl of [3H]Dex-mes (27 pmol) was added, and incubation was continued for an additional 60 min. [3H]Dex-mes bound to protein was separated from unbound steroid by gel filtration, and radioactivity in the protein fraction was determined.
Stoichiometry and Kinetics of Reaction of Thiol Groups of Keap1 with Dipyridyl Disulfides.
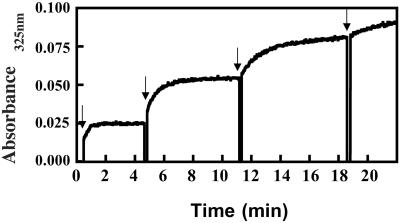
2,2′-Dipyridyl disulfide and 4,4′-dipyridyl disulfide were chosen as spectroscopic titration reagents for the thiol groups of Keap1, because their reductions by thiols are unidirectional and relatively pH-insensitive. The resulting thiones have distinct UV absorptions and are produced in stoichiometric amounts (21). Thus, sequential titration of Keap1 with multiple 1-equivalent (per cysteine residue) additions of each dipyridyl disulfide revealed that at pH 7.0 two thiol groups were titrated at each of the first three steps, whereas only a single thiol group was titrated at the next step (Fig. 6). The reaction rates for the first three steps, calculated with GRAFIT software, showed pseudo-first order kinetics, with observed rate (k) values of 0.3607, 0.2097, and 0.1306 min−1 for the reaction with 2,2′-dipyridyl disulfide and 0.9858, 0.4533, and 0.2019 min−1 for reaction with 4,4′-dipyridyl disulfide, respectively. Standard errors were <5%. Rates of reaction of the two disulfides with 2-mercaptoethanol under identical conditions showed similar differences. Reaction of Keap1 with a 1,000-fold excess of 4,4′-dipyridyl disulfide was complete in <1 min, and 21–24.5 equivalents (depending on the method of protein determination) of pyridinethione were formed, suggesting that most of the cysteines were titrated and not initially involved in disulfide linkages in DTT-treated Keap1. Assuming that these measurements reflect sequential reactivities of cysteine residues, the experiments demonstrate that (i) a single cysteine residue in Keap1 is most reactive, and there is a hierarchy of less reactive cysteines, and (ii) the addition of a single equivalent of dipyridyl disulfide leads to formation of two molecules of the pyridinethione. This finding suggests that once the Keap1-pyridyl mixed disulfide is formed, the linkage is attacked rapidly by another cysteine thiolate ion of the protein to form either an intra- or intermolecular protein disulfide. The 25 cysteines of Keap1 provide many opportunities for sulfhydryl–disulfide interchange.
Figure 6.
Reaction of thiol groups of Keap1 with 4,4′-dipyridyl disulfide. To a solution of 0.5 μM Keap1 in 0.3 ml of 5 mM potassium phosphate buffer, pH 7.0, at 25°C, was added 0.5 μM (1 equivalent) 4,4′-dipyridyl disulfide in four sequential steps at times indicated with the arrows. Immediately after mixing, the increase in absorbance at 325 nm was monitored at 0.1-s intervals.
Spectroscopic Evidence for Reactivity of Thiol Groups of Keap1 with Sulforaphane, 2-HBA, and 4-HBA.
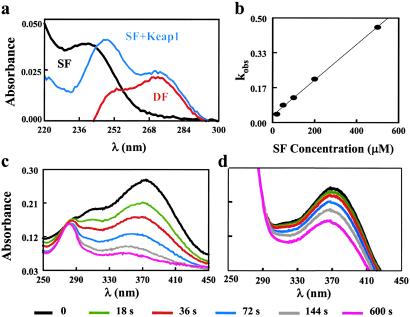
The chemistry of the reaction of the thiol groups of Keap1 with inducers was established by spectroscopic observation with two types of inducers: an isothiocyanate and two Michael reaction acceptors. Isothiocyanates such as sulforaphane (λmax 240 nm, ɛ 950 M−1⋅cm−1) react with mercaptans to give rise to dithiocarbamates with characteristic absorptions near 270 nm (ɛ ≈ 10,000 M−1⋅cm−1). When the potent inducer sulforaphane (CD value 0.2 μM) was added to Keap1, absorbance at 270 nm was increased. Although overlapping the aromatic residues of the protein, this absorption was sufficiently intense to be quantified easily (Fig. 7a), and the initial reaction velocity was proportional to the concentration of sulforaphane (Fig. 7b). No reaction was observed if Keap1 was boiled or treated with excess of the potent inducer 2-HBA.
