Abstract
Retroviruses are able to cross species barriers and have done so many times throughout evolution. Perhaps as a consequence, dominant mechanisms have arisen to block infection by murine retroviruses in mice (restriction factor Fv1) and humans (restriction factor Ref1), as well as in other mammals. Here we describe a block to HIV and simian immunodeficiency virus in monkeys. Like previously described restrictions the block is saturable and gives rise to multiple-hit infection kinetics. Furthermore, like restriction of murine leukemia virus in humans, the block is before reverse transcription. Intriguingly, African green monkey cells are able to block both HIV and simian immunodeficiency virus, and each virus is able to saturate and abrogate the restriction of the other, suggesting that a common factor is responsible.
Recent phylogenetic analysis of retroviral sequences has suggested that interspecies retroviral infection between mammals may have been frequent during their evolution (1). Certainly HIV types 1 and 2 are derived from simian immunodeficiency virus from chimpanzees (SIVcpz) and sooty mangabees (SIVsm), respectively (2, 3). Despite these viruses' ability to cross species, several nonimmunological blocks to replication in foreign hosts have been demonstrated. Such a block exists for HIV type 1 (HIV-1) in rhesus macaques, the major HIV-1 primate experimental model (4, 5). Much work from several groups has focused on the reasons for the block in macaques. Experiments in macaque PBL and simian MAGI cells have shown that the block is generally, although not always, before completion of reverse transcription (4, 6, 7). However, the block is not due to an inability of HIV-1 env to direct entry into macaque cells as shown by the efficient replication of SHIVs, chimeras of HIV-1 and simian immunodeficiency virus from macaques (SIVmac) encoding HIV-1 envelope, tat, rev, and vpu proteins and SIVmac gag-pro-pol.
Attempts to map the determinant for the block more closely have been hampered by the difficulty of making functional HIV-SIV gag-pol fusions. However, a SHIV has been produced with an HIV-1 capsid-p2 domain replacing the equivalent SIVmac sequence (8). This virus was able to replicate in human cells but not macaque PBL, strongly implying a determinant in capsid. The virus incorporated cyclophilin and was inhibited by a cyclosporin analogue, further demonstrating the functional capsid phenotype of HIV-1. Unfortunately, the converse SHIV, an HIV-1 with an SIVmac capsid-p2 region, was noninfectious, demonstrating the difficulty of obtaining such chimeras (8). Further experiments on HIV-1 infection of macaque have shown that the coreceptor may somehow be involved. Although expression of human CD4 allows entry of HIV-1 into simian MAGI cells, the infection is nonproductive with a block before or after reverse transcription depending on the HIV-1 strain used. Expression of human coreceptor, however, facilitates their infection by HIV-1, suggesting that certain entry pathways may be able to bypass postentry restrictions (7).
We have recently characterized a block to murine leukemia virus in a range of mammals, which seems to be due to expression of a saturable factor in the resistant cells (9, 10). This block to infection resembles Fv1-mediated restriction in mice in its saturable nature and its target specificity. Characterization of these restrictions allowed us to develop an assay based on abrogation or saturation of restriction, depending on the ability of a sensitive virus to soak up and overcome the restriction factor. Whereas previous assays have been dependent on measuring relative infection by a pair of viruses, one restricted and one not, this assay uses a single restricting virus. We have used this assay to examine blocks to HIV-1- and SIVmac251-derived vectors in monkey cell lines. Although rhesus and owl monkey cells have been described as being refractory to HIV-1 infection (4, 11), it has been unclear whether these cells possess factors preventing HIV-1 infection or whether HIV-1 is unable to interact with cell factors supporting virus infection. Here we demonstrate the presence of a saturable factor that is specifically able to block infection of lentiviruses.
Materials and Methods
Cell Lines.
FRhK4 and LLC-MK2 were obtained from the Centro Substrati Cellulari, Brescia, Italy; and OMK, TE671, and SIRC cells were from the European Collection of Cell Cultures, Porton Down, U.K. CV1 cells were a kind gift of P. Jat, Ludwig Institute for Cancer Research, London. Cells were maintained as recommended by suppliers.
Viral Vector Preparation.
