Abstract
Many nonhuman primate cells are unable to support the replication of HIV-1, whereas others are nonpermissive for infection by simian immunodeficiency virus from macaques (SIVmac). Here, we show that restricted HIV-1 and SIVmac infection of primate cell lines shares some salient features with Fv1 and Ref1-mediated restriction of murine retrovirus infection. In particular, the nonpermissive phenotype is most evident at low multiplicities of infection, results in reduced accumulation of reverse transcription products, and is dominant in heterokaryons generated by fusion of permissive and nonpermissive target cells. Moreover, in nonpermissive primate cells, HIV-1 and SIVmac infection is cooperative, and enveloped HIV-1 virus-like particles, minimally containing Gag and protease, abrogate restriction. In African green monkey cells, HIV-1 virus-like particles ablate restrictions to HIV-1 and SIVmac, suggesting that both are restricted by the same factor. Finally, a virus that contains an HIV-1 capsid-p2 domain in an SIVmac background exhibits a tropism for primate cells that is HIV-1-like rather than SIVmac-like. These data indicate the existence of one or more saturable inhibitors that are polymorphic in primates and prevent HIV and SIV infection by targeting the capsid of the incoming lentivirus particle.
Primate lentiviruses exhibit a highly restricted species tropism. HIV-1, for example, is unable to replicate in cells derived from several nonhuman primate species (1), including rhesus macaques, which limits the usefulness of this species as a vaccine or treatment model for human AIDS. The block to HIV-1 replication in macaques occurs primarily during the early steps of the viral life cycle, such that reverse transcription does not proceed to completion (2, 3). Because engineered simian immunodeficiency viruses from macaques (SIVmac) that contain envelope genes from HIV-1 can replicate in macaques, inefficient HIV-1 replication in this nonpermissive species cannot be explained by envelope–receptor incompatibility (4–9). Although postentry restrictions may be modulated, in part, by envelope–receptor interactions (10), cells of diverse primate species clearly exhibit large and opposing differences in their ability to support preintegration steps of the HIV-1 and SIVmac life cycles, even when virions are pseudotyped with the pan-tropic vesicular stomatitis virus envelope glycoprotein (VSV-G; ref. 11). These discrepancies are apparently determined by the primate species, not the tissue of origin, of the target cells, and cells from many other mammalian species, including rodents, can support the formation of an integrated provirus, provided that entry restrictions are overcome by pseudotyping (11, 12). Collectively, these observations indicate that cellular factors modulate HIV and SIV infection at a postentry, preintegration step. It is unknown whether inefficient HIV-1 or SIVmac infection of nonpermissive primate cells is caused by the absence of a necessary activity or the presence of an inhibitor that blocks infection.
Postentry, preintegration restrictions have long been known to limit murine leukemia virus (MuLV) tropism (13–17). In this case, the product of a murine gene, Fv1, inhibits MuLV infection. Fv1 encodes retroviral Gag-like protein (18), different forms of which (e.g., Fv1N, Fv1B) permit or restrict infection by reciprocal N- and B-tropic MuLV strains. Fv1-restricted MuLV tropism is determined by sequences within the capsid (CA) protein indicating that it is the target for inhibition (19). Remarkably, many nonmurine mammalian cells restrict infection by N-tropic MuLV (20), even though nonmurine cells lack a gene that is closely related to Fv1 (18). Because the same single amino acid substitution that governs N- versus B-tropism in murine cells also determines tropism for nonmurine cells (20, 21), it is reasonably assumed that nonmurine cells express an inhibitor of N-tropic MuLV infection, termed Ref1, that acts in a mechanistically similar manner to Fv1.
Here, we demonstrate that primate cells express an Fv1-like activity that, remarkably, restricts lentivirus tropism. The resistant phenotype is envelope-independent, results in defective accumulation of reverse transcription products, and is dominant in interspecies heterokaryons. Like Fv1 and Ref1 (22–25), inhibition is not absolute and can be saturated at high levels of inoculum or by the addition of virus-like particles during infection. Moreover, the inhibitor(s) apparently exists in alternative forms that restrict infection by HIV-1, SIVmac, or both. Finally, we show that the target for restriction is contained within the CA-p2 region of the HIV/SIV Gag protein.
Materials and Methods
Plasmid Construction.
HIV-1 and SIVmac239 proviral plasmids that express green fluorescent protein (GFP) in place of, or appended to, Nef (pHIV/GFP and pSIV/GFP) have been described (26). Env-defective versions of these clones were used in this study. In some experiments, envelope-defective HIV-1 (pNL4–3) or SIVmac239 proviral plasmids that lack a reporter gene were used.