Figure 7.
Reaction of Keap1 with sulforaphane. (a) Absorption spectra of 50 μM sulforaphane (SF), the reaction mixture of 50 μM sulforaphane after it has been incubated with 0.7 μM Keap1 for 90 min in 5 mM potassium phosphate buffer, pH 8.0, at 25°C against a Keap1 blank (SF+Keap1), and their difference spectrum (DF). (b) Plot of the reaction rates of sulforaphane with 0.7 μM Keap1 under the same reaction conditions as a function of the concentration of sulforaphane. (c and d) Absorption spectrum kinetics of the reaction of 2-HBA with Keap1 in the absence (c) or presence (d) of 20 mM sulforaphane.
The favorable absorption characteristics of 2-HBA and 4-HBA were used next to measure the reactivity of thiol groups of Keap1 with these phase 2 enzyme inducers. Whereas 2-HBA is a powerful inducer (CD value 0.14 μM) and its absorbance at 370 nm (ɛ 16,000 M−1⋅cm−1 in this buffer) is bleached by the addition of thiols, 4-HBA is a much weaker inducer (CD value 14 μM) and absorbs maximally at 320 nm. We monitored the decrease in absorbance at 370 nm after the addition of Keap1 to 2-HBA (Fig. 7c). The rate of this reaction was reduced markedly if Keap1 was incubated first with sulforaphane (Fig. 7d). In contrast, when 4-HBA was used as a ligand under identical conditions, absorbance at 320 nm was not decreased, in agreement with its much lower reactivity with thiol reagents and its much weaker NQO1 inducer potency (9).
Dex-mes Binding to Cysteine Residues of Keap1.
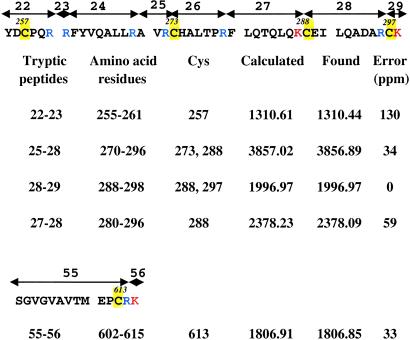
Initially Keap1 (0.5 μM) was labeled extensively with [3H]Dex-mes (75 μM or 150-fold molar excess) at 25°C, pH 8.0, for 21 h. Measurements of the incorporated radioactivity showed that ≈23 cysteine residues had been labeled with the ligand. In contrast, a much smaller excess (10-fold) of [3H]Dex-mes and shorter incubation time (2 h) resulted in labeling of only 3–4 cysteines. Therefore, we then attempted to identify the most reactive cysteine residues of Keap1. The protein was incubated with a 33-fold excess of Dex-mes for 1 h at pH 8.0 and then treated with N-ethylmaleimide to alkylate the unreacted thiols. After tryptic digestion and peptide separation by reversed-phase HPLC, the resulting fractions were analyzed by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry. All except three of the predicted tryptic peptides (i.e., 39, 51, and 53; Fig. 2) were recovered in the fractions. Most important, 18 of the 19 cysteine-containing tryptic peptides were observed. The following Dex-mes-labeled cysteine residues were identified in peptides that increased in mass by 374.19 (the mass added by the steroid; Table 1): C257 (peptides 22–23), C273 (peptides 25–28), C288 (peptides 25–28, 28–29, and 27–28), C297 (peptides 28–29), and C613 (peptides 55–56). Peptides containing C23, C38, C77, C151, C226, C241, C249, C319, C395, C406, C434, C489, C513, and C518 were alkylated by N-ethylmaleimide, thereby increasing the cysteine mass by 125.05. Peptides containing C171, C196, C622, and C624 were unmodified. Only cysteine-containing peptide 53 was not detected. The sequences and positions of Dex-mes-modified cysteines in peptides 22–23 and 27–28 were confirmed by liquid chromatography tandem mass spectrometry coupled with electrospray ionization and fragmentation by collision-induced dissociation. The predicted b (100%) and y (88%) ions of these peptides, including the Dex-mes-modified cysteine-containing ions, matched 71 and 77%, respectively, of the most intense masses in the fragmentation spectra. Fragmentation patterns of dexamethasone-modified peptides were more complex than those of unmodified peptides probably because of partial fragmentation of Dex-mes itself during ionization.