Viral vectors were prepared by transfection of 293T cells by using Fugene-6 (Roche Molecular Biochemicals) as follows. To make HIV-1 vectors, confluent 293T cells were transfected on a 10-cm plate with a mixture of 18 μl of Fugene-6 in 200 μl of OptiMEM (GIBCO/BRL) with 1 μg of pMDG [vesicular stomatitis virus envelope protein (VSV-G) expression vector (12)], 1 μg of p8.91 HIV-1 gag-pol expression vector (12), and 1.5 μg of retroviral expression vector encoding enhanced green fluorescent protein (eGFP), SIN CSGW, a kind gift of A. Thrasher, Institute of Child Health, University College London, U.K. or SIN CSPW encoding puro (puromycin resistance). To make SIVmac vectors 1 μg of SIV3+ (SIVmac gag-pol expression vector) and 1.5 μg of SIV-eGFP or SIV-β-galactosidase (LacZ) (SIV vectors encoding eGFP and LacZ, respectively) were used (13). SIVmac plasmids were a kind gift of François-Loic Cosset, Ecole Normale Superieure de Lyon, France. Murine leukemia virus (MLV) vectors were made by using plasmids as described (9). VSV-G expression vector pMDG was used to supply envelope for most SIV and MLV preparations, whereas a cytomegalovirus promoter-driven expression plasmid, a gift of F.-L. Cosset, was used to produce an HIV-eGFP vector bearing amphotropic MLV Env. Viral supernatant was collected at 48, 72, and 96 h after transfection and stored at −80°C. When required, virus was concentrated by centrifugation at 17,000 rpm in an SW28 rotor for 2 h. The pellet was resuspended in 1/10 volume media.
Viral Titer Determination.
All HIV-1 and SIVmac vector titers were measured on the permissive human cell line TE671, and titers are described as TE671 infectious units (i.u.)/ml. HIV-eGFP titers were determined 48 h after infection by a fluorescence-activated cell sorter (FACS) as described (14). HIV-puro titer was measured by infection of TE671 cells and colony counting after selection in 1 μg/ml puromycin. SIV-LacZ titers were determined 48 h after infection and LacZ staining as described (15).
Abrogation Assays.
Abrogation assays were performed in six-well plates on 105 cells in 1 ml containing 5 μg/ml Polybrene. Cells were exposed to HIV-puro or SIV-LacZ for 4 h. Cells were washed and exposed to HIV-eGFP or SIV-eGFP. eGFP fluorescence was measured 48 h later by FACS analysis on a Becton Dickinson FACScan or LSR by using CELLQUEST software as described (14). In titration experiments in Figs. 2 and 4, 50,000 cells were analyzed by FACS to increase sensitivity; in all other experiments 10,000 cells were analyzed.
Figure 2.
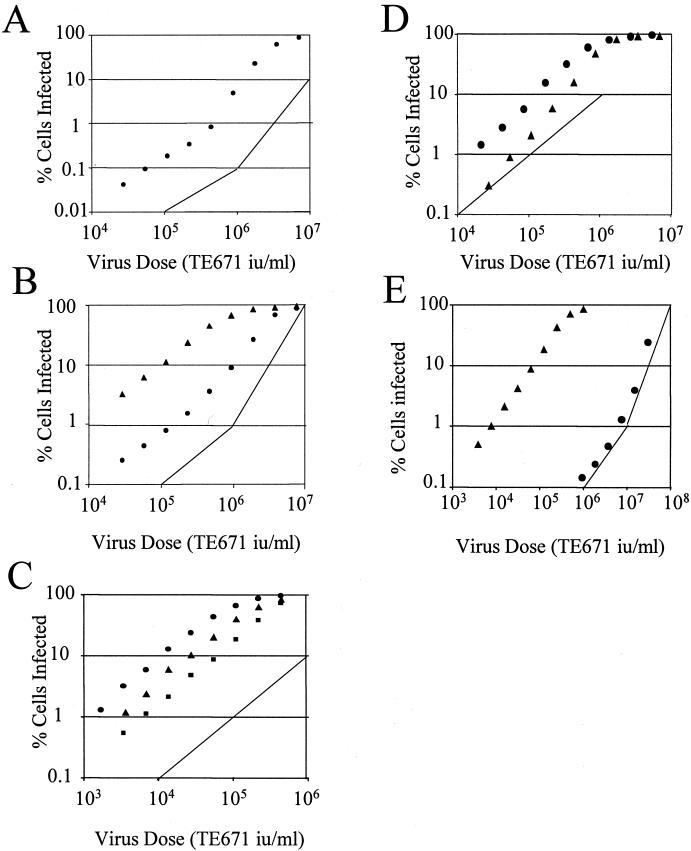
Titration of lentiviral vectors onto restricting cells. Two-fold serial dilutions of HIV-eGFP were titrated onto 105 OMK cells (A), LLC-MK2 (▴) and FRhK4 (●) cells from rhesus macaque (B). (C) Two-fold serial dilutions of SIV-LacZ were titrated onto OMK (●), FRhK4 (▴), and LLC-MK2 (■) cells. Two-fold serial dilutions of HIV-eGFP (●) or SIV-eGFP (▴) were titrated onto African green monkey CV1 cells (D) or rabbit SIRC cells (E). Results are representative of two independent experiments. Lines are guides for slopes of 1 and 2.