A retroviral vector, pLNC/CAT/IRES/GFP, was constructed by inserting a cDNA encoding the ecotropic MuLV receptor, CAT (amplified from murine cDNA), and a fragment of pIRES2/GFP into pLNCX2 (CLONTECH). Upon transduction, pLNC/CAT/IRES/GFP expresses a bicistronic RNA that encodes CAT and GFP separated by an internal ribosome entry site.
A codon-optimized HIV-1 Gag-Pol expression plasmid, pSYN-GP (27) was a gift from Kyriacos Mitrophanous (Oxford Biomedica, Oxford, U.K.). Plasmids that express Gag only and Gag-protease were derived by insertion of PCR-amplified fragments of pSYN-GP into pCR3.1 (Invitrogen).
To express the HTLV-I envelope protein, an HIV-1-based expression vector, pV1, was used that lacks HIV genes except Tat, Rev, and Vpu, but retains cis-acting sequences necessary for the expression of Rev-dependent genes. In addition, a CMV promoter replaces the HIV-1 5′ U3 region. The HTLV-I env gene was inserted into pV1 to generate pV1/HTLV.
Cells and Viruses.
Cell lines of human (HeLa, 293T), Owl monkey (OMK), and African green monkey (CV-1) origin were obtained from the American Type Culture Collection. The rhesus monkey (Rh.F) and squirrel monkey (Pindak) fibroblast cell lines were gifts from Ron Desrosiers (New England Regional Primate Research Center, Southborough, MA) and Jonathan Scammell (University of South Alabama, Mobile, AL).
Reporter viruses (HIV/GFP and SIV/GFP) expressing GFP and pseudotyped with VSV-G were used throughout this study and were generated by transfecting 293T as described (12), as were similar viruses lacking reporter genes. In some cases, “bald” virus particles [denoted (E-)] were generated by omitting the VSV-G expression plasmid. Alternatively, HIV(MuLV-E) (lacking a reporter gene) was generated by transfecting HIV-1 proviral and ecotropic MuLV envelope expression plasmids. HIV-1 Gag-Pol, Gag-Protease (Gag-Pr), and Gag virus-like particles (VLPs) were made by transfecting cells with the codon-optimized expression plasmids described above. Enveloped (VSV) or ‘bald’ (E-) VLPs were obtained by the inclusion or omission of a VSV-G expression plasmid. All virus and VLP stocks were filtered (0.22 μm) and viral protein content determined by p24 (HIV-1) or p27 (SIV) ELISA.
Infection Assays.
Cells (2 × 104 per well in 24-well plates) were inoculated with fixed or varying amounts of reporter virus, as indicated in Results, in the presence of 5 μg/ml Polybrene. In some experiments, additional virions that lacked a reporter gene or VLPs were added simultaneously. Infected (GFP-positive) cells were enumerated by fluorescence-activated cell sorter (FACS) analysis 48–72 h later. For PCR analyses, 1.5 × 105 cells in six-well trays were inoculated with a DNase-treated HIV/GFP stock. Cells were harvested at various times after inoculation, DNA extracted by using a commercial kit (Qiagen, Chatsworth, CA), and HIV-1 reverse transcription products quantitated by using primers spanning the 5′ primer binding site in a real-time PCR assay that will be described elsewhere (T.C. and M.AM., unpublished work).
Generation and Infection of Interspecies Heterokaryons.
The pLNC/CAT/IRES/GFP retroviral vector was packaged into VSV-G-pseudotyped MuLV particles as described (12). After transduction, GFP-positive OMK and HeLa cells were sorted by FACS to generate pure GFP- and CAT-expressing populations. These cells were seeded in 24-well trays (4 × 104 per well) and overlaid with 293T cells transfected with pV1/HTLV the previous day. After OMK × 293T and HeLa × 293T heterokaryon formation (24 h), fused cells were inoculated with serially diluted HIV(MuLV-E). Forty-eight hours later, cells were fixed and HIV(MuLV-E) infection revealed by immunofluorescence, as described (12) [except that an Alexafluor 594-conjugated secondary antibody (red) was used to distinguish infected GFP-positive cells]. Infected single GFP-positive cells (or foci of several single cells) were counted separately from infected heterokaryons (GFP-positive multinucleated cells) at an appropriate dilution (20–100 infection events per well).
Results
Nonlinear Titration Curves upon Infection of Primate Cells with HIV-1 or SIVmac.