Table 1.
Masses of cysteine-containing tryptic peptides of Keap1 modified by reaction with Dex-mes and detected by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry
Dex-mes labeled cysteine residues (C257, C273, C288, and C297) of Keap1 are located in the intervening region (IVR, between the BTB and double glycine or Kelch repeat domains) and close (C613) to the C-terminal domain. The identity of peptides 22–23 and 27–28 was confirmed by liquid chromatography tandem mass spectrometry coupled with electrospray ionization and fragmentation by collision-induced dissociation.
Binding of Keap1 and Neh2 Domain of Nrf2: Dissociation by Inducers.
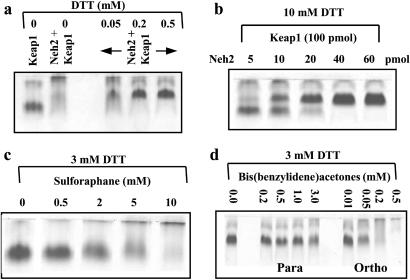
When Keap1 was incubated with the Neh2 domain of Nrf2, native PAGE revealed a band migrating more slowly than Keap1; its formation depended on the DTT concentration and increased in intensity with the Neh2 concentration (Fig. 8 a and b). This band presumably is a complex between the two proteins. If solutions containing the Keap1–Neh2 complexes were exposed to increasing concentrations of sulforaphane, a concentration-dependent loss of this complex was observed (Fig. 8c). Similar experiments with 2-HBA and 4-HBA revealed that the ortho isomer, a 100-fold more potent inducer of NQO1 and more than 100-fold more reactive with glutathione (9) than the para isomer, also much more potently disrupted the complex between the two proteins (Fig. 8d). These experiments establish that the formation of complexes between Keap1 and Nrf2 and their disruption by inducers in vivo can be modeled in isolated systems with pure Keap1 and the Neh2 domain of Nrf2.
Figure 8.
Native gel electrophoresis showing complex formation between Keap1 (100 pmol) and the Neh2 domain (50 pmol) of Nrf2, and their dissociation. Concentration-dependent effects of DTT (a), Neh2 (b), sulforaphane (c), and 2-(ortho) and 4-(para) hydroxylated bis-(benzylidene)acetones (d) are shown. The binding reactions and native gel electrophoresis were carried out as described in Experimental Procedures. The 11.9-kDa Neh2 fragment migrates rapidly as a single band and is not shown in these gels.