Figure 4.
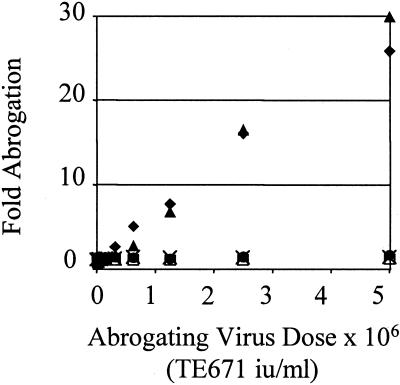
Abrogation of restriction of SIV-eGFP by SIV-LacZ and HIV-puro in African green monkey CV1 cells. TE671 (□), LLC-MK2 (●), OMK (×), or CV1 cells (▴) were exposed to 5 × 106 TE671 i.u. of SIV-LacZ or 5 × 106 TE671 i.u HIV-puro (♦) (CV1 only) for 4 h, washed, and exposed to 5 × 104 i.u. of SIV-eGFP. Percentage infection was measured by analysis of eGFP expression 48 h later by FACS. Fold abrogation is calculated by dividing the increased percentage infection after abrogation by unabrogated control HIV-eGFP infection. Approximately 1% of unabrogated monkey controls and 10% of human TE671 controls typically were infected. Data are representative of two independent experiments.
Mutation of Reverse Transcriptase Active Site.
Quikchange site-directed mutagenesis (Stratagene) was performed according to manufacturer's protocols by using p8.91 HIV-1 gag-pol expression vector and oligonucleotides forward GT141 CAATACATGGAAGATTTGTATGTAGGATC and reverse GT142 GATCCTACATACAAATCTTCCATGTATTG. The conserved YMDD motif was changed to YMED.
TaqMan Quantitative PCR of Viral DNA.
As in the abrogation assays, 105 cells were infected in six-well plates in triplicate. Four hours after the second infection total DNA was extracted by using a DNeasy kit (Qiagen, Chatsworth, CA) from two samples. The third sample was subjected to FACS analysis 48 h later to measure infection. DNA (100 ng) was subjected to TaqMan quantitative PCR essentially as described (16) by using TaqMan 2× quantitative PCR buffer (Applied Biosystems) with primers and probe at 300 nM and 150 nM, respectively. Primer and probe sequences were homologous to GFP and are as follows: GFP forward CAACAGCCACAACGTCTATATCAT, GFP reverse, ATGTTGTGGCGGATCTTGAAG, Probe 5′-FAM-CCGACAAGCAGAAGAACGGCATCAA-3′TAMRA (17).
Results
To measure infection by retrovirus we made high-titer retroviral vectors encoding marker genes. These vectors were pseudotyped with the pantropic VSV-G unless otherwise stated, and are designated by a code describing the virus, HIV and SIVmac, and the marker gene, eGFP, LacZ, and puro.
Saturation of HIV-1 Restriction in Monkey Cells.