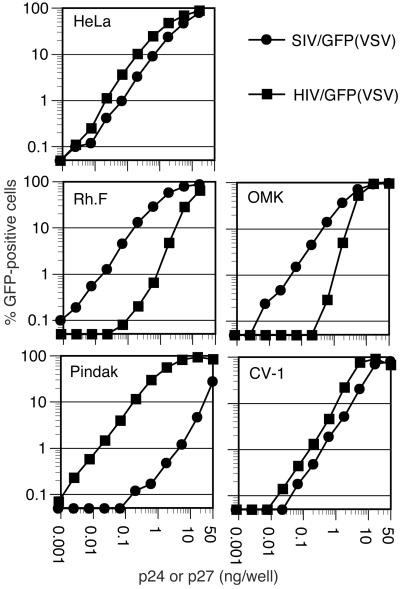
We first inoculated several primate cell lines with increasing amounts of VSV-G-pseudotyped HIV-1 and SIVmac reporter viruses. As reported (11), and shown in Fig. 1, large differences exist in the susceptibility of some of the cell lines to HIV-1 versus SIVmac infection at low multiplicity of infection (MOI). Specifically, whereas the human cell line HeLa was approximately equivalently infected by HIV/GFP and SIV/GFP, Rhesus monkey (Rh.F) and Owl monkey (OMK) cells were infected at an almost 100-fold greater frequency by SIV/GFP than by HIV/GFP. In contrast, the squirrel monkey cell line, Pindak, exhibited precisely the opposite phenotype. The African green monkey (CV-1) cell line was almost equivalently infected by each of the viruses, although both appeared less infectious on CV-1 cells at low MOI as compared with HeLa cells.
Figure 1.
Titration of HIV-1 and SIVmac reporter viruses on primate cells. The indicated cells lines were inoculated with increasing amounts of VSV-G-pseudotyped GFP reporter viruses. Infected cells were enumerated by FACS.
If ‘one-hit’ kinetics is assumed, the frequency of infected cells should increase in a linear manner proportional to inoculum size at nonsaturating levels of infection. Remarkably, however, and in contrast to previous findings (11), we noted that the differences among the various cell lines in susceptibility to HIV-1 and SIVmac infection were decreased as the MOI was increased. Put another way, the titration curves were nonlinear in cells that were less permissive. In the logarithmic plots shown in Fig. 1, this nonlinearity results in a slope value of >1 at nonsaturating infection levels. This finding was especially evident when HIV/GFP was titrated on OMK cells, where the slope of the titration curve was ≈2.2 (i.e., a 10-fold increase in MOI resulted in a >100-fold higher frequency of infected cells). Unusual titration curves were also evident for HIV/GFP infection of Rh.F cells (slope = 1.7) and to a lesser extent when SIV/GFP was titrated on Pindak cells (slope = 1.4 at its steepest point). Both HIV/GFP and SIV/GFP titration curves had slopes slightly greater than 1 (1.3 and 1.1, respectively) on CV-1 cells. Where cells were highly permissive to either HIV/GFP or SIV/GFP, the slopes of the titration curves were ≈1 (0.94–0.98).
Reduced Levels of Reverse Transcription Products in Nonpermissive Cells.
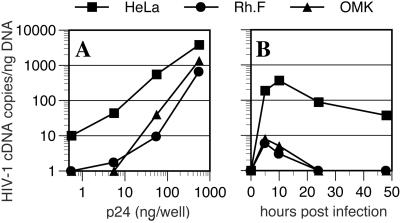
To determine at what step HIV-1 infection was blocked in nonpermissive primate cells, we performed a real-time PCR analysis of late reverse transcription products after inoculation of target cells with varying amounts of HIV-1/GFP. As is shown in Fig. 2A, a greatly reduced level (20- to 80-fold) of HIV-1 cDNA was detected after inoculation of OMK or Rh.F cells as compared with HeLa cells. Again, this phenotype was suppressed as the MOI was increased. A kinetic analysis, done at low MOI, revealed a large defect in the accumulation of reverse transcripts in OMK and Rh.F cells that was evident within a few hours of inoculation (Fig. 2B). These data indicate that restricted HIV-1 infection of these primate cells is due either to a failure to complete reverse transcription or to an instability of the reverse transcription products, specifically at low MOI.
Figure 2.
Reduced accumulation of HIV-1 cDNA in nonpermissive primate cells at low MOI. (A) HeLa, Rh.F, and OMK cells were inoculated with varying amounts of VSV-G-pseudotyped HIV/GFP and reverse transcripts measured by real-time PCR 48 h later. Alternatively, (B) cells were inoculated with 25 ng of HIV/GFP, and reverse transcripts were measured at the indicated times after infection.
Infection of Nonpermissive Primate Cells at High MOI Is Cooperative.