Discussion
Direct demonstration on native gels of the formation of complexes between Keap1 and the Neh2 domain of Nrf2 and their disruption by inducers leaves little doubt that the behavior of these proteins in purified systems is consistent with that inferred from studies in vivo (11). Participation of other cellular components or metabolic modifications therefore is not required for these processes. Furthermore, because all inducers react with thiol groups, and Keap1 is rich in cysteine residues, whereas the Neh2 domain lacks cysteine, we conclude that Keap1 is the long-sought sensor that recognizes and reacts with inducer molecules, causes dissociation of the Keap1-Nrf2 complex, releasing Nrf2 to migrate to the nucleus, bind to ARE elements, and activate phase 2 gene transcription. Our probing of the interaction of inducers with Keap1 by radiometric and UV spectroscopic techniques suggests that 4 of the 25 cysteines of Keap1 are especially reactive. These reactive residues (C257, C273, C288, and C297) were identified by modification with Dex-mes and mass spectrometric analyses of tryptic peptides. It is of particular interest that these cysteines are located in the intervening region between the two protein-binding domains (BTB and double glycine or Kelch repeat) of Keap1, and that this is consistent with their function as sensors for inducer ligands. Although these four cysteines may not be the only ones that are most reactive in vivo, their modification could lead to substantial conformational changes in Keap1, resulting in dissociation from Nrf2. The effects of modification of C613 are unclear, because it is located in the C-terminal domain of Keap1. Notably, C273, C288, C297, and C613 are adjacent to basic residues and are expected to have lower pKa values (27).
Because of the extraordinary chemical diversity of inducers, their interactions with the reactive thiol groups of Keap1 are probably of several types (Fig. 1). (i) Reaction of a single inducer molecule, e.g., phenylarsine oxide, with two cysteine residues that are in spatial proximity. (ii) Reaction of a disulfide, e.g., dipyridyl disulfide, initially with a single cysteine, followed by the attack of another cysteine thiolate ion to form a disulfide bridge in the protein. Direct oxidation may have the same outcome. (iii) Irreversible reaction with an inducer, e.g., Dex-mes, leading to cysteine alkylation. These modifications all could lead to conformational changes in Keap1, resulting in dissociation of the Keap1–Nrf2 complex.
“Redox switches” involving critical cysteines participate in regulation of a number of transcription factors (26, 28–30) and molecular chaperones (31, 32). The Keap1–Nrf2 interaction resembles the RsrA-σR system of Streptomyces coelicolor, which controls induction of the thioredoxin reductase/thioredoxin operon in response to oxidants (29). Under basal conditions Keap1 and RsrA bind Nrf2 and σR, respectively, anchoring them in the cytoplasm. After induction, Keap1 and RsrA lose their ability to bind their partners, allowing their migration to the nucleus and transcription of phase 2 and antioxidant genes. Both Nrf2 knockout mice and σR deletion mutants are more sensitive to electrophiles and oxidants, which correlates with lower basal levels and the inability to induce phase 2 and antioxidant enzymes. In the Keap1 knockout mouse Nrf-2-dependent transcription is increased (unpublished data), and the RsrA null mutant shows high σR-dependent transcription. The capacity of both Keap1 and RsrA to bind Nrf2 and σR is regulated by critical cysteine residues. Increased synthesis of thioredoxin enhances the regeneration of reduced RsrA (29). Increased levels of glutathione, thioredoxin, glutathione reductase, and thioredoxin reductase, as part of the phase 2 response, could provide regeneration system(s) for reduced Keap1. We conclude that the function of Keap1 is under oxidation/reduction (and alkylation) control via its highly reactive thiol groups that are modified by inducers.
Acknowledgments
We thank our colleagues Philip A. Cole, Thomas W. Kensler, and James T. Stivers for important and perceptive scientific advice, Pamela Talalay for valuable editorial consultation, Richard E. Bozak and Ronald J. Hicks for samples of the benzylideneacetones, and Tania O'Connor for preparation of a construct. The AB-Mass Spectrometry Facility at The Johns Hopkins School of Medicine is funded by National Center for Research Resources Shared-Instrumentation Grant 1S10-RR14702, The Johns Hopkins Fund for Medical Discovery, and the Institute for Cell Engineering. This work was supported by generous gifts from the Lewis B. and Dorothy Cullman Foundation, the Barbara Lubin Goldsmith Foundation, and the McMullan Family Fund, and by grants from the National Cancer Institute, Department of Health and Human Services (CA 94076), and from the American Institute for Cancer Research (Washington, DC).