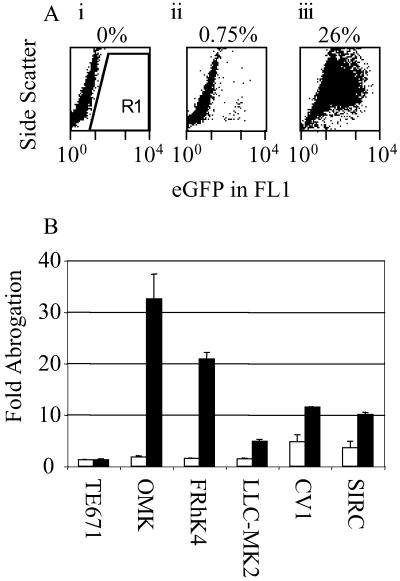
Abrogation assays were performed on cells from rhesus macaque (FRhK4 and LLC-MK2) and owl monkey (OMK) because cells from these species have been shown to be refractory to HIV-1 infection (4, 11). We also tested African green monkey CV1 cells and human TE671 cells, which are relatively permissive, and rabbit SIRC cells, which are highly resistant to VSV-pseudotyped HIV-eGFP vector. We were testing for an increase in HIV-eGFP titer after an initial exposure to HIV-puro or SIV-LacZ had saturated the available restriction factor. Cells were exposed to high doses of HIV-puro or SIV-LacZ, and then exposed to a low dose of HIV-eGFP. The titer of HIV-eGFP was increased by 10 times (CV1), even though these cells are relatively permissive, 20 times (FRhK4), 3 times (LLC-MK2), or 30 times (OMK) by preexposure to HIV but not when preexposed to SIVmac vectors (Fig. 1). HIV-eGFP titer was increased by a small amount (5-fold) on CV1 cells after exposure to SIVmac and also on SIRC cells after exposure to either SIVmac or HIV vectors (4- and 10-fold, respectively).
Figure 1.
(A) Abrogation of HIV-1 restriction in owl monkey cells. FACS plots of side scatter versus GFP fluorescence are shown. Owl monkey cells are uninfected (i), infected with 105 TE671 i.u. of HIV-eGFP (ii), or exposed to 107 TE671 i.u. of HIV-puro for 4 h, washed, and then infected with 105 TE671 i.u. of HIV-eGFP. Region of GFP-positive cells (R1) and percentages of positive cells are indicated. (B) Abrogation of HIV restriction in monkey and rabbit cells. Cells (105) were exposed to 5 × 106 TE671 i.u. of either HIV-puro (■) or SIV-LacZ (□), incubated for 4 h, washed, and then exposed to 104 (TE671), 105 (OMK, FRhK4), 5 × 104 (LLC-MK2, CV1), and 5 × 106 (SIRC) TE671 i.u. of HIV-eGFP. Percentage infection was measured by analysis of eGFP expression 48 h later by FACS. Fold abrogation is calculated by dividing the increased percentage infection after abrogation by unabrogated control HIV-eGFP infection. Typically 1–5% of unabrogated controls were infected. Errors are standard error of the mean of three independent experiments.
Kinetics of Strong Restriction Are Multihit at High Virus Dose.
Restriction of MLV in mice by Fv1 and in humans by Ref1 has been shown to result in two-hit kinetics of infection at high virus doses (10, 18–20), which means that the chance of a cell being infected is related to the square of the virus concentration and the slope of a log(% infection) vs. log(virus dose) plot is 2. To investigate infection kinetics of HIV restriction, serial dilutions of HIV-eGFP were titrated onto permissive and nonpermissive cells. We were seeking a characteristic bend in the titration curve, which appears as a restricting virus begins to soak up factors that inhibit infection, and a slope of 2 or greater at high virus dose.
Fig. 2A shows a titration curve of HIV-eGFP on OMK, and Fig. 2B shows FRhK4 and LLC-MK2 cells. The characteristic bend, indicating a switch from one- to multiple-hit kinetics, can be seen as between 1% and 10% cells are infected. Rhesus macaque LLC-MK2 cells are infected 10 times more efficiently than FRhK4 cells, show no bend in the titration curve (Fig. 2), and are not made significantly more permissive by abrogation (Fig. 1). Two-hit kinetics was also seen when FRhK4 and OMK cells were infected with HIV-eGFP pseudotyped with MLV amphotropic envelope, demonstrating that restriction is not specific to VSV-G (data not shown). Fig. 2C shows titration of SIV-eGFP onto OMK, LLC-MK2, and FRhK4 cells. These cells do not restrict SIVmac and show single-hit kinetics of infection.