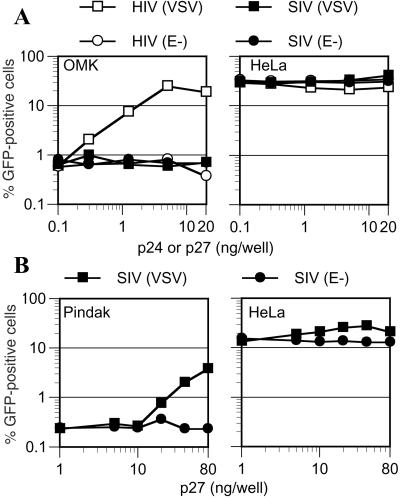
The nonlinear relationship between the inoculum size and the resulting levels of infection in nonpermissive cells are suggestive of cooperativity. In other words, infection of an otherwise nonpermissive target cell by a given vision particle is facilitated by other virion particles. To test this notion, we inoculated permissive (HeLa) and nonpermissive (OMK) target cells with a fixed amount of VSV-G-pseudotyped HIV/GFP reporter virus in the presence of virions that lacked a reporter gene. The reporter virus inoculum (1 ng of p24 per well) was chosen to give low but readily detectable levels of infection in nonpermissive OMK cells (≈0.8%), whereas about 30% of permissive HeLa cells became infected. As seen in Fig. 3A, the inclusion of additional HIV-1 virions dramatically enhanced infection of OMK cells (up to 40-fold) by the HIV/GFP reporter virus, in a dose-dependent manner. In the presence of saturating levels of HIV-1 virions (>5 ng per well) the reporter virus infected OMK cells as efficiently as it did HeLa cells. This enhancing effect was specific; SIVmac virions did not enhance infection by HIV/GFP. Moreover, ‘bald’ HIV-1 particles were also inactive, suggesting that entry of the ‘rescuing’ virus is required to restore HIV/GFP infection. Infection of permissive HeLa cells by HIV/GFP was unaffected by the presence of excess enveloped or bald HIV-1 or SIVmac particles.
Figure 3.
Cooperative infection of nonpermissive cells at high MOI. (A) OMK and HeLa cells were inoculated with 1 ng of HIV/GFP(VSV) in the presence of increasing amounts of pseudotyped or bald HIV-1 or SIVmac virions. (B) Pindak or HeLa cells were inoculated with 1 ng of SIV/GFP(VSV) in the presence of the indicated amounts of VSV-G pseudotyped SIVmac virions. For both A and B, reporter virus infection was measured by FACS.
High MOI SIVmac infection of nonpermissive Pindak cells also appeared cooperative (Fig. 3B). In this case, SIVmac(VSV) but not SIVmac(E-) particles were able to enhance infection by the SIV/GFP reporter virus by 10-fold. SIVmac infection of Pindak cells seemed to be more profoundly restricted than was HIV-1 infection of OMK cells, in that larger quantities of rescuing virions were required to measurably enhance SIV/GFP infection, and the restoration of reporter virus infection was incomplete.
Restricted Infection of Owl Monkey Cells by HIV-1 Is Due to the Presence of an Endogenous Inhibitor.
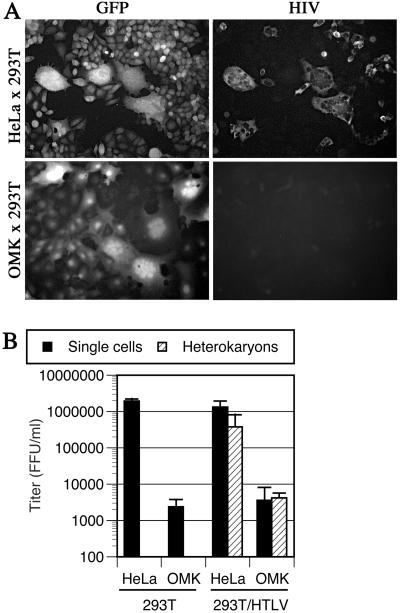
The apparently ‘saturable’ nature of the restriction to HIV-1 or SIVmac infection in some primate cells is difficult to reconcile with the notion that nonpermissive cells lack a necessary activity that is required for infection. Nevertheless, to determine whether inefficient infection of primate cells by HIV-1 is caused by the presence of an inhibitor or, alternatively, by the lack of a necessary activity, we first generated OMK and, as a control, HeLa cells that express the ecotropic MuLV receptor (CAT) as well as GFP. These cells were mixed with 293T cells that were, or were not, transfected with an HTLV-I envelope expression plasmid. The HTLV-I envelope mediates efficient fusion between the 293T cells and either OMK or HeLa target cells, and in both cases efficient heterokaryon formation was observed as the appearance of green-fluorescent multinucleated cells, as seen in Fig. 4A. After heterokaryon formation, the cultures were infected with HIV(MuLV-E). Because nonmurine cells lack endogenous MuLV-E receptors, only the CAT- and GFP-expressing HeLa or OMK cells (and heterokaryons) permit entry of HIV(MuLV-E). Infection of unfused cells and heterokaryons, quantitated by counting immunostained foci of HIV-1-infected cells that contained either single or multiple nuclei, is shown in Fig. 4 A and B. HIV(MuLV-E) was approximately 100-fold less infectious on unfused OMK/CAT/GFP cells than on HeLa/CAT/GFP, specifically at low MOI (Fig. 4 A and B). Thus, the restriction to HIV-1 infection in OMK cells is preserved by using this alternative route of virus entry. Moreover, HeLa × 293T heterokaryons were infected almost as efficiently as unfused HeLa cells, indicating that prior fusion of two permissive cell lines does not inhibit HIV-1 infection per se. OMK × 293T heterokaryons were infected as poorly as were unfused OMK cells. Thus, in the context of a heterokaryon formed between permissive and nonpermissive cells, the nonpermissive phenotype is dominant. This finding strongly suggests that the nonpermissive cell, in this case OMK, expresses at least one factor that inhibits HIV-1 infection.