Abbreviations
- ARE
antioxidant response element
- Dex-mes
dexamethasone 21-mesylate
- NQO1
NAD(P)H:quinone acceptor oxidoreductase (EC 1.6.99.2)
- CD
concentration of an inducer that doubles specific activity of NQO1 in Hepa1c1c7 murine hepatoma cells
- 2-HBA
bis(2-hydroxybenzylidene)acetone
- 4-HBA
bis(4-hydroxybenzylidene)acetone
References
- 1.Kensler T W. Environ Health Perspect. 1997;105:965–970. doi: 10.1289/ehp.97105s4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talalay P. Biofactors. 2000;12:5–11. doi: 10.1002/biof.5520120102. [DOI] [PubMed] [Google Scholar]
- 3.Talalay P, Fahey J W, Holtzclaw W D, Prestera T, Zhang Y. Toxicol Lett. 1995;82/83:173–179. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- 4.Talalay P, De Long M J, Prochaska H J. Proc Natl Acad Sci USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prestera T, Holtzclaw W D, Zhang Y, Talalay P. Proc Natl Acad Sci USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prestera T, Spencer S R, Wilczak C, Zhang Y, Talalay P. Adv Enzyme Regul. 1993;33:281–296. doi: 10.1016/0065-2571(93)90024-8. [DOI] [PubMed] [Google Scholar]
- 7.Hayes J D, Ellis E M, Neal G M, Harrison D J, Manson M M. Biochem Soc Symp. 1999;64:141–168. [PubMed] [Google Scholar]
- 8.Hayes J D, McMahon M. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova A T, Massiah M A, Bozak R E, Hicks R J, Talalay P. Proc Natl Acad Sci USA. 2001;98:3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chui D H K, Tang W, Orkin S H. Biochem Biophys Res Commun. 1995;209:40–46. doi: 10.1006/bbrc.1995.1467. [DOI] [PubMed] [Google Scholar]
- 11.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel J D, Yamamoto M. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 13.Rushmore T H, Pickett C B. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 14.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 15.Chan K, Kan Y W. Proc Natl Acad Sci USA. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho H Y, Jedlicka A E, Reddy S P, Kensler T W, Yamamoto M, Zhang L Y, Kleeberger S R. Am J Respir Cell Mol Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Gomez M, Kwak M K, Dolan P M, Itoh K, Yamamoto M, Talalay P, Kensler T W. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahey J W, Haristoy X, Dolan P M, Kensler T W, Schollus I, Stephenson K K, Talalay P, Lozniewski A. Proc Natl Acad Sci USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams J, Kelso R, Cooley L. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 20.Simons S S, Jr, Pons M, Johnson D F. J Org Chem. 1980;45:3084–3088. [Google Scholar]
- 21.Brocklehurst K. Int J Biochem. 1979;10:259–274. doi: 10.1016/0020-711x(79)90088-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Talalay P, Cho C-G, Posner G H. Proc Natl Acad Sci USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fahey J W, Zhang Y, Talalay P. Proc Natl Acad Sci USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 26.Gostick D O, Green J, Irvine A S, Gasson M J, Guest J R. Microbiology. 1998;144:705–717. doi: 10.1099/00221287-144-3-705. [DOI] [PubMed] [Google Scholar]
- 27.Snyder G H, Cennerazzo M J, Karalis A J, Field D. Biochemistry. 1981;20:6509–6519. doi: 10.1021/bi00526a001. [DOI] [PubMed] [Google Scholar]
- 28.Zheng M, Storz G. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 29.Kang J-G, Paget M S, Seok Y-J, Hahn M-Y, Bae J B, Hahn J-S, Kleanthous C, Buttner M J, Roe J-H. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Oberley L W. Free Radical Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 31.Barbirz S, Jakob U, Glocker M O. J Biol Chem. 2000;275:18759–18766. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- 32.Yan B, Smith J W. Biochemistry. 2001;40:8861–8867. doi: 10.1021/bi002902i. [DOI] [PubMed] [Google Scholar]