Fig. 2D shows titration of HIV-eGFP and SIV-eGFP onto CV1 cells. Although HIV-eGFP shows single-hit kinetics, the slope for the SIV-eGFP titration curve was higher than 1, suggesting stronger restriction for SIVmac infection. Abrogation of this restriction by both HIV and SIVmac is further examined below (see Fig. 4).
Low Permissivity to HIV-eGFP Is Partly Due to Saturable Restriction in Rabbit SIRC Cells.
Rabbit SIRC cells are 3–4 logs less infectable by HIV vectors compared with human TE671 cells. In Fig. 2E, these cells are infected with SIV-eGFP and HIV-eGFP. Rabbit SIRC cells are permissive for SIV-eGFP and single-hit kinetics are seen. At very high doses of HIV-eGFP, a switch to two-hit kinetics occurs, but SIRC cells remain at least 3 logs less permissive to HIV than SIVmac. We conclude that the block in SIRC cells is only partly due to a saturable restriction factor.
Requirement for DNA Synthesis and Decay of Abrogation.
Abrogation of restriction should be mediated by the incoming virus components, most likely viral proteins. Restriction could be associated with the process of reverse transcription of the viral DNA because it seemed to occur at or before DNA synthesis (see Fig. 5). To test whether DNA synthesis was required in the abrogating vector we prepared p8.91 HIV-1 gag-pol expression vector with a D185E change in the highly conserved YMD185D motif of the reverse transcriptase active site. This mutation completely blocks the ability of the virus to reverse transcribe but is unlikely to have any physical effect on retroviral core structure (21). HIV-eGFP was prepared with this construct and used to abrogate restriction in OMK and FRhK4 cells. This virus produced no infection on TE671 cells at high dose (data not shown). Fig. 3A shows that this virus was able to abrogate infection to a similar degree as wild-type HIV-puro (Fig. 3B). Fig. 3B also shows that restriction is about 20- and 30-fold for FRhK4 and OMK cells, respectively, and that 107 TE671 i.u./ml of HIV-puro are required for maximum abrogation of restriction. We have measured the ratio of infectious dose to physical particles in vector preparations similar to those used here (17) and estimate that 2 × 104 particles per cell are required to abrogate HIV-1 restriction.
Figure 5.
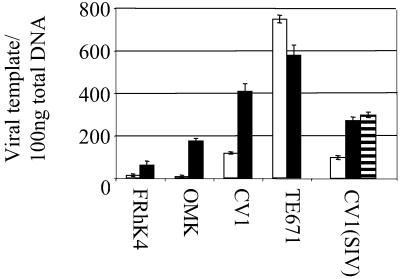
Quantitative PCR of restricted and unrestricted infection. FRHK4, OMK, CV1, or TE671 cells (105) were infected preexposed to 107 TE671 i.u. of HIV-puro for 4 h (■) or left unexposed (□), washed, and then infected with 5 × 104 TE671 i.u. of HIV-eGFP. For SIV on CV1 cells 106 TE671 i.u. of HIV-puro (■) or 106 TE671 i.u. of SIV-LacZ (striped bar) was used to abrogate and then 5 × 104 i.u. of SIV-eGFP was used to infect cells. Four hours after the second round of infection, total DNA was extracted and 100 ng was subjected to quantitative PCR (see Materials and Methods). Viral template copy number per 100 ng of total DNA was calculated by reference to a standard curve. Approximately 1–10% of unabrogated monkey control cells were infected, and 35% of TE671 controls. Data are representative of two independent experiments with duplicate PCR. Errors are standard error of the mean. Parallel samples were analyzed by FACS, and levels of restriction were similar to those in Figs. 3 and 4.