Figure 4.
Resistance to HIV-1 infection is dominant in heterokaryons. OMK or HeLa cells expressing CAT and GFP were fused with 293T cells expressing an HTLV-I envelope protein and inoculated with HIV(MuLV-E), and infected cells were revealed by immunofluorescence. (A) Representative photomicrographs showing GFP expression (Left) and HIV immunofluorescence (Right) in the same fields of cells. (B) Titers of HIV(MuLV-E) measured on CAT and GFP expressing OMK or HeLa cells cocultivated with 293T cells that did or did not express HTLV-I envelope. Infected heterokaryons were counted separately from infected unfused cells.
HIV-1 Virus-Like Particles Can Ablate the Infection Resistance of Primate Cells.
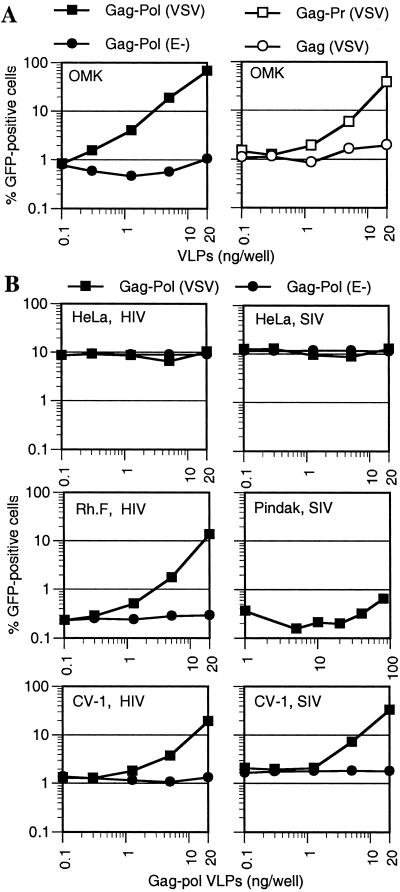
We next determined what components of a ‘rescuing’ particle are necessary to ablate the restriction to HIV-1 infection. We used an HIV-1 Gag-Pol expression plasmid that generates VLPs which lack an HIV-1 genome or any other HIV-1 gene product (27). As shown in Fig. 5A, VSV-enveloped (but not ‘bald’) Gag-Pol VLPs were able to enhance HIV/GFP infection of OMK cells as efficiently as infectious HIV-1 virions (Fig. 3A). Thus, no virion component other than Gag-Pol and an envelope is required to ablate the inhibitory activity in OMK cells. A deletion analysis revealed that enveloped VLPs containing only Gag and protease also enhanced infection, although these VLPs were less potent than Gag-Pol VLPs. In contrast, enveloped particles containing Gag alone were inactive (Fig. 5A), indicating that either protease itself or particle maturation is required to form a structure that can ablate the inhibitory activity expressed by OMK cells.
Figure 5.
HIV-1 VLPs enhance HIV and SIVmac infection in a target-cell-dependent manner. (A) OMK cells were inoculated with 1 ng HIV/GFP(VSV) in the presence of the indicated amounts of VSV-G enveloped or bald Gag-Pol VLPs (Left). Alternatively, enveloped Gag-Pr or Gag VLPs were used (Right). (B) The indicated cell lines were inoculated with 0.2 ng of HIV/GFP(VSV) (Left) or 1 ng SIV/GFP(VSV) (Right) reporter viruses in the presence of increasing levels of enveloped or bald HIV-1 Gag-Pol VLPs. Infected cells were enumerated by FACS.
We next examined whether restricted HIV-1 or SIVmac infection of other primate cell lines was affected by HIV-1 VLPs. As shown in Fig. 5B, neither HIV/GFP nor SIV/GFP infection of HeLa cells was influenced by HIV-1 VLPs. However, HIV/GFP infection of the rhesus monkey cell line Rh.F was dramatically enhanced (approximately 50-fold) by HIV-1 VLPs to a level similar to that in HeLa cells. In contrast, restricted SIV/GFP infection of squirrel monkey (Pindak) cells that was enhanced by saturating SIVmac virions (Fig. 3B) was unaffected by the same levels of HIV-1 VLPs (Fig. 5B). Thus, it appears likely that Rh.F and OMK cells express phenotypically similar inhibitors that block HIV-1 but not SIVmac infection (Figs. 1 and 5), whereas Pindak cells express an inhibitory activity that recognizes incoming SIVmac but not HIV-1 virions.
HIV-1 VLPs significantly enhanced infection by both HIV/GFP and SIV/GFP when African green monkey (CV-1) cells were used as targets (Fig. 5B). Virtually identical levels of HIV-1 VLPs were required to enhance infection by either reporter virus. This result suggests that CV-1 cells restrict both HIV-1 and SIVmac infection and, because HIV-1 VLPs can suppress resistance to SIVmac, that both restrictions are mediated by the same saturable factor or factors.
The CA-p2 Domain of Gag Is a Major Determinant of Tropism Restriction.
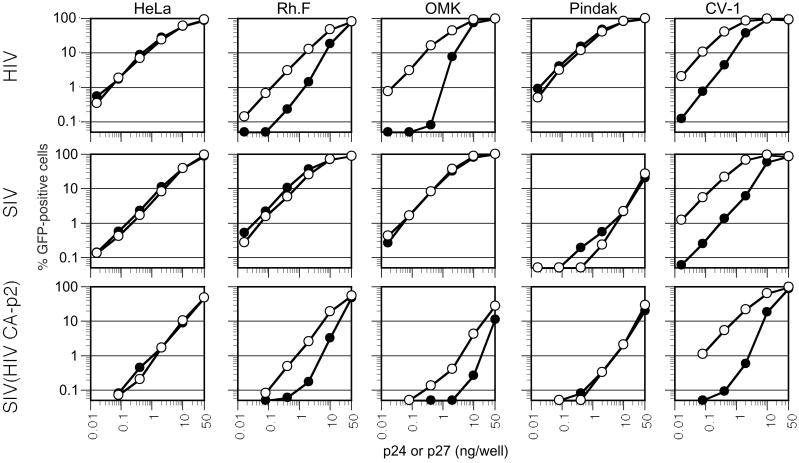
It has previously been shown that a chimeric SIVmac containing an HIV-1 CA-p2 domain replicates in human but not macaque lymphocytes (28), for unknown reasons. The ability of enveloped VLPs to enhance HIV-1 infection in restrictive cells raised the possibility that the inhibitory activity described herein targets the incoming virion core, perhaps the CA or p2 proteins, to prevent infection. To address this, we titrated a GFP reporter virus that contains an HIV-1 CA-p2 protein in an otherwise SIVmac background as well as the parental SIV/GFP and HIV/GFP viruses on the various primate cell lines. To determine whether each virus was restricted by a saturable inhibitory activity that recognizes HIV-1 components, we simultaneously performed titrations in the presence of HIV-1 VLPs. As shown in Fig. 6, HIV-1 VLPs enhanced the infectivity of HIV/GFP in restrictive Rh.F, OMK, and CV-1 cells, by 10- to 100-fold at low MOI, but did not affect HIV/GFP infection of nonrestrictive HeLa or Pindak cells at any level of inoculum. In contrast, SIV/GFP infection was unaffected by HIV-1 VLPs in any cell line except CV-1, which restricts both HIV and SIVmac infection (Figs. 1, 3B, and 6). The chimeric SIV(HIV CA-p2) reporter virus was somewhat less infectious than either HIV/GFP or SIV/GFP in HeLa cells, which do not restrict either parental virus, suggesting that it has a modest, nonspecific replication defect. Nevertheless, the SIV(HIV CA-p2) virus exhibited an HIV-1- rather than SIVmac-like phenotype. Specifically, HIV-1 VLPs enhanced SIV(HIV CA-p2)/GFP infection of HIV-1-restricting (Rh.F, OMK, and CV-1) cells, but not nonrestricting (HeLa or Pindak) cells (Fig. 6). This result suggests that the transfer of an HIV-1 CA-p2 domain into SIVmac renders it sensitive to an inhibitor in Rh.F and OMK cells that can be saturated by HIV-1 VLPs.