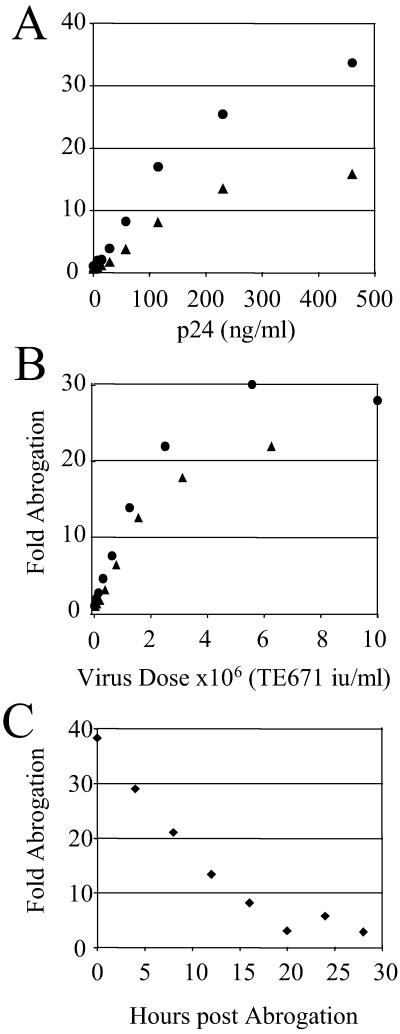
Figure 3.
Abrogation by reverse transcriptase mutant and time course of abrogation decay. OMK (●) or FRhK4 (▴) cells (105) were exposed to serial dilutions of reverse transcriptase-defective HIV-eGFP D185E (A) or wild-type HIV-puro (B) for 4 h. Cells were washed and exposed to 105 TE671 i.u. of HIV-eGFP. Percentage infection was measured by analysis of eGFP expression 48 h later by FACS. Fold abrogation is calculated by dividing the increased percentage infection after abrogation by unabrogated control HIV-eGFP infection. About 1% of unabrogated controls typically were infected. p24 concentrations of wild-type and mutant supernatants were similar with around 500 ng/ml corresponding to a titer of 107 TE671 i.u. wild-type HIV-puro. OMK cells (105) were exposed to 107 TE671 i.u. of HIV-puro for 4 h, washed, and exposed to 105 TE671 i.u. of HIV-eGFP at 4-h intervals (C). Percentage infection and fold abrogation were determined as above. Data are representative of two independent experiments.
To investigate the stability of the abrogation of restriction we performed the following experiment. OMK cells were exposed to high-dose HIV-puro for 4 h, washed, and then exposed to 105 TE671 i.u. of HIV-eGFP at various later time points. HIV-eGFP titer after abrogation linearly decreases with a half-life of around 9 h (Fig. 3C).
SIV Restriction and Its Abrogation by Both HIV and SIV in African Green Monkey CV1 Cells.
CV1 data in Fig. 1B, showing that exposure to SIV-LacZ was able to increase the titre of HIV-eGFP, implied that there is a factor in CV1 cells restricting both HIV1 and SIVmac. Furthermore, a steep titration curve suggested stronger restriction of SIVmac than HIV in these cells (Fig. 2). To examine this possibility further we titrated HIV-puro and SIV-LacZ onto CV1 cells and then infected them with a fixed dose of SIV-eGFP. High doses of either HIV or SIVmac vector were able to increase the titer of SIV-eGFP by up to 30-fold (Fig. 4).
Reverse Transcription Is Blocked in Restricted Cells.
Ref1 restriction of mouse viruses in human cells occurs at or before reverse transcription. To determine whether reverse transcription of HIV and SIV is inhibited in restricting monkeys we performed quantitative PCR to measure viral DNA synthesis on extracts from cells 4 h after infection. Equal virus doses (105 TE671 i.u.) were used to infect a fixed number of cells (105) with and without preexposure to an abrogating virus HIV-puro or SIV-LacZ. This assay indicates the difference between DNA synthesis between restricted and nonrestricted infection in the same cell line and differences in reverse transcription between species. In agreement with previous studies (4, 6, 7) the block to infection in macaque cells is before reverse transcription (Fig. 5).