Figure 6.
The CA-p2 domain of Gag is a major target for restriction. The indicated cell lines were inoculated with varying amounts (as indicated) of VSV-G-pseudotyped HIV/GFP, SIVmac/GFP, or a chimeric virus, SIV(HIV CA-p2)/GFP. Infections were done in the absence (black symbols) or presence (white symbols) of 20 ng per well HIV-1 Gag-Pol VLPs. Infected cells were enumerated by FACS.
In the absence of VLPs, SIV(HIV CA-p2)/GFP was 10- to 50-fold less infectious on Rh.F and OMK cells than on HeLa cells. Moreover, the titration curves of SIV(HIV CA-p2)/GFP were nonlinear in these monkey cell lines. Again these results are similar to those obtained with HIV/GFP, and contrast with those obtained with SIV/GFP (Fig. 6). Furthermore, whereas SIV/GFP was approximately 30-fold less infectious on Pindak versus HeLa target cells, SIV(HIV CA-p2) was only 4- to 5-fold less infectious on Pindak cells. Therefore, although we cannot formally exclude a role for other virion components in influencing HIV-1/SIVmac tropism, it is evident that primate lentivirus tropism is restricted by interactions between components of the target cell and the viral CA-p2 Gag domain.
Discussion
These findings strongly suggest that the genomes of several primate species encode a specific, saturable inhibitor of lentivirus infection that targets components of the incoming virion core, specifically CA and/or p2. This activity is remarkably similar to that of Fv1, and in keeping with the nomenclature previously established for this factor, we propose to name the new inhibitor Lv1 (lentivirus susceptibility factor 1). Thus, our findings suggest that Lv1 in Rh.F and OMK cells restricts infection by HIV-1 but not SIVmac, whereas Lv1 in Pindak cells restricts SIVmac but not HIV-1, and that the CV-1 form of Lv1 restricts both viruses.
An Fv1-like activity (Ref1) that restricts N-tropic MuLV infection of nonmurine cells has been described (20), and it is formally possible that Lv1 is, in fact, allelic with Ref1. In this case, it would be necessary to speculate that certain forms of Lv1/Ref1 can block infection by both HIV/SIV and N-tropic MuLV. The ‘infection-rescue’ assays in cells that restrict each of these viruses should resolve whether this is the case. It will also be interesting to determine whether the Lv1 phenotype shown here is related to the redistribution of nuclear body components to the cytoplasm that was recently reported to occur upon HIV-1 entry (29).
Although the data presented here and previously (11) suggest that Lv1 exists in species-specific forms, it is possible that intraspecies variation occurs in this phenotype. The documented variation in the ability of human cells to support HIV-1 replication (30–32) could conceivably be influenced by Lv1. Although surveys have not been exhaustive (11; data not shown), it seems that postentry, preintegration blocks to HIV-1 infection are largely confined to primates, the possible exception being rabbits. Both new- and old-world primate species express an Lv1 phenotype, although only the latter have documented lentivirus infections. Although it is possible that as-yet-unidentified lentiviruses exist in South American primates, it is also reasonable to speculate that this postentry, preintegration block and other inhibitory activities have contributed to the extinction of primate lentiviruses in new-world species.
Lv1 is the second cell-autonomous and dominant phenotype in primates that inhibits lentivirus replication. Some primate cells also express a dominant inhibitor that acts late in the virus life cycle (33–35), whose activity can attenuate the infectivity of divergent HIV and SIV strains. However, all characterized HIVs and SIVs encode a gene product, Vif, whose sole known function is to counteract the effect of the cellular inhibitor (36). The existence of Lv1 and other lentivirus inhibitors is likely to influence dramatically the propensity of HIV/SIV strains to cause zoonotic infections. Although several widely divergent genetic lineages exist within this group of viruses, only two (HIV-1/SIVcpz and HIV-2/SIVsm) have caused epidemics in humans, with evidence for multiple zoonoses (37–40). Other, more common SIVs have, thus far, failed to colonize human hosts effectively. Both viruses used in this study are apparently not subject to Lv1 restriction in human cells and represent lineages that have infected human populations. It will be important to determine whether human cells exhibit an Lv1-like phenotype that protects or, more importantly, does not protect, from infection by other SIV lineages, because this may suggest potential sources of future immunodeficiency virus epidemics in humans (38).