Discussion
Saturable factors that restrict MLV in mice (Fv1) (22) and humans (Ref1) (9) have been described. Although rhesus macaques have long been known to have very low permissivity for HIV-1, with a viral determinant in gag-pol, the mechanism of the block to infection has remained unclear. These data demonstrate the presence of a saturable factor or factors able to restrict HIV-1 and SIVmac251 in simian cell lines. Fig. 1 shows that the titer of HIV-eGFP vector can be increased in cells from rhesus macaque, owl monkey, and African green monkeys by preexposure to HIV-puro. Rhesus macaques remain the major primate experimental model, essential in the production of an HIV vaccine. The presence of a restriction factor against HIV-1 in macaque may explain the inability of HIV-1 to cause disease in these monkeys.
Where a virus is restricted more than 20-fold, multiple-hit kinetics are seen at high virus dose (Fig. 2), which indicates the factor is being soaked up by high-titer incoming virus. Two-hit kinetics implies that exposure to an initial restricted virion facilitates infection by a second restricted virion. In this case the probability of infection is related to the square of the concentration of the virus, and the slope of a log(infection) versus log(virus dose) is 2. These properties of restriction of retroviral infection are reminiscent of MLV restriction by Ref1 in human cells and Fv1 in mouse cells. The abrogation of HIV restriction does not require reverse transcription of abrogating virus particles (Fig. 3A), and the activity after exposure to abrogating virus has a half-life of 9 h (Fig. 3C).
Rhesus macaque LLC-MK2 cells only weakly restrict HIV-1 (Figs. 1 and 2), and consequently, HIV-eGFP has a 10-fold higher titer on these cells than on FRhK4. Both FRhK4 cells, which restrict, and LLC-MK2, which do not, are derived from Macaca mulatta kidney. LLC-MK2 cells may have lost expression of the restriction factor. A precedent exists for cell lines from restricting mice lacking restriction in SC1 cells and 3T3FL cells (23, 24). Further analysis of factor expression will require isolation of the gene or genes responsible.
Rabbit SIRC cells are resistant to HIV-eGFP but not SIV-eGFP (Fig. 2D). The titer of HIV-eGFP is increased 10-fold by exposure to high-dose HIV-puro (Fig. 1), and HIV-eGFP infection has multiple-hit kinetics at a high virus dose (Fig. 2). Poor HIV infection in SIRC cells is therefore partly due to a saturable restricting factor(s). However, the low titer of HIV vector on SIRC cells may be largely due to an inability of HIV to use rabbit host cofactors. Difference in retroviral titer between cells from different species are likely to be dependent to some degree on cellular cofactor compatibility. Poor infection because of lack of compatibility is not likely to be saturable by preexposure to retrovirus.
The data presented here show that restriction factors are more common than previously thought (22). The target for restriction is likely to reside in the capsid of the lentivirus. It has been shown that an SIVmac with an HIV-1 capsid-p2 domain has the restriction phenotype of HIV-1 rather than SIVmac and was able to replicate in human cells and not macaque peripheral blood mononuclear cells (8). Furthermore, the Fv1/Ref1 target sequence is in the capsid of MLV (9, 25). Cells from African green monkey are able to restrict SIVmac as well as HIV, and reciprocal saturation of the factor by SIVmac or HIV indicates that a single factor is responsible. The fact that a restriction factor can hit more than one virus suggests that the selecting virus, which forced the host to acquire this restriction factor, may be at least as unrelated to HIV or SIVmac as they are to each other. Although Fv1 has approximately 60% sequence homology with gag from HERV-L and MuERV-L (22, 26), it is only distantly related to MLV, which suggests that lentiviral restriction is a function of retrovirus-like sequences unrelated to lentiviruses themselves. We have shown that cells from African green monkey are also able to restrict N tropic MLV (9), and it will be interesting to investigate the relationship between restrictions against C-type retrovirus and lentivirus. The large amount of retroviral sequence in mammalian genomes could serve as a pool from which protective sequence can be selected by pressure from pathogenic retrovirus. Some common aspects of the life cycle of distantly related retroviruses may present a capsid target susceptible to interference from gag-related molecules, which depend on their gag-like nature for their protective function. The further characterization and cloning of these factors will provide insight into early postentry events in the retroviral life cycle and reveal opportunities for therapeutic intervention.
Acknowledgments
We thank Stephen Goff, Columbia University, New York, and Robin Weiss, University College London, for helpful discussion; Yasuhiro Ikeda, University College London, for HIV-1 Ampho pseudotype; François-Loic Cosset, Ecole Normale Superieure de Lyon, France, and Adrian Thrasher, Institute of Child Health, University College London, for plasmids; the Centro Substrati Cellulari, Brescia, Italy, and Parmjit Jat, Ludwig Institute for Cancer Research, London, for cell lines; and Ben Strange and Ian Gerrard for technical assistance. This work was funded by Career Development Fellowship 064257 (to G.T.) from the Wellcome Trust and by a grant from the Medical Research Council United Kingdom (to Y.T.).
Abbreviations
- MLV
murine leukemia virus
- SIV
simian immunodeficiency virus
- SIVmac
SIV from macaques
- eGFP
enhanced green fluorescent protein
- LacZ
β-galactosidase
- FACS
fluorescence-activated cell sorter
- puro
puromycin resistance
- VSV-G
vesicular stomatitis virus envelope protein
- i.u.
infectious units
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 11549.
References
- 1.Martin J, Herniou E, Cook J, O'Neill R W, Tristem M. J Virol. 1999;73:2442–2449. doi: 10.1128/jvi.73.3.2442-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Hahn B H. Nature (London) 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 3.Weiss R A, Wrangham R W. Nature (London) 1999;397:385–386. doi: 10.1038/17008. [DOI] [PubMed] [Google Scholar]
- 4.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. J Virol. 1991;65:3514–3520. doi: 10.1128/jvi.65.7.3514-3520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. J Gen Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 6.Himathongkham S, Luciw P. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian B, Long E M, Luciw P A, Overbaugh J. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman T, Gottlinger H G. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towers G, Collins M, Takeuchi Y. J Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 13.Negre D, Mangeot P E, Duisit G, Blanchard S, Vidalain P O, Leissner P, Winter A J, Rabourdin-Combe C, Mehtali M, Moullier P, et al. Gene Ther. 2000;7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 14.Bock M, Bishop K, Towers G, Stoye J P. J Virol. 2000;74:7422–7430. doi: 10.1128/jvi.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K L. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towers G J, Stockholm D, Labrousse-Najburg V, Carlier F, Danos O, Pages J C. J Gene Med. 1999;1:352–359. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<352::AID-JGM57>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda Y, Collins M K L, Radcliffe P A, Mitrophanous K A, Takeuchi Y. Gene Ther. 2002;9:932–938. doi: 10.1038/sj.gt.3301708. [DOI] [PubMed] [Google Scholar]
- 18.Decleve A, Niwa O, Gelmann E, Kaplan H S. Virology. 1975;65:320–332. doi: 10.1016/0042-6822(75)90038-0. [DOI] [PubMed] [Google Scholar]
- 19.Pincus T, Rowe W P, Lilly F. J Exp Med. 1971;133:1234–1241. doi: 10.1084/jem.133.6.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tennant R W, Otten J A, Brown A, Yang W K, Kennel S J. Virology. 1979;99:349–357. doi: 10.1016/0042-6822(79)90014-x. [DOI] [PubMed] [Google Scholar]
- 21.Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J, Pandey V N. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 22.Best S, Le Tissier P, Towers G, Stoye J P. Nature (London) 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 23.Hartley J W, Rowe W P. Virology. 1975;65:128–134. doi: 10.1016/0042-6822(75)90013-6. [DOI] [PubMed] [Google Scholar]
- 24.Gisselbrecht S, Bassin R H, Gerwin B I, Rein A. Int J Cancer. 1974;14:106–113. doi: 10.1002/ijc.2910140113. [DOI] [PubMed] [Google Scholar]
- 25.Boone L R, Glover P L, Innes C L, Niver L N, Bondurant M C, Yang W K. J Virol. 1988;62:2644–2650. doi: 10.1128/jvi.62.8.2644-2650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benit L, De Parseval N, Casella J-F, Callebaut I, Cordonnier A, Heidmann T. J Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]