Acknowledgments
We thank Ronald Desrosiers, Kyriacos Mitrophanous, and Jonathan Scammell for gifts of reagents. This work was supported by the Donald A. Pels Charitable Trust, The Aaron Diamond AIDS Research Center, The Mark Bertuch Memorial Fund, and Grants RO1AI47054 and RO1AI50111 from the National Institute of Allergy and Infectious Diseases (to M.A.M. and P.D.B., respectively).
Abbreviations
- MuLV
murine leukemia virus
- VSV
vesicular stomatitis virus
- VLP
virus-like particle
- MOI
multiplicity of infection
- CA
capsid
- SIVmac
simian immunodeficiency virus from macaques
- FACS
fluorescence-activated cell sorter
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 11549.
References
- 1.Gartner S, Liu Y, Polonis V, Lewis M G, Elkins W R, Hunter E A, Miao J, Corts K J, Eddy G A. J Med Primatol. 1994;23:155–163. doi: 10.1111/j.1600-0684.1994.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 2.Himathongkham S, Luciw P A. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 3.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. J Gen Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 4.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L J, Adany I, Pinson D M, McClure H M, Narayan O. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. J Acquired Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 6.Li J T, Halloran M, Lord C I, Watson A, Ranchalis J, Fung M, Letvin N L, Sodroski J G. J Virol. 1995;69:7061–7067. doi: 10.1128/jvi.69.11.7061-7067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luciw P A, Pratt-Lowe E, Shaw K E, Levy J A, Cheng-Mayer C. Proc Natl Acad Sci USA. 1995;92:7490–7494. doi: 10.1073/pnas.92.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. J Virol. 1991;65:3514–3520. doi: 10.1128/jvi.65.7.3514-3520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reimann K A, Li J T, Voss G, Lekutis C, Tenner-Racz K, Racz P, Lin W, Montefiori D C, Lee-Parritz D E, Lu Y, et al. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian B, Long E M, Luciw P A, Overbaugh J. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bieniasz P D, Cullen B R. J Virol. 2000;74:9868–9877. doi: 10.1128/jvi.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff S P. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 14.Jolicoeur P. Curr Top Microbiol Immunol. 1979;86:67–122. doi: 10.1007/978-3-642-67341-2_3. [DOI] [PubMed] [Google Scholar]
- 15.Lilly F. J Natl Cancer Inst. 1970;45:163–169. [PubMed] [Google Scholar]
- 16.Pincus T, Hartley J W, Rowe W P. J Exp Med. 1971;133:1219–1233. doi: 10.1084/jem.133.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pryciak P M, Varmus H E. J Virol. 1992;66:5959–5966. doi: 10.1128/jvi.66.10.5959-5966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Best S, Le Tissier P, Towers G, Stoye J P. Nature (London) 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 19.DesGroseillers L, Jolicoeur P. J Virol. 1983;48:685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak C A, Chakraborti A. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 22.Towers G, Collins M, Takeuchi Y. J Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tennant R W, Otten J A, Brown A, Yang W K, Kennel S J. Virology. 1979;99:349–357. doi: 10.1016/0042-6822(79)90014-x. [DOI] [PubMed] [Google Scholar]
- 24.Boone L R, Innes C L, Heitman C K. J Virol. 1990;64:3376–3381. doi: 10.1128/jvi.64.7.3376-3381.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decleve A, Niwa O, Gelmann E, Kaplan H S. Virology. 1975;65:320–332. doi: 10.1016/0042-6822(75)90038-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Hatziioannou T, Zang T, Braaten D, Luban J, Goff S P, Bieniasz P D. J Virol. 2002;76:6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotsopoulou E, Kim V N, Kingsman A J, Kingsman S M, Mitrophanous K A. J Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorfman T, Gottlinger H G. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Mol Cell. 2001;7:1245–1254. doi: 10.1016/s1097-2765(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 30.Spira A I, Ho D D. J Virol. 1995;69:422–429. doi: 10.1128/jvi.69.1.422-429.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams L M, Cloyd M W. Virology. 1991;184:723–728. doi: 10.1016/0042-6822(91)90442-e. [DOI] [PubMed] [Google Scholar]
- 32.Eisert V, Kreutz M, Becker K, Konigs C, Alex U, Rubsamen-Waigmann H, Andreesen R, von Briesen H. Virology. 2001;286:31–44. doi: 10.1006/viro.2001.0940. [DOI] [PubMed] [Google Scholar]
- 33.Simon J H, Miller D L, Fouchier R A, Soares M A, Peden K W, Malim M H. EMBO J. 1998;17:1259–1267. doi: 10.1093/emboj/17.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 35.Madani N, Kabat D. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Nature (London) 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 38.Hahn B H, Shaw G M, De Cock K M, Sharp P M. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 39.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, et al. Nature (London) 